Abstract
The replicative DNA polymerases are generally blocked by template DNA damage. The resulting replication arrest can be released by one of two post-replication repair (PRR) pathways, translesion DNA synthesis (TLS) and template switching by homologous recombination (HR). The HERC2 ubiquitin ligase plays a role in homologous recombination by facilitating the assembly of the Ubc13 ubiquitin-conjugating enzyme with the RNF8 ubiquitin ligase. To explore the role of HERC2 and RNF8 in PRR, we examined immunoglobulin diversification in chicken DT40 cells deficient in HERC2 and RNF8. Unexpectedly, the HERC2-/- and RNF8-/- cells and HERC2-/-/RNF8-/- double mutant cells exhibit a significant reduction in the rate of immunoglobulin (Ig) hypermutation, compared to wild-type cells. Further, the HERC2-/- and RNF8-/- mutants exhibit defective maintenance of replication fork progression immediately after exposure to UV while retaining proficient post-replicative gap filling. These mutants are both proficient in mono-ubiquitination of PCNA. Taken together, these results suggest that HERC2 and RNF8 promote TLS past abasic sites and UV-lesions at or very close to stalled replication forks.
Keywords: HERC2, RNF8, Translesion DNA Synthesis, Post-replication repair, Ubiquitination
1. Introduction
Replication involves a complex and fragile enzymatic reaction that can readily be disrupted by template DNA damage. To restart arrested replication, cells have evolved two post-replicational repair (PRR) pathways, translesion synthesis (TLS) and template switching by homologous recombination (HR). HR facilitates transient switching of replication from the damaged template strand to the newly synthesized sister chromatid [1–6]. In addition to template switching by HR, replication blockage is released by employing specialized TLS polymerases such as DNA polymerases η and ζ (Polη and Polζ) [7–9]. The deployment of the Y-family TLS polymerases is controlled by PCNA ubiquitination at K164 and by the non-catalytic function of the Y-family polymerase REV1 [10–13]. RAD18 ubiquitin ligase is responsible for the mono-ubiquitination in Saccharomyces cerevisiae, the chicken DT40 B lymphocyte cell line and mammalian cells [10–14].
HERC2 is a HECT domain E3 ubiquitin ligase and one of the largest genes in the vertebrate genome. It has been shown to play a role in control of nucleotide excision repair by ubiquitinating and degrading XPA [15,16]. It also plays a role in double strand break repair by facilitating the assembly of the Ubc13 ubiquitin-conjugating enzyme with the ubiquitin ligase RNF8 [17,18]. HERC2 also associates with RNF168, another ubiquitin ligase operating downstream of RNF8 in DSB repair [17,19–21]. RNF168 amplifies the RNF8-dependent histone ubiquitination by targeting H2A-type histones and by promoting the formation of lysine 63-linked ubiquitin conjugates [19,20]. These modifications orchestrate the accumulation of 53BP1 and BRCA1 to DNA lesions [19,20]. A recently published study showed that RNF8 but not RNF168 is responsible for the K63-linked ubiquitination of H1-type linker histones [22]. While the contribution of HERC2 to the ubiquitination response to DNA double strand breaks and HR-mediated DSB repair has been established, its role in restoring stalled replication forks has not been explored. Indeed, it remains unclear whether RNF8 is involved in DNA damage response other than DSB repair.
The chicken DT40 B lymphocyte cell line provides a unique opportunity to specifically analyze the involvement of DNA damage repair proteins in PRR pathways (Fig. S1) through examination of the immunoglobulin variable (Ig V) gene diversification during in vitro culture [23]. This diversification is driven by release of abasic site-mediated replication blocks either by TLS, which results in non-templated point mutations, or by HR, which drives gene conversion with a set of homeologous pseudogenes [9,24]. These diversification processes are initiated by deamination of deoxycytidine by activation-induced deaminase (AID) to generate uracil [25,26], which is subsequently excised, leaving an abasic site [27– 29]. TLS releases replication forks stalling at abasic sites, leading to Ig V non-templated mutations at G/C pairs (Ig hypermutation) [30–34]. Both gene conversion tracts and the spectrum of non-templated point mutations can be evaluated by identifying Ig V nucleotide sequence variations during clonal expansion of DT40 cells.
In this study, we show that HERC2-/-, RNF8-/- and HERC2-/-/RNF8-/- cells all exhibit a several-fold decrease in the rate of Ig hypermutation. The HERC2-/- and RNF8-/- mutants are also defective in the maintenance of replication fork progression immediately after exposure of cells to UV. These data are consistent with HERC2 and RNF8 promoting TLS past abasic sites at or very close to stalled replication forks.
2. Materials and Methods
2.1. Cell culture
RAD18-/-, RAD18-/-/RNF8-/-, RNF8-/-, RNF8-/C398F, RAD18-/-/RNF8-/C398F DT40 cells [21,35] and HERC2-/-, HERC2-/-/RNF8-/- DT40 cells [21] were previously generated. Cells were cultured in the same manner as described previously [36].
2.2. Cell survival assay
Cells were treated with each DNA-damaging agent in 1 ml of medium using 24-well plates and incubated at 39.5°C for 48 h. We transferred 100 μl of cell culture to 96-well plates and measured the amount of ATP using Cell Titer Glo (Promega), according to the manufacturer’s instructions. The P-value of figure 1B and 1C were calculated by Student’s t-test [37].
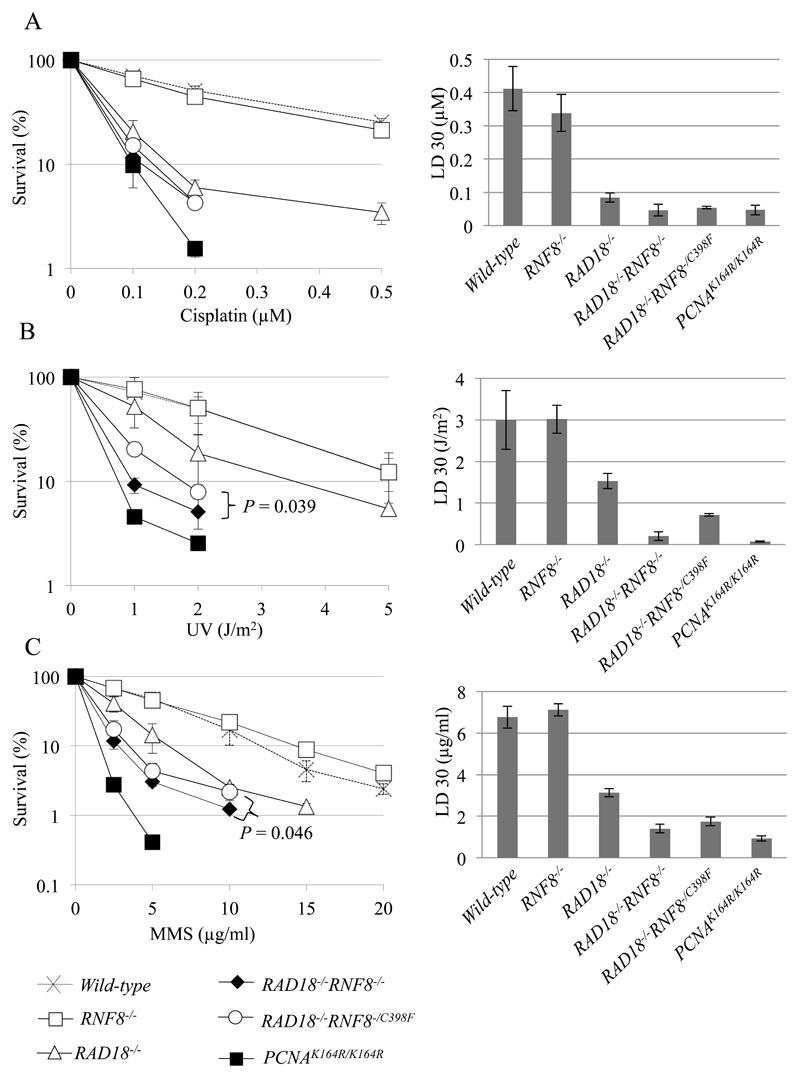
Fig. 1. Cellular Sensitivity to DNA-damaging agents.
(A–C) Cellular sensitivity to cisplatin (A), UV (B), and MMS (C) was analyzed. Survival rate was calculated as the percentage of surviving cells treated with DNA-damaging agents relative to the untreated surviving cells. The concentration or dose is displayed on the x-axis on a linear scale, while the survival rate is displayed on the y-axis on a logarithmic scale. P-values were calculated by Student’s t-test. Lethal dose 30% (LD30) is the concentration of DNA damaging agents that reduces cellular survival to 30% relative to cells non-treated with DNA damaging agents. LD30 was calculated by the statistics software, R. Error bars show the standard deviation of the mean of at least three independent experiments.
2.3. Ectopic expression of AID by retroviral infection
An AID-encoding retrovirus was used to infect DT40 as described before [32,33,38]. Briefly, the recombinant plasmid encoding mono-cistronic AID and GFP genes, was co-transfected with pCL-Ampho in human 293T cells in order to produce a retrovirus capable of infecting DT40 cells. Following retroviral infection of DT40 cells, we evaluated ectopic expression of AID by flow cytometric measurement of GFP expression, and chose five clones from each genotype, expressing comparable levels of GFP. Individual cells were grown clonally following limiting dilution, and expanded for two weeks to analyze Ig Vλ diversification.
2.4. Analysis of Ig Vλ diversification
Genomic DNA was extracted from independent clones and the Vλ segment was amplified by PCR using primers 5’-CAGGAGCTCGCGGGGCCGTCACTGATTGCCG-3’ and 5’-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG-3’ to facilitate cloning into pCR2.1. The Vλ segment was then sequenced using M13 forward and reverse primers. The method for classification of gene conversion and non-templated single base substitution was previously described [9,30,33]. Since the rate of Ig V diversification in wild-type cells slightly differed in two sets of experiments due to a variation of ectopic AID expression, we calculated Ig V diversification rate in each genotype relative to that in wild-type cells.
2.5. Analysis of PCNA mono-ubiquitination
For the detection of PCNA mono-ubiquitination, 1×106 cells were UV irradiated (30 J/m2), lysed 1 h post-treatment with 50 μl of Laemmli Sample buffer (Bio-lad), boiled for 5 min and kept on ice. 20 μl of the sample was run in a 12% gradient gel, after which the proteins were electroblotted onto a PVDF membrane. The following antibodies were used: mouse monoclonal PCNA PC10 (Santa Cruz), monoclonal anti-β-actin (Sigma), and anti-mouse IgG HRP-linked (GE-Healthcare). Proteins were visualized using Super Signal West Pico Chemiluminescent Substrate (Thermo).
Ubiquitinated PCNA was quantified from the image of the western blot using ImageJ software. First, the background noise was subtracted from all other values. In the second step, the signals for each sample on the anti-PCNA blot at the positions of ubiquitinated PCNA were normalized according to the signals obtained from the anti-β-actin blot, to account for sample loading variations. In the last step, all values were normalized to the value of ubiquitinated PCNA in untreated wild-type cells, and set to 1.00.
2.6. Measurement of the size of newly synthesized DNA strands following UV irradiation
To analyze the size of the nascent DNA following UV irradiation, cells were irradiated with UV light (5 J/m2), incubated for 20 min, and then pulse-labeled with [methyl-14C] thymidine (10 μCi/ml) for 20 min. In a pulse-chase experiment, the pulse-labeled cells were incubated 3 h further in the chase medium. Samples were sedimented on 5–20% alkaline sucrose gradients.
Cells were harvested and layered on the alkaline sucrose density gradient as follows. Cell-lysis solution (0.6 M KOH, 2.0 M KCl, 10 mM EDTA, and 1% N-lauroylsarcosine), 1% sucrose in Ca2+, Mg2+-free phosphate- buffered saline (CMF-PBS), and the cell suspension were layered in series on top of a pre-formed alkaline 5–20% sucrose gradient (0.3 M KOH, 2.0 M KCl, 1 mM EDTA, and 0.1% N-lauroylsarcosine) with alkaline 80% sucrose as a cushion at the bottom. The gradient was centrifuged at 6,000 rpm (4,320×g) for 15.6 h at 15°C in a Beckman SW50.1 rotor. The gradient was fractionated from the bottom by drop-counting onto 3 MM paper circles (Whatman) using a peristaltic pump. The paper circles were dried, immersed in cold 5% trichloroacetic acid, washed with ethanol and then with acetone, dried, and then tested for radioactivity in a toluene-based scintillator. These experiments were repeated at least twice, and equivalent profiles were obtained.
2.7. Dynamic molecular combing and immunofluorescent detection
Cells were sequentially labeled for 15 min with 25 µM IdU and for 15 min with 25 µM CIdU. UV treated cells were irradiated at 20 J/m2 prior to the CldU treatment. At the end of the labeling period (30 min), cells were placed in ice cold 1× PBS and centrifuged at 250xg for 5 min at 4ºC, washed and resuspended in PBS. The cell suspension was spotted onto Superfrost slides and lysed with 0.5 % SDS in 200 mM Tris-HCl (pH 5.5) and 50 mM EDTA (5 min, at room temperature). Slides were tilted at 15° to horizontal, allowing the DNA to run slowly down the slide. Slides were then air dried and fixed in 3:1 methanol/acetic acid, and stored at 4°C before immunolabeling. IdU, CldU, DNA revelations and analysis were performed as described [39], with the following minor modifications: the DNA was denatured for 55 minutes in 2.5 N HCl, and CldU was detected using rat anti-BrdU (ABD Serotec, Raleigh, NC) at a 1/750 dilution. A stretching factor of 2.6 for conversion from µm to kb was applied, as previously described [40]. Slides were mounted in 10% 1X PBS and 90% glycerol, kept at -20°C and imaged using a Nikon C1-si confocal microscope.
3. Results
3.1. The loss of RNF8 increases sensitivity of RAD18-/- cells to MMS and UV
HERC2-/- and RNF8-/- DT40 cells are sensitive to camptothecin, a topoisomerase 1 poison, indicating that the two ubiquitin ligases are involved in repair of DSB repair by HR [21,35]. We extended this analysis by measuring the sensitivity of HERC2-/-, RNF8-/- and HERC2-/-/RNF8-/- cells to cisplatin, methyl methanesulfonate (MMS) and UV (Fig. S2). We failed to detect any significant sensitivity of the mutant cells to these three DNA-damaging agents. Disruption of RNF168, another ubiquitin ligase operating downstream of RNF8 and HERC2 in DSB repair [21], also showed no sensitivity to these agents (Fig. S2). We then tested whether the loss of RNF8 increased cellular sensitivity to these DNA damaging agents in the RAD18-deficient background. RAD18-/- cells are hyper sensitive to a variety of DNA damaging agents, which is consistent with the critical role of RAD18 in TLS as well as HR [12,13,35,41]. RAD18-/-/RNF8-/- cells are more sensitive to cisplatin, MMS and UV than are RAD18-/- cells (Fig. 1). Thus, significant contribution of RNF8 to DNA damage tolerance is masked by RAD18 in DT40 cells. A previous report indicates that a non-catalytic role of RNF8 may contribute to cellular responses to DSBs including DSB repair by HR [35,42]. To address this non-catalytic role, we analyzed the effect of the RNF8-/C398F mutation, which inactivates the ubiquitin ligase activity of RNF8. RAD18-/-/RNF8-/- cells show more sensitivity to MMS and UV in comparison with RAD18-/-/RNF8-/C398F cells (Fig. 1), supporting the contribution of the non-catalytic role to DNA damage tolerance. More importantly, RAD18-/-/RNF8-/C398F cells show significantly higher sensitivity to the DNA-damaging agents in comparison to RAD18-/- cells (Fig. 1). We conclude that the ubiquitin ligase activity of RNF8 contributes to DNA damage tolerance possibly through promotion of TLS.
3.2. Significant decreases in the rate of Ig V non-templated single base substitutions in the HERC2-/- and RNF8-/- mutants
To explore the role of HERC2 and RNF8 in PRR, we examined TLS-dependent Ig hypermutation in the DT40 clones. We ectopically expressed an AID transgene by retrovirus infection. The resulting extensive deamination of deoxycytidine by AID allows for very sensitive detection of TLS events in the Ig V segment [32]. We then examined nucleotide sequences of Ig Vλ segments in AID-expressing clones derived from wild-type, RNF8-/-, RAD18-/-, RAD18-/-/RNF8-/-, RAD18-/-/RNF8-/C398F, HERC2-/-, HERC2-/-/RNF8-/- and RNF168-/- cells (Fig. S3). The rate of Ig gene conversion was decreased four to five fold in both RNF8-/- and HERC2-/- mutants compared to wild-type cells (Fig. 2A and B). HERC2-/-/RNF8-/- cells showed virtually the same phenotype as any single mutants (Fig. 2A and B). This observation agrees with the fact that these two ubiquitin ligase enzymes work together to promote HR [17,18].
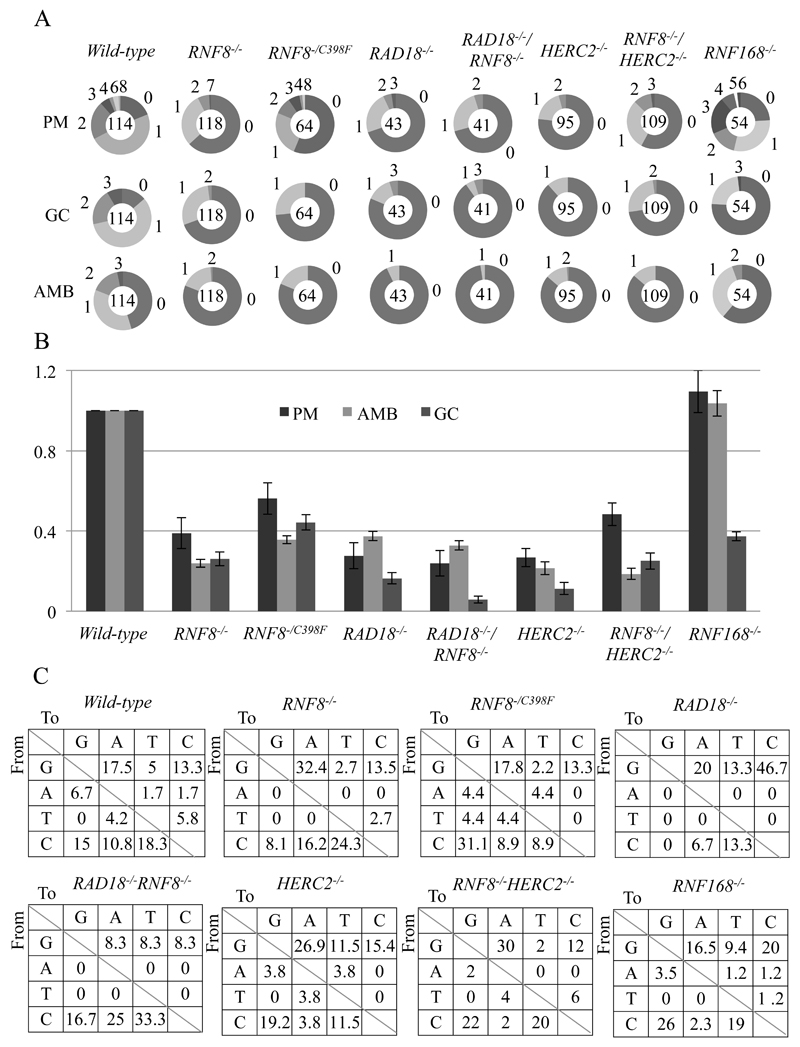
Fig. 2. The important role of HERC2 and RNF8 in TLS past abasic sites during Ig Vλ hypermutation.
(A) Clones over-expressing AID were expanded for two weeks before isolating genomic DNA and sequencing the Vλ segment. Charts displaying the frequency of each type of mutation. Segment size indicates the proportion of cells in which the number of mutations stated outside the chart had occurred. The total number of sequences analyzed is shown in the centre of the chart. PM = Point mutation/non-templated single base substitution; AMB = Ambiguous; GC = Gene conversion. (B) The rate of point mutation/non-templated single base substitution (PM) events in wild-type cells were set to 1.00 and all values were normalized to the value of wild-type cells and are indicated with standard error. (C) Pattern of point mutation in the indicated cell lines. Tables showing the pattern of mutation in each line, given a percentage of mutations observed.
Next we analyzed the rate of TLS dependent Ig V nontemplated mutation. The rate of the nontemplated mutagenesis was significantly decreased in HERC2-/- and RNF8-/- mutants, both alone and in combination, compared to wild type (Fig. 2A and B), with no impact on the mutation spectrum (Fig. 2C). RNF8-/C398F cells exhibited a similar decrease in the rate of Ig hypermutation (Fig. 2A and B). Thus, HERC2 and RNF8 appear to also play a role in releasing replication blockage by promoting TLS in DT40 cells.
3.3. Intact PCNA mono-ubiquitination in HERC2-/- and RNF8-/- mutants
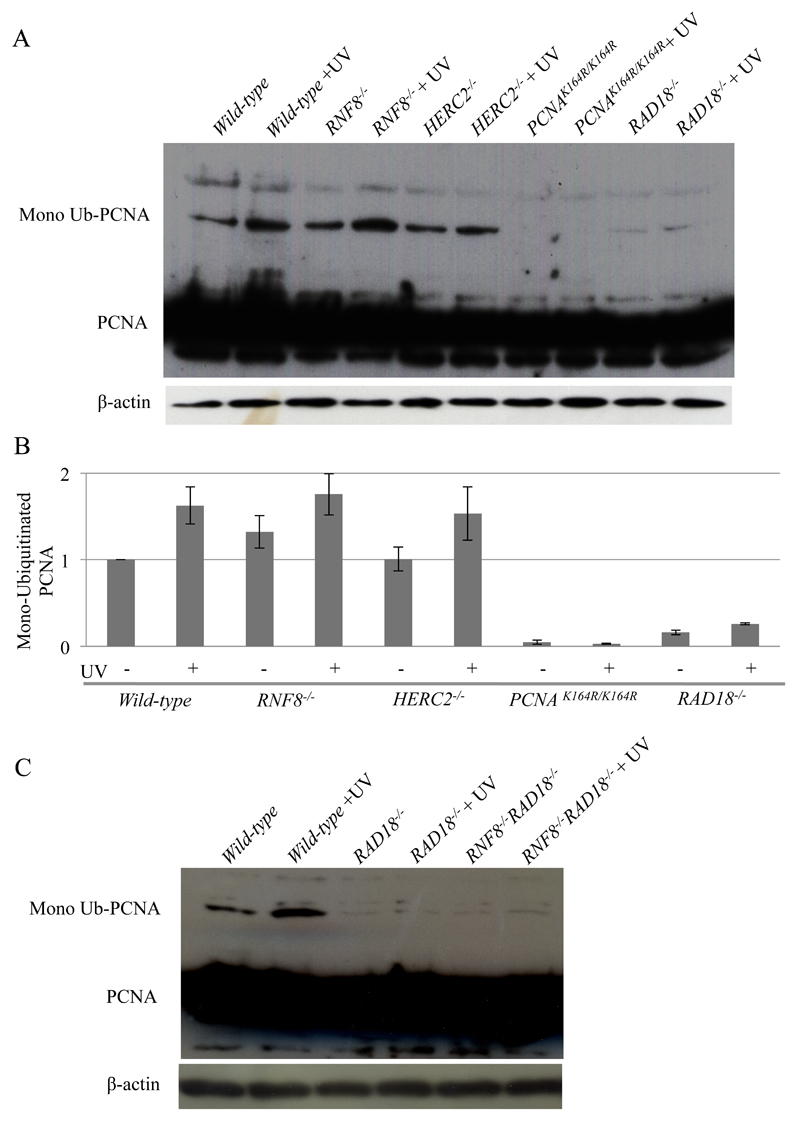
We analyzed the mono-ubiquitination of PCNA following UV irradiation. PCNAK164R/K164R cells were analyzed as a negative control. As expected, the loss of Rad18 resulted in a substantial decrease in UV-induced PCNA mono-ubiquitination, but not complete loss as previously reported [43], whereas the loss of RNF8 or HERC2 had no prominent effect on mono-ubiquitination (Fig. 3A, B and S4). Moreover, the loss of RNF8 had no detectable impact on the PCNA mono-ubiquitination even in RAD18-/- cells (Fig. 3C). These observations indicate that neither RNF8 nor HERC2 contributes to the mono-ubiquitination of PCNA. This conclusion does not agree with that of a previous report [44]. Thus, RNF8 and HERC2 promote TLS past abasic sites in the Ig V diversification most likely by ubiquitinating molecule(s) other than PCNA. It remains elusive why PCNAK164R/K164R cells showed higher sensitivity than RAD18-/- cells (Fig. 1).
Fig. 3. Neither HERC2 nor RNF8 is required for the mono-ubiquitination of PCNA in UV-irradiated cells.
(A) Analysis of ubiquitination of PCNA. Cells were irradiated with 30 J/m2 UV and then incubated for 1 h. Protein extracts were prepared in native conditions for mono-ubiquitination, and analyzed by gel-electrophoresis followed by western blotting using an anti-PCNA antibody. (B) Quantification of ubiquitinated PCNA by immunoblotting. The values for ubiquitinated PCNA were calculated as described in the Materials and Methods. Error bars indicate standard deviation (SD). (C) Analysis of ubiquitination of PCNA in RAD18-/- and RAD18-/-/RAD18-/- cells as in fig. 3A.
3.4. HERC2 and RNF8 are not required to promote the filling of post-replicative gaps
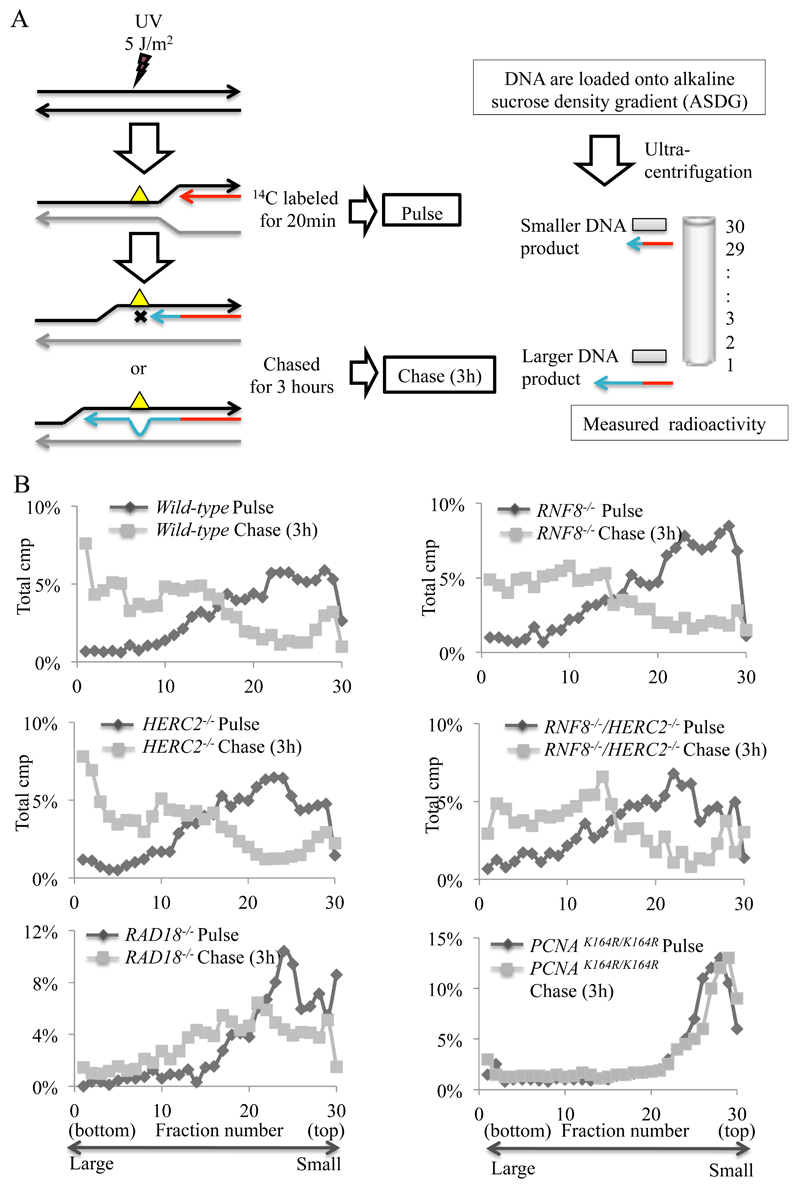
We analyzed the requirement of HERC2 and RNF8 for post-replicative gap filling by alkaline sucrose gradient sedimentation of 3H pulse-labeled DNA (Fig. 4A) [7,45]. In the absence of UV irradiation, newly synthesized DNA in HERC2-/- and RNF8-/- clones was comparable to that of wild-type cells (data not shown). However, following UV irradiation, neither HERC2-/-, RNF8-/- nor HERC2-/-/RNF8-/- cells exhibited any detectable reduction in the joining of the labeled nascent fragments into high molecular weight replicons in comparison with wild-type cells (Fig. 4B: pulse-label, grey line; 3 h chase, black circle). Similarly, REV1-/- and POLZ-/- (REV3-/-) cells can proficiently reconstitute higher molecular weight DNA following UV irradiation [46,47]. These data contrast with the defective post-replicative gap filling observed in POLH-/-, RAD18-/- and PCNAK164R/K164R DT40 cells [7]. Thus, unlike RAD18-/- and PCNAK164R/K164R cells, neither HERC2-/- nor RNF8-/- cells are deficient in the filling of post-replicative gaps at UV-damage sites.
Fig. 4. Intact post-replicative gap filling in HERC2-/- cells.
(A) Schematic diagram of post-replicative gaps filling at UV-damage sites. (B) Indicated cells were irradiated with UV (5 J/m2) and pulse-labelled with [methyl-14C] thymidine for 20 min and either lysed immediately or chased for 3 h in fresh medium containing 10 μM unlabeled thymidine before lysis. Samples were separated on 5–20 % alkaline sucrose gradient sedimentation.
3.5. HERC2-/- and RNF8-/- mutant cells have normal velocity of unperturbed DNA synthesis and comparable unidirectional fork movement
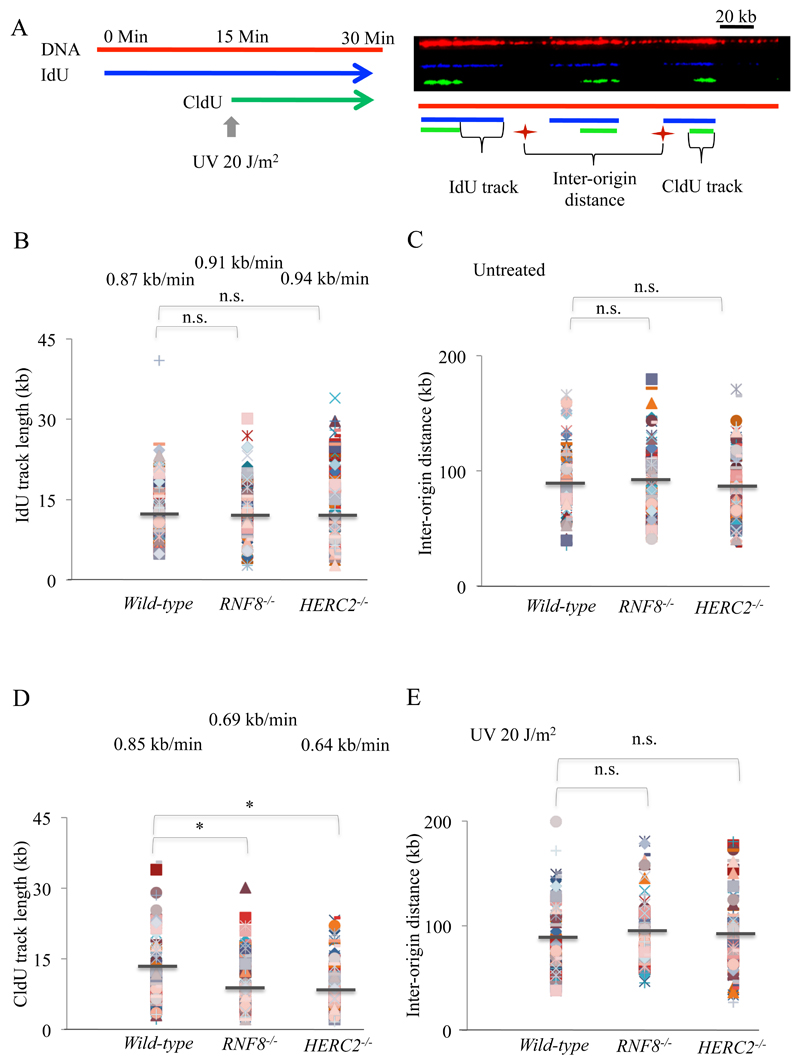
In order to determine the background levels, we examined the kinetics of unperturbed DNA replication using molecular combing (Fig. 5). The replication rate for wild-type, HERC2-/- and RNF8-/- cells in unperturbed conditions was not significantly different at 0.87 ± 0.04 kbp/min, 0.91 ± 0.04 kbp/min and 0.94 ± 0.05 kbp/min respectively (Fig. 5B). This is consistent with the observation that HERC2 and RNF8 are not essential for unperturbed DNA replication (Fig. 4). Thus, loss of RNF8 or HERC2 does not attenuate global progression of DNA replication forks on undamaged DNA templates. Moreover, both HERC2-/- and RNF8-/- mutant cells did not exhibit significant increases in the proportion of unidirectional forks (16.35% in HERC2-/- and 14.95% in RNF8-/- cells as comparable to 14.02% in wild-type cells), in which one of the two forks heading away from a presumed origin is missing (Table 1) [47] and exhibited the same inter-origin distances (Fig. 5C). Thus, RNF8 and HERC2 appear not be play any major role in maintaining DNA replication during a physiological cell cycle.
Fig. 5. HERC2 and RNF8 are dispensable to maintain the velocity of unperturbed DNA synthesis.
(A) Schematic of treatment with UV and pulse labeling with IdU and CldU are shown. Representative image showing stained DNA fibres. (B) Replication fork lengths were obtained by converting the IdU track sizes in µM to kb and analyzed in the indicated cell types. IdU track lengths are calculated by dividing the track lengths by the labeling time and depicted above the track lengths. More than 100 forks were calculated in each cell types. The P-values were calculated by Student’s t-test (C) Indicated cell lines were left untreated and DNA fibres were analyzed for inter-origin distances (IODs). IODs were measured as the distance between two adjacent initiation sites during IdU pulse, and average values are indicated in kb. Over 50 fibers were analyzed in measurement of IODs. The P-values were calculated by Student’s t-test. (D) CldU track lengths were analyzed in the indicated cell types as previous. The P-values for Student’s t-test were *p<0.001. (E) Indicated cell lines were treated with UV and DNA fibres were analyzed for inter-origin distances (IODs) as previous.
Table 1.
Proportion of unidirectional forks. Proportion of unidirectional forks in indicated cells with and without UV irradiation was analyzed by molecular combing.
| Cell Type | UV | % of unidirectional forks |
|---|---|---|
| Wild-type | - | 14 |
| + | 18 | |
| RNF8-/- | - | 15 |
| + | 21 | |
| HERC2-/- | - | 16 |
| + | 25 |
3.6. HERC2 and RNF8 are required to maintain replication fork progression on UV damaged DNA
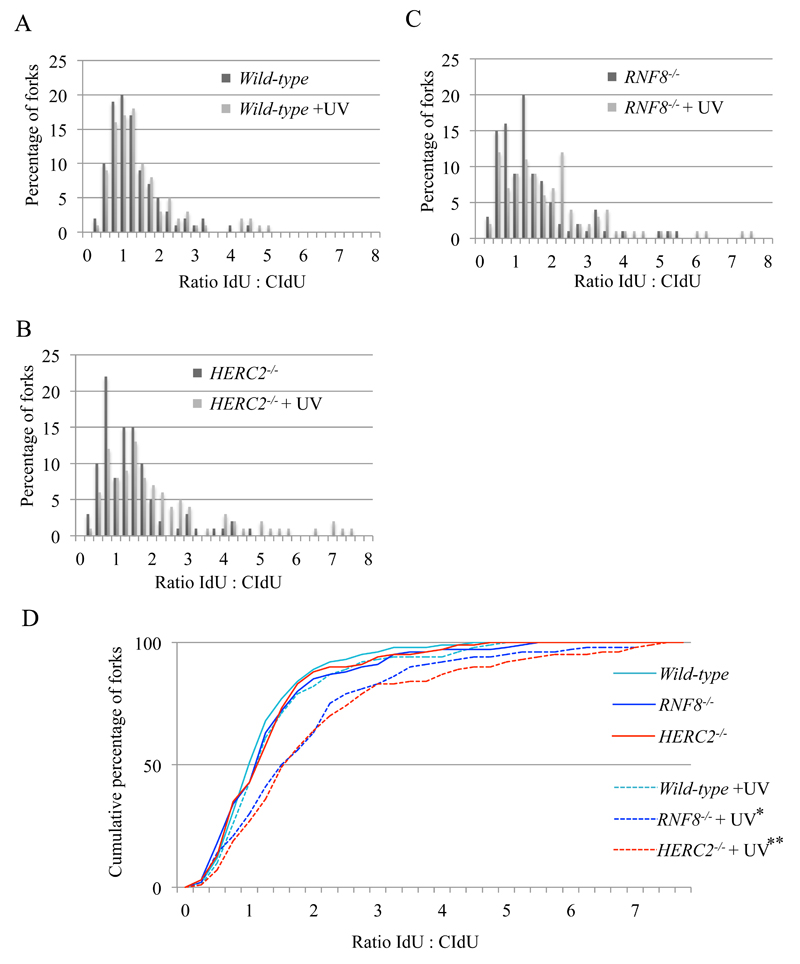
There are two temporally-separated modes of lesion bypass in DT40 cells [7]. One is responsible for filling post-replicative gaps at UV-damage sites, while the other functions at or very close to stalled replication forks and maintains normal fork progression on UV-damaged template DNA strands. Both modes contribute to TLS dependent Ig V hypermutation. The former and latter modes can be evaluated by the alkaline sucrose gradient sedimentation of newly replicated DNA (Fig. 4) and DNA molecular combing (Fig. 6), respectively. The mono-ubiquitination of PCNA by Rad18 is required for the post-replicative gap filling mode, while loss of REV1 predominantly affects replication fork progression on damaged templates [7,48]. Having shown above (Figure 4) that neither HERC2 nor RNF8 are required for post-replicative filling of gaps opposite UV lesions, we examined their impact on fork progression in isolated DNA fibres. We labeled nascent strands with IdU for 15 minutes, irradiated the cells with 20 J/m2 UV, and then continued labeling the nascent strands with CldU (Fig. 5A). After DNA combing, we measured the tracts of CldU and IdU and calculated the ratio between them to allow comparison of the total DNA synthesized before and after UV exposure on a fork-by-fork basis. We plotted the data as a percentage (Fig. 6A, B, and C) and cumulative percentage (Fig. 6D) of forks at each ratio. In contrast with RAD18-/- and PCNAK164R/K164R cells [7], HERC2-/- and RNF8-/- clones are unable to maintain DNA replication fork progression to the same extent as wild-type cells. Consistent with this result, following UV irradiation, HERC2-/- and RNF8-/- clones showed significantly reduced average fork speeds (Fig. 5D) and increased proportion of unidirectional forks (Table 1). We therefore conclude that HERC2 and RNF8 are required for the release of replication blockage by TLS in DT40 cells. Unlike Rad18, RNF8 and HERC2 are both required to maintain replication fork progression at UV damaged DNA but not for the filling of post-replicative gaps.
Fig. 6. RNF8 and HERC2 work together to maintain replication fork progression on UV damaged DNA.
(A-C) Replication stalling in response to 20 J/m2 UV (red bars) or sham irradiation (blue bars) in WT (A), HERC2-/-(B) and RNF8-/- (C) cell lines. Each data set is derived from measurement of at least 100 forks. (D) The data for cells carrying the indicated genotypes was plotted as a cumulative percentage (y-axis) of forks at each ratio (x-axis). The P-values of the Kolmogorov–Smirnov test for ratio distribution of each mutant for UV compared to sham treatment are *p<0.002 and **p<0.001.
4. Discussion
In this study, we have uncovered a role for HERC2 and RNF8 in the promotion of TLS in the chicken DT40 cell line. Thus, HERC2 and RNF8 are required for efficient PRR in addition to DSB repair. RNF8 functions independently of the RAD18 ubiquitin ligase in DNA damage response, since RAD18-/-/RNF8-/- cells showed significantly higher sensitivity to cisplatin, MMS and UV in comparison with RAD18-/- and RNF8-/- cells (Fig. 1). The different roles of RAD18 and HERC2/RNF8 in TLS are substantiated by the observations that RAD18 ubiquitinates lysine 164 of PCNA and thereby facilitates post-replicative gap filling (Fig. 4), while HERC2 and RNF8 play the essential role in efficient TLS by operating at or close to replication forks (Fig. 6) without affecting the ubiquitination of PCNA (Fig. 3).
Given this apparent role in TLS and the prominent defect in maintenance of replication fork progression after UV irradiation (Fig. 6) exhibited by the HERC2-/- and RNF8-/- mutants, it is rather unexpected that neither exhibit significant sensitivity to the DNA-damaging agents (Fig. S1). In fact, the phenotype of these two mutants is in marked contrast with that of REV1-/- cells, which are hypersensitive to cisplatin, MMS and UV, deficient in maintaining replication fork progression after UV irradiation and proficient in post-replicative gap filling [7,49]. Thus, the hypersensitivity of REV1-/- cells may not be simply attributable to a defect in maintaining replication fork progression. In other words, even when replication fork blockage at UV lesions is not restored within the 15 minutes CldU-labeling time after UV irradiation (Fig. 5A), the vast majority of the blockage might be eventually restored during the G2 phase. This idea is supported by the previous reports that although TLS normally operates during the S phase in Saccharomyces cerevisiae, limiting the functionality of TLS to the G2/M phase does not decrease the efficiency of TLS or cellular tolerance to exogenous DNA damage [50,51]. Accordingly, the significantly higher sensitivities of RAD18-/-/RNF8-/- cells to cisplatin, MMS and UV in comparison with RAD18-/- cells (Fig. 1) can be explained by combined defects in both TLS operating immediately after DNA replication and TLS for filling post-replicative gap during the G2 phase.
Both HERC2-/- and RNF8-/- mutants showed a several fold decrease in the rate of Ig hypermutation (Fig. 2), indicating that they promoted TLS past abasic sites. Since RNF8 has both catalytic and non-catalytic roles in DNA damage responses [35,42], we also examined Ig hypermutation in RNF8-/C398F cells, where the C398F mutation specifically inactivated the ubiquitination activity of RNF8 [35]. The phenotypic similarity between RNF8-/C398F and RNF8-/- cells indicated the catalytic role of RNF8 is responsible for the promotion of TLS past abasic sites. Deficiency of RNF8 in HERC2-/- cells did not further reduce the rate of Ig hypermutation, indicating that HERC2 and RNF8 might operate in the same pathway, consistent with their role in HR [17]. Nonetheless, we must interpret the data very carefully to conclude an epistatic relationship between different ubiquitination enzymes. This is because an excess amount of DNA damage at the Ig V segment can cause cell death in TLS-deficient cells, possibly leading to an over-estimation of the actual number of non-templated mutations by analyzing only surviving cells. In summary, HERC2 and RNF8 promote TLS in the DT40 cell line probably through a collaboration of the two ubiquitination enzymes, as HERC2 activates RNF8 in DSB repair [17,18].
Similar to REV1, HERC2 and RNF8 are important for maintaining fork progression on damaged DNA but not for the filling of post-replicative gaps [31,52]. It is thus attractive to speculate that the two proteins may cooperate to restart replication forks by recruiting REV1 to stalled replication forks. One possible scenario is that HERC2 and RNF8 might facilitate the ubiquitination of REV1, since at least two studies have indicated that the direct conjugation of REV1 to ubiquitin [53,54]. Another possible scenario is that HERC2 and RNF8 might facilitate the ubiquitination of POLD3 [55], which is subjected to ubiquitination. Furthermore, similar to REV1, the non-essential subunit of replicative polymerase δ, POLD3, significantly contributes to maintenance of fork progression on damaged DNA but not for the filling of post-replicative gaps [47]. However, the phenotype of HERC2-/- and RNF8-/- mutants in Ig V diversification is somewhat different from that of REV1-/- or POLD3-/- mutants. The point mutation spectrum in HERC2-/- and RNF8-/- mutants is comparable to that in wild-type cells (Fig. 2C), while inactivation of REV1 strongly reduces point mutation from C/G to G/C [56]. HERC2-/- and RNF8-/- mutants are deficient also in Ig gene conversion (Fig. 2B), while POLD3-/- cells show the up-regulation of Ig gene conversion [47]. In summery, the regulation of REV1 or POLD3 by HERC2/RNF8 does not fully explain Ig V diversification shown in Fig. 2. Future studies will clarify substrates for HERC2 and RNF8 in promotion of TLS.
Supplementary Material
Acknowledgments
This work was supported by the JSPS Core-to-Core Program, A. Advanced Research Networks (to S.T.), a Grant-in-Aid for Scientific Research on Innovative Areas, and the Core-to-core Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to S.T.). Work in the Sale lab is supported by a core grant from the MRC to the Laboratory of Molecular Biology (U1051178808).
Abbreviations
- AID
activation-induced deaminase
- DSB
double strand break
- HR
homologous recombination
- Ig V gene
immunoglobulin variable gene
- MMS
methyl methanesulfonate
- PCNA
proliferating cell nuclear antigen
- XPA
xeroderma pigmentosum complementation group A
- PRR
post-replication repair
- TLS
translesion DNA synthesis
Footnotes
Conflict of interest statement
None declared.
References
- [1].Torres-Ramos CA, Prakash S, Prakash L. Requirement fo RAD5 and MMS2 for Postreplication Repair of UV-Damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Blastyák A, Pintér L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 Protein Required for Postreplication Repair Has a DNA Helicase Activity Specific for Replication Fork Regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Blastyák A, Hajdú I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- [5].Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, Branzei D. Visualization of recombination-mediated damage bypass by template switching. Nat Struct Mol Biol. 2014;21:884–893. doi: 10.1038/nsmb.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Izhar L, Goldsmith M, Dahan R, Geacintov N, Lloyd RG, Livneh Z. Analysis of Strand Transfer and Template Switching Mechanisms of DNA Gap Repair by Homologous Recombination in Escherichia coli: Predominance of Strand Transfer. J Mol Biol. 2008;381:803–809. doi: 10.1016/j.jmb.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Edmunds CE, Simpson LJ, Sale JE. PCNA Ubiquitination and REV1 Define Temporally Distinct Mechanisms for Controlling Translesion Synthesis in the Avian Cell Line DT40. Mol Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- [8].Hirota K, Sonoda E, Kawamoto T, Motegi A, Masutani C, Hanaoka F, et al. Simultaneous disruption of two DNA polymerases, Polη and Polζ, in avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet. 2010;6:e1001151. doi: 10.1371/journal.pgen.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- [11].Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- [12].Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- [14].Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e0040366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kang T-H, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kang TH, Reardon JT, Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 2011;39:3176–3187. doi: 10.1093/nar/gkq1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol. 2010;12:80–86. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- [18].Zhao Y, Brickner JR, Majid MC, Mosammaparast N. Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair. Trends Cell Biol. 2014;24:426–434. doi: 10.1016/j.tcb.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE Syndrome Protein Mediates a Ubiquitin-Dependent Signaling Cascade at Sites of DNA Damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- [20].Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- [21].Oestergaard VH, Pentzold C, Pedersen RT, Iosif S, Alpi A, Bekker-Jensen S, et al. RNF8 and RNF168 but not HERC2 are required for DNA damage-induced ubiquitylation in chicken DT40 cells. DNA Repair (Amst) 2012;11:892–905. doi: 10.1016/j.dnarep.2012.08.005. [DOI] [PubMed] [Google Scholar]
- [22].Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527:389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- [23].Buerstedde JM, Reynaud CA, Humphries EH, Olson W, Ewert DL, Weill JC. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sale JE. Immunoglobulin diversification in DT40: A model for vertebrate DNA damage tolerance. DNA Repair (Amst) 2004;3:693–702. doi: 10.1016/j.dnarep.2004.03.042. [DOI] [PubMed] [Google Scholar]
- [25].Arakawa H, Hauschild J, Buerstedde J-M. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- [26].Harris RS, Sale JE, Petersen-Mahrt SK, Neuberger MS. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr Biol. 2002;12:435–438. doi: 10.1016/s0960-9822(02)00717-0. [DOI] [PubMed] [Google Scholar]
- [27].Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- [28].Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- [29].Saribasak H, Saribasak NN, Ipek FM, Ellwart JW, Arakawa H, Buerstedde J-M. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J Immunol. 2006;176:365–371. doi: 10.4049/jimmunol.176.1.365. [DOI] [PubMed] [Google Scholar]
- [30].Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- [31].Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Saberi A, Nakahara M, Sale JE, Kikuchi K, Arakawa H, Buerstedde J-M, et al. The 9-1-1 DNA clamp is required for immunoglobulin gene conversion. Mol Cell Biol. 2008;28:6113–6122. doi: 10.1128/MCB.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nakahara M, Sonoda E, Nojima K, Sale JE, Takenaka K, Kikuchi K, et al. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig V in chicken B lymphocytes. PLoS Genet. 2009;5:e1000356. doi: 10.1371/journal.pgen.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kohzaki M, Nishihara K, Hirota K, Sonoda E, Yoshimura M, Ekino S, et al. DNA polymerases ν and θ are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol. 2010;189:1117–1127. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kobayashi S, Kasaishi Y, Nakada S, Takagi T, Era S, Motegi A, et al. Rad18 and Rnf8 facilitate homologous recombination by two distinct mechanisms, promoting Rad51 focus formation and suppressing the toxic effect of nonhomologous end joining. Oncogene. 2015;34:4403–4411. doi: 10.1038/onc.2014.371. [DOI] [PubMed] [Google Scholar]
- [36].Mohiuddin, Keka IS, Evans TJ, Hirota K, Shimizu H, Kono K, et al. A novel genotoxicity assay of carbon nanotubes using functional macrophage receptor with collagenous structure (MARCO)-expressing chicken B lymphocytes. Arch Toxicol. 2014;88:145–160. doi: 10.1007/s00204-013-1084-7. [DOI] [PubMed] [Google Scholar]
- [37].Keka IS, Mohiuddin, Maede Y, Rahman MM, Sakuma T, Honma M, et al. Smarcal1 promotes double-strand-break repair by nonhomologous end-joining. Nucleic Acids Res. 2015;43:6359–6372. doi: 10.1093/nar/gkv621. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, et al. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- [39].Guilbaud G, Rappailles A, Baker A, Chen CL, Arneodo A, Goldar A, et al. Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput Biol. 2011;7:e1002322. doi: 10.1371/journal.pcbi.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Huang J, Huen MSY, Kim H, Leung CCY, Glover JNM, Yu X, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Luijsterburg MS, Acs K, Ackermann L, Wiegant WW, Bekker-Jensen S, Larsen DH, et al. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 2012;31:2511–2527. doi: 10.1038/emboj.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Simpson LJ, Ross A-L, Szüts D, Alviani CA, Oestergaard VH, Patel KJ, et al. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang S, Chea J, Meng X, Zhou Y, Lee EYC, Lee MYWT. PCNA is ubiquitinated by RNF8. Cell Cycle. 2008;7:3399–3404. doi: 10.4161/cc.7.21.6949. [DOI] [PubMed] [Google Scholar]
- [45].Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, et al. A Critical Role for the Ubiquitin-Conjugating Enzyme Ubc13 in Initiating Homologous Recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- [46].Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, et al. Multiple roles of Rev3, the catalytic subunit of polζ in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hirota K, Yoshikiyo K, Guilbaud G, Tsurimoto T, Murai J, Tsuda M, et al. The POLD3 subunit of DNA polymerase can promote translesion synthesis independently of DNA polymerase. Nucleic Acids Res. 2015;43:1671–1683. doi: 10.1093/nar/gkv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yamashita YM, Okada T, Matsusaka T, Sonoda E, Zhao GY, Araki K, et al. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25:6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- [51].Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Guo C, Tang T-S, Bienko M, Parker JL, Bielen AB, Sonoda E, et al. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim H, Yang K, Dejsuphong D, D’Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol. 2012;19:164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu G, Warbrick E. The p66 and p12 subunits of DNA polymerase?? are modified by ubiquitin and ubiquitin-like proteins. Biochem Biophys Res Commun. 2006;349:360–366. doi: 10.1016/j.bbrc.2006.08.049. [DOI] [PubMed] [Google Scholar]
- [56].Ross AL, Sale JE. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.