Abstract
Ribosomes stall when they encounter the end of mRNA without an in-frame stop codon. In bacteria, these nonstop complexes can be rescued by alternative ribosome-rescue factor A (ArfA). Here, using electron cryomicroscopy, we have determined structures of ArfA bound to the ribosome with 3′-truncated mRNA to resolutions from 3.0 to 3.4 Å. ArfA binds within the ribosomal mRNA channel and substitutes for the absent stop codon in the A site by specifically recruiting release factor 2 (RF2), initially in a compact pre-accommodated state. A similar conformation of RF2 may occur on stop codons, suggesting a general mechanism for release-factor-mediated translational termination in which a conformational switch leads to peptide release only when the appropriate signal is present in the A site.
In bacteria, 2–4% of mRNA transcripts lack an in-frame stop codon due to faulty transcription or nucleolytic cleavage (1). When translated, the inability to recruit release factors causes ribosomes to stall at the 3′ end of these nonstop transcripts. Translating ribosomes also stall at the 3′ end of intact transcripts when a stop codon is either read through or bypassed by translational frameshifting (2, 3). As the accumulation of stalled ribosomes is potentially lethal (4), bacteria have evolved various mechanisms to rescue these complexes (2, 3). Trans-translation is the primary rescue mechanism, present in nearly all sequenced bacterial species, and redirects ribosomes to resume translation on transfer-messenger RNA (tmRNA). The reading frame of tmRNA encodes a degradation signal with a stop codon, which results in both recycling of the stalled ribosome and proteolysis of the aberrant polypeptide. Trans-translation is a promising target for antibiotic development (4); however, any therapeutic approach would need to circumvent the back-up mechanisms of alternative ribosome-rescue factors A (ArfA) and B (ArfB) which can allow some species of bacteria to survive in the absence of a functional trans-translation system (2, 3).
ArfA in particular acts as a fail-safe for trans-translation in many bacterial species (5). In Escherichia coli, ArfA can support continued growth in the absence of trans-translation with few phenotypic consequences (5, 6). Under normal conditions, the arfA transcript is cleaved by RNase III to produce a nonstop mRNA substrate for trans-translation, resulting in the truncated ArfA protein being tagged for degradation (7–9), although a small constitutively expressed population of full-length ArfA may result from the translation of uncleaved arfA transcripts (7). When trans-translation is impaired or overwhelmed, full-length ArfA may rescue the synthesis of its truncated form. ArfA relieves stalled ribosomes through a mechanism that requires peptidyl-tRNA hydrolysis by release factor 2 (RF2) but not the paralogous release factor 1 (RF1) (10, 11). This is in contrast to ArfB, which shares homology with the catalytic domains of these release factors and is able to directly hydrolyze peptidyl-tRNA within stalled ribosomes (12).
To understand how RF2 functions with ArfA instead of a stop codon, we solved the structure of the E. coli ribosome programmed with a 3′-truncated mRNA (Fig. S1, S2 and table S1), and in complex with ArfA and RF2 (Fig. 1A) or ArfA(A18T) and RF2 (Fig. 1B), by electron cryomicroscopy (cryo-EM) to resolutions between 3.0 and 3.4 Å. The amino-acid substitution A18T abolishes the ability of ArfA to support peptidyl-tRNA hydrolysis (5), although the mutant can still bind and recruit RF2 to stalled ribosomes (5, 11). The structures reveal two distinct conformations of RF2 on the ribosome and explain how ArfA specifically recognizes nonstop complexes.
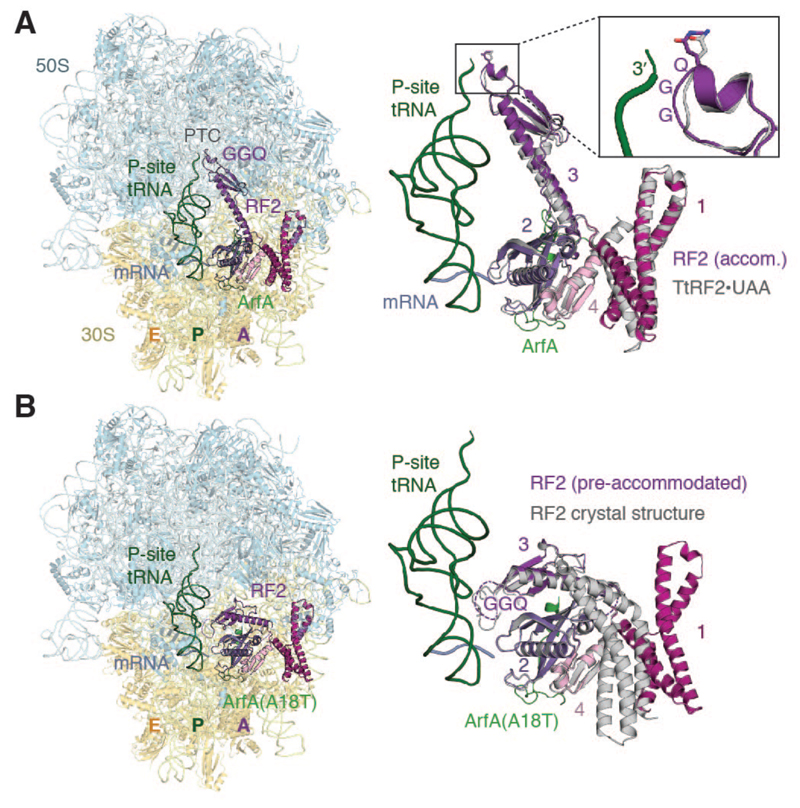
Fig. 1. Structures of nonstop complexes recognized by ArfA and ArfA(A18T).
(A) Overview of the bacterial ribosome with a 3′-truncated mRNA in complex with ArfA and RF2 (left). ArfA and RF2 occupy the A site with a nonhydrolyzable aminoacyl-tRNA in the P site. RF2 adopts a similar conformation on the ribosome with ArfA as with a UAA stop codon (right). The catalytic GGQ motif of RF2 domain 3 is accommodated within the PTC (inset). (B) Overview of a nonstop complex recognized by ArfA(A18T) (left). Pre-accommodated RF2 resembles the isolated RF2 crystal structure (PDB accession code 1GQE) (right). The GGQ motif faces the P-site tRNA. Movement of domain 1 results from contacts with the L7/L12 stalk base.
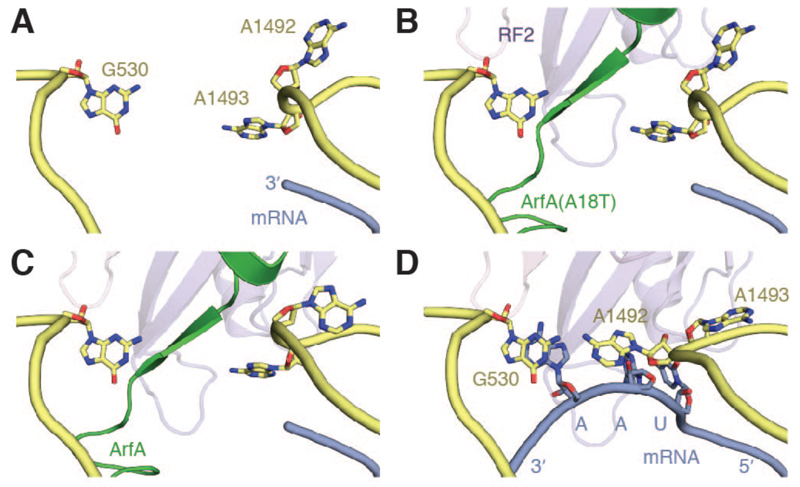
In both structures, ArfA interacts with the decoding-center nucleotides G530 and A1492 of the 16S rRNA (Fig. 2). A1492 is stacked within helix 44, interacting with ArfA via the rRNA backbone, whereas A1493 is flipped out and disordered. These nucleotides are unchanged from their positions in an unoccupied A site (Fig. 2A), suggesting that ArfA specifically recognizes a vacant decoding center. ArfA does not induce the remodeling of the decoding center, in particular the flipping-out of A1492, which occurs during stop-codon recognition (Fig. 2D).
Fig. 2. Conformations of the decoding nucleotides.
(A) Conformation of the decoding center with an unoccupied A site. A1493 is disordered in this and all ArfA-containing structures. (B) ArfA(A18T) recognizes a vacant A site and does not remodel the decoding center. (C) Closer contacts between wild-type ArfA, RF2 and the ribosome cause A1492 to switch from a syn to an anti configuration. (D) Canonical termination on a UAA stop codon involves remodeling of the decoding center that does not occur with ArfA.
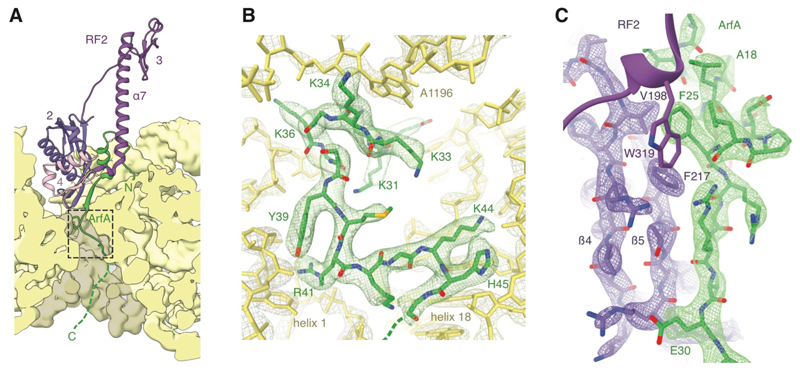
The C-terminus of ArfA protrudes into the otherwise-unoccupied mRNA channel downstream of the decoding center and, in agreement with hydroxyl-radical-probing data (13), contacts the rRNA that lines it (Fig. 3A). The conserved KGKGS motif (Fig. S3A, residues 34–37) forms an expansive helical turn within the channel that may help to anchor ArfA to the ribosome (Fig. 3B). Below the turn, ArfA binds first to helix 1 of the 16S rRNA and then to helix 18 on the opposite wall of the mRNA channel (Fig. 3B). The C-terminus beyond Gly46 exits through the channel entrance and makes few contacts with the ribosome. This is consistent with the last ~18 residues being truncated as a result of RNase-III cleavage and dispensable for ribosome rescue (8).
Fig. 3. Interactions between ArfA, RF2 and the ribosome.
(A) The N-terminal half of ArfA binds RF2 while the C-terminal tail occupies the mRNA channel. The path of ArfA as it emerges from the channel entrance, which can be traced in unfiltered maps, is indicated with a dashed line. (B) Interactions between ArfA and the rRNA lining the mRNA channel for the region highlighted in panel C as viewed from the channel entrance. (C) Both wild-type ArfA and ArfA(A18T) form an antiparallel β-sheet with domain 2 of RF2. F25 packs against an RF2-specific hydrophobic pocket formed by V198 and F217. With wild-type ArfA, this pocket is also recognized by W319 from the switch loop of accommodated RF2.
The positioning of the C-terminal tail suggests that ArfA may discriminate against translating ribosomes by recognizing vacant mRNA channels. A similar hypothesis has been proposed for small protein B (SmpB), which is delivered to the ribosome with tmRNA (14), and ArfB (12). SmpB and ArfB both have C-termini that insert into the mRNA channel, although these tails and their interactions with the ribosome are distinct from ArfA (Fig. S4). A discriminatory role for the ArfA C-terminus is consistent with the dramatic decline of ArfA-dependent peptide release as the 3′ length of mRNA increases (11, 15). However, recent data have shown that ArfA can bind ribosomes, and even recruit RF2, irrespective of mRNA in the channel (13). This suggests that simultaneous occupancy of the channel may be possible, but only in the absence of mRNA can ArfA position RF2 in a productive conformation for peptidyl-tRNA hydrolysis.
The basis for RF2 recruitment is provided by ArfA residues 25–30, which form a β-addition motif with the β5-strand of RF2 domain 2 (Fig. 3C). Phe25 of ArfA protrudes into a conserved hydrophobic pocket of RF2 formed by Val198 from the β4-strand and Phe217 from the β5-strand (Fig. 3C). This pocket is absent in RF1 from all known ArfA-containing species (Fig. S3B,C), likely explaining the exclusivity of ArfA for RF2 (10, 11). The RF2 recognition loop between the β4- and β5-strands that confers stop-codon specificity (16) faces solvent and does not interact with ArfA.
With wild-type ArfA, RF2 adopts an extended conformation that resembles the crystal structures of release factors bound to stop codons (16–20) (Fig. 1A). Consistent with a shared mechanism of catalysis for canonical and ArfA-mediated termination, the catalytic GGQ motif of domain 3 is accommodated within the peptidyl-transferase center (PTC) (Fig. 1A, inset). By contrast, RF2 adopts a compact conformation with ArfA(A18T) that resembles the crystal structures of isolated release factors (21–23), the only difference being the position of domain 1. In this “pre-accommodated” conformation, domain 3 lies across an eight-stranded β-sheet formed by domains 2 and 4, with the GGQ motif facing the anticodon arm of the P-site tRNA approximately 60 Å from the PTC (Fig. 1B).
A pre-accommodated conformation of release factors on ribosomes has previously been proposed as an initial codon-sampling state during stop-codon recognition (17, 18). This hypothesized state rationalizes the compact release-factor form seen in three different crystal structures (21–23) and in solution by small-angle X-ray scattering (SAXS) (23), although another SAXS study observed only the extended conformation (24). Further support for a physiological role of the compact form comes from hydroxyl-radical-probing (25) and Förster-resonance-energy-transfer (26) experiments that show discrete release-factor conformations on the ribosome depending on the identity of the A-site codon. However, due to its presumably transient nature, structural data for a codon-sampling state are lacking.
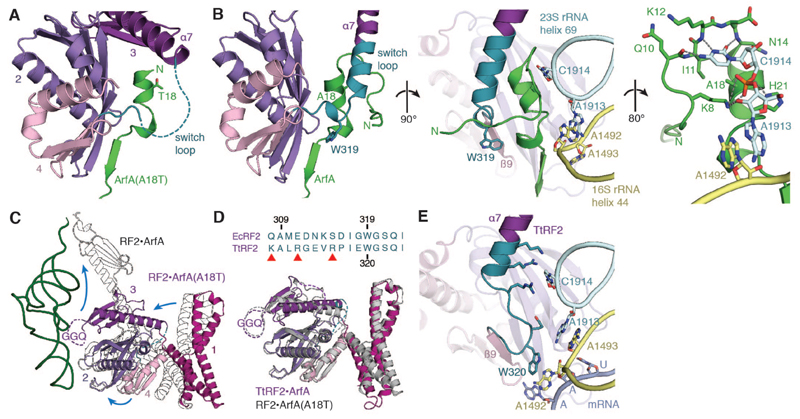
Here, the pre-accommodated conformation results from the A18T substitution, which prevents the interdependent folding of the ArfA N-terminus with the switch loop of RF2. The switch loop connects the α7-helix of domain 3 with the β9-strand of domain 4 and has been proposed to mediate accommodation during canonical termination by undergoing a disorder-to-order transition (19). In the mutant structure, the RF2 switch loop and the first 14 residues of ArfA are disordered. A18T is located within the α-helix of ArfA that packs against the central β-sheet of RF2, but faces away from the interface (Fig. 4A). However, the ordered N-terminus of wild-type ArfA turns tightly to run antiparallel to this α-helix (Fig. 4B). Compared to alanine, a polar threonine would be unable to pack against I11 in this conformation (Fig. 4B). By contrast, a hydrophobic A18C substitution has little effect on ArfA activity (13).
Fig. 4. Switch-loop stabilization and RF2 accommodation.
(A) In the structure of the nonstop complex recognized by ArfA(A18T), the first 14 residues of ArfA(A18T) and the switch loop between domains 3 and 4 of RF2 are disordered. (B) In the structure with wild-type ArfA, the ordered N-terminus of ArfA helps to stabilize the switch loop of RF2, which extends the α7-helix. A1913 from helix 69 of the 23S rRNA stacks with A1492 from helix 44 of the 16S rRNA, while C1914 from helix 69 stabilizes residues 10–14 of ArfA. (C) RF2 recruited by ArfA(A18T) adopts a compact conformation with the GGQ loop disordered and facing the P-site tRNA. Superposition with accommodated RF2 (outlined) reveals that all four domains move during accommodation. (D) TtRF2 has a switch-loop composition (top) incompatible with ArfA and adopts a pre-accommodated conformation on the ribosome (bottom) similar to E. coli RF2 recruited by ArfA(A18T). (E) During stop-codon recognition, the switch loop of TtRF2 is stabilized by interactions that are dependent on the stop-codon-induced remodeling of the decoding center.
The ordered N-terminus of wild-type ArfA induces conformational changes in the RF2 switch loop. As also occurs in stop-codon recognition (16, 19), the switch loop becomes partially α-helical, extending the α7-helix (Fig. 4B). This is followed by a ~100° kink as the conserved Trp319 interacts with ArfA at the RF2-specific hydrophobic pocket (Fig. 3C). An additional α-helical turn leads into the β9-strand. Compression of the switch loop draws the α7-helix, together with the rest of domain 3, from its interface with domain 2 such that the GGQ motif rises and accommodates into the PTC (Fig. 4C).
Upon accommodation, there is also a ~10° rotation of RF2 domains 2 and 4 (Fig. 4C) which increases the interface with the ribosome by 25%. Together with a movement of the ArfA α-helix, these changes result in a tighter fit between ArfA, RF2 and the decoding center. The decoding nucleotide A1492 of the 16S rRNA switches from a syn to an anti configuration within helix 44 (Fig. 2C) and stacks with A1913 at the apex of helix 69 of the 23S rRNA. This may help to reposition helix 69, allowing C1914 to coordinate the turn adopted by ArfA residues 11–14 (Fig. 4B). The movement of domain 2 appears to pull domain 1, together with the L7/L12 stalk base, into closer contact with the small subunit, possibly stabilizing RF2 in its accommodated state (Fig. S5A).
To further explore the interaction between ArfA and the switch loop, we examined the compatibility of wild-type ArfA from E. coli with Thermus thermophilus RF2 (TtRF2), which has a distinct switch-loop composition (Fig. 4D). The structure shows that, although TtRF2 is recruited to stalled ribosomes through the conserved hydrophobic pocket with ArfA, it adopts a compact conformation similar to that of E. coli RF2 recruited by ArfA(A18T) (Fig. 4D) and the N-terminus of ArfA remains disordered. The inability to accommodate presumably results from clashes between ArfA and the longer side chains of the TtRF2 switch loop. Taken together, our structures demonstrate that accommodation of RF2 is dependent on switch-loop stabilization by the ArfA N-terminus.
These switch-loop interactions appear to emulate the interactions between RF2 and the ribosome that result from stop-codon-dependent rearrangement of the decoding center (Fig. 4E). During canonical termination, the conserved tryptophan of the switch loop stacks with flipped-out A1492 while the α7-helix extension interacts directly with helix 69, which adopts a different conformation when A1492 is flipped out (16, 20).
The delivery of RF2 in a pre-accommodated state followed by a conformational switch that depends on specific changes in the decoding center shares many parallels with the universal elongation (27) and eukaryotic termination pathways (28) (Fig. S5B,C). Both aminoacyl-tRNAs and the structurally unrelated eukaryotic release factor 1 (eRF1) adopt pre-accommodated conformations during codon sampling that prevent premature engagement of reactive groups with the PTC before accommodating into the PTC upon codon recognition. A similar pathway for bacterial release factors is consistent with a 4,000-fold difference in dissociation rates between stop and sense codons despite similar association rates (29) and may explain the remarkable accuracy of termination in bacteria (~10−5), which is comparable to the fidelity of aminoacyl-tRNA selection (30).
In summary, our structures reveal the mechanism of ArfA-mediated ribosome rescue on 3′-truncated mRNA and provide new insights into how a conserved conformational switch might maintain the accuracy of translational termination in bacteria.
Supplementary Material
One Sentence Summary.
Structures reveal how ArfA rescues stalled ribosomes and suggest a general mechanism for release-factor-mediated translational termination.
Acknowledgements
We thank Y. Shimizu for providing the arfA clone, A.C. Kelley for providing tRNA, R.S. Hegde for providing E. coli RF2, S.H.W. Scheres for help with RELION-2.0, J. Grimmett and T. Darling for computing support, and C.D. Rae and S. Shao for comments. The work was supported by grants to V.R. from the UK Medical Research Council (MC_U105184332), the Wellcome Trust (WT096570), the Agouron Institute and the Louis-Jeantet Foundation. The maps have been deposited with the Electron Microscopy Data Bank under the accession codes EMD-3489, EMD-3490, EMD-3492, and EMD-3493. Atomic coordinates have been deposited with the Protein Data Bank under the accession codes 5MDV, 5MDW, 5MDY, and 5MDZ.
References
- 1.Ito K, et al. PLOS ONE. 2011;6:e28413. doi: 10.1371/journal.pone.0028413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keiler KC. Nat Rev Micro. 2015;13:285–297. doi: 10.1038/nrmicro3438. [DOI] [PubMed] [Google Scholar]
- 3.Giudice E, Gillet R. Trends Biochem Sci. 2013;38:403–411. doi: 10.1016/j.tibs.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Ramadoss NS, et al. Proc Natl Acad Sci USA. 2013;110:10282–10287. doi: 10.1073/pnas.1302816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chadani Y, et al. Mol Microbiol. 2010;78:796–808. doi: 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- 6.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. Proc Natl Acad Sci USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chadani Y, et al. Genes Genet Syst. 2011;86:151–163. doi: 10.1266/ggs.86.151. [DOI] [PubMed] [Google Scholar]
- 8.Garza-Sánchez F, Schaub RE, Janssen BD, Hayes CS. Mol Microbiol. 2011;80:1204–1219. doi: 10.1111/j.1365-2958.2011.07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaub RE, Poole SJ, Garza-Sanchez F, Benbow S, Hayes CS. J Biol Chem. 2012;287:29765–29775. doi: 10.1074/jbc.M112.374074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chadani Y, Ito K, Kutsukake K, Abo T. Mol Microbiol. 2012;86:37–50. doi: 10.1111/j.1365-2958.2012.08190.x. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu Y. J Mol Biol. 2012;423:624–631. doi: 10.1016/j.jmb.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. Science. 2012;335:1370–1372. doi: 10.1126/science.1217443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurita D, Chadani Y, Muto A, Abo T, Himeno H. Nucleic Acids Res. 2014;42:13339–13352. doi: 10.1093/nar/gku1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neubauer C, Gillet R, Kelley AC, Ramakrishnan V. Science. 2012;335:1366–1369. doi: 10.1126/science.1217039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng F, Jin H. RNA. 2016;22:49–60. doi: 10.1261/rna.053082.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weixlbaumer A, et al. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rawat UBS, et al. Nature. 2003;421:87–90. [Google Scholar]
- 18.Klaholz BP, et al. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 19.Laurberg M, et al. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 20.Korostelev A, et al. Proc Natl Acad Sci USA. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vestergaard B, et al. Mol Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 22.Shin DH, et al. J Mol Biol. 2004;341:227–239. doi: 10.1016/j.jmb.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 23.Zoldák G, et al. Nucleic Acids Res. 2007;35:1343–1353. doi: 10.1093/nar/gkl696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vestergaard B, et al. Mol Cell. 2005;20:929–938. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 25.He SL, Green R. Nat Struct Mol Biol. 2010;17:465–470. doi: 10.1038/nsmb.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trappl K, Joseph S. J Mol Biol. 2016;428:1333–1344. doi: 10.1016/j.jmb.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 27.Voorhees RM, Ramakrishnan V. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 28.Shao S, et al. Cell. 2016 doi: 10.1016/j.cell.2016.10.046. [DOI] [Google Scholar]
- 29.Hetrick B, Lee K, Joseph S. Biochemistry. 2009;48:11178–11184. doi: 10.1021/bi901577d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. Proc Natl Acad Sci USA. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.