Abstract
This study describes the development of a gene-specific DNA microarray coupled with multiplex PCR for the comprehensive detection of pathogenic vibrios that are natural inhabitants of warm coastal waters and shellfish. Multiplex PCR with vvh and viuB for Vibrio vulnificus, with ompU, toxR, tcpI, and hlyA for V. cholerae, and with tlh, tdh, trh, and open reading frame 8 for V. parahaemolyticus helped to ensure that total and pathogenic strains, including subtypes of the three Vibrio spp., could be detected and discriminated. For DNA microarrays, oligonucleotide probes for these targeted genes were deposited onto epoxysilane-derivatized, 12-well, Teflon-masked slides by using a MicroGrid II arrayer. Amplified PCR products were hybridized to arrays at 50°C and detected by using tyramide signal amplification with Alexa Fluor 546 fluorescent dye. Slides were imaged by using an arrayWoRx scanner. The detection sensitivity for pure cultures without enrichment was 102 to 103 CFU/ml, and the specificity was 100%. However, 5 h of sample enrichment followed by DNA extraction with Instagene matrix and multiplex PCR with microarray hybridization resulted in the detection of 1 CFU in 1 g of oyster tissue homogenate. Thus, enrichment of the bacterial pathogens permitted higher sensitivity in compliance with the Interstate Shellfish Sanitation Conference guideline. Application of the DNA microarray methodology to natural oysters revealed the presence of V. vulnificus (100%) and V. parahaemolyticus (83%). However, V. cholerae was not detected in natural oysters. An assay involving a combination of multiplex PCR and DNA microarray hybridization would help to ensure rapid and accurate detection of pathogenic vibrios in shellfish, thereby improving the microbiological safety of shellfish for consumers.
The warm coastal waters of temperate regions, including the Gulf of Mexico (gulf water), support a diverse aquatic biota and a robust seafood industry. This environment, however, also provides suitable conditions for the growth of pathogenic bacteria that pose problems for human health. Molluscan shellfish, especially oysters, concentrate microorganisms from surrounding waters during the filter-feeding process. Consequently, filter feeders are recognized as reservoirs for various microbial pathogens, including Vibrio spp. (36). Among the various Vibrio spp. that inhabit marine and estuarine environments, Vibrio vulnificus, V. parahaemolyticus, and V. cholerae are primarily involved in causing gastrointestinal illnesses or, in some cases, septicemia. Infections arise due to the consumption of raw, undercooked, or improperly processed shellfish or other seafood containing significant levels of bacteria.
Infections caused by V. vulnificus often result in fatal consequences, with a mortality rate of up to 60%, primarily in individuals who are immunocompromised or have liver disease (27, 31). Due to an increased number of disease incidents, the state of California in 2003 released emergency restrictions on the sale of all oysters harvested between April and October from the Gulf of Mexico unless they are treated with a scientifically validated process to reduce V. vulnificus to a nondetectable level (<30 CFU/g of oyster) (California Department of Health Services, April 2003, [http://www.dhs.ca.gov]). Besides illnesses caused by V. vulnificus, sporadic outbreaks of gastrointestinal illnesses have also been caused by V. parahaemolyticus. The first reported outbreak in the United States occurred in 1982, affecting 10 individuals, followed by the occurrence of a relatively larger outbreak in the Pacific Northwest, affecting 209 people and including 1 death (14). Recently, a newly emerging serotype of V. parahaemolyticus, O3:K6, marked the largest reported food-borne outbreak in the United States, affecting 416 individuals (19, 20).
Although V. vulnificus and V. parahaemolyticus are considered the two major causative agents for shellfish-associated bacterial illnesses, incidents of cholera in the United States have also been reported in recent years (11-13, 41). Sporadic cases of V. cholerae-associated illnesses in the United States from the consumption of raw or undercooked shellfish suggest the reintroduction of this pathogen into the U.S. marine and estuarine environments, primarily from South America and Asia (10, 30). Pathogenic strains of V. parahaemolyticus and V. cholerae have been involved in epidemics worldwide (16, 29, 33, 39, 45) The bacterial load in shellfish, including pathogenic strains, increases in coastal waters during summer months and thereby increases opportunities for outbreaks of food-borne illnesses, which are a cause for concern for the seafood industry. Recently, the Centers for Disease Control and Prevention estimated a 126% increase in the incidence of Vibrio-associated infections between 1996 and 2002, despite efforts directed at seafood consumers (especially high-risk consumers) to warn them of the potential hazards of eating raw shellfish (15). Targeted efforts to reduce the rate of food-borne illnesses should therefore include steps to decrease the prevalence of pathogenic bacteria in seafood as well as the development of rapid, specific, and sensitive detection methods.
Conventional methods of pathogen detection, such as the most-probable-number technique in association with biochemical tests or colony blot hybridization with gene-specific probes, are limited by the time and labor involved in analyzing a large number of samples (25). Detection of pathogens by PCR has been shown to be highly specific and relatively less time-consuming than conventional methods. However, discrimination of potentially virulent strains by PCR demands the targeting of multiple genes. During the last decade, several reports have described PCR detection and discrimination of total and pathogenic V. vulnificus, V. parahaemolyticus, and V. cholerae strains, including selected serotypes (4, 23, 34, 38). However, comprehensive detection of all of the aforementioned pathogenic vibrios by the targeting of at least 10 genes would be unreliable by conventional PCR. Moreover, the Taqman probe-based real-time PCR is limited by the ability to detect only 4-fluorophores in a single reaction. In most situations, postamplification detection methods for each of the pathogens must be applied to ensure that the amplicons were in fact generated from targeted gene segments during the PCR process. Comprehensive detection of the amplicons from all of the pathogenic vibrios in a single assay would significantly increase the speed of detection and thereby improve the microbiological safety of seafood.
Recently, the DNA microarray hybridization approach was shown to be effective for the detection of amplicons generated by PCR from multiple food-borne microorganisms, including pathogenic strains (17, 46). In this format, the DNA array involves the immobilization of numerous oligonucleotide DNA probes at high densities on a solid support to which fluorescence-labeled PCR-amplified DNAs are hybridized. It allows for the identification of microbial pathogens that are present in clinical as well as environmental samples, such as water, soil, or food products, and that could be hazardous to human health. The application of a DNA microarray hybridization approach for the detection of total and pathogenic Listeria, Campylobacter, Shigella, and Escherichia coli strains has been reported (8, 26, 42, 43). In these studies, various serovars, pathotypes, and virulence genes of each of these pathogens were successfully identified by multigene hybridizations on a DNA array. In separate studies, PCR-amplified DNAs in various combinations from 18 microbial pathogens, including bacteria, viruses, and eukaryotes, were detected on a single DNA array with high sensitivity and accuracy; in addition, 15 bacterial species were discriminated based on 16S ribosomal DNA polymorphisms (44, 46).
With the increased incidence of seafood-related illnesses caused by vibrios, rapid detection and discrimination of total and pathogenic strains by multiplex PCR amplification followed by analysis of all amplicons together on a single DNA array would help to ensure a steady supply of oysters that are safe to eat and thereby protect consumer health and financially benefit the seafood industry. In this study, we coupled multiplex PCR with a DNA array for the comprehensive detection of V. vulnificus, V. parahaemolyticus, and V. cholerae in shellfish.
MATERIALS AND METHODS
Bacterial strains and media.
The following bacterial strains were used in the present study: V. cholerae 154 serovar O:1 Inaba Tox (ATCC 14100), V. vulnificus MO6-24 (O), V. parahaemolyticus F113-A, and V. parahaemolyticus serotype O3:K6 BAC 98-03372. V. vulnificus was maintained on 0.5× (18.7 g/liter) marine agar (Becton Dickinson, Franklin Lakes, N.J.), V. cholerae was maintained on Luria-Bertani agar (Difco), and V. parahaemolyticus was maintained on nutrient agar (Becton Dickinson) supplemented with 3% (wt/vol) NaCl. All cultures were grown at 37°C. The seeded oyster tissue homogenates were enriched with gulf water (salinity, 18 ppt) supplemented with 0.2% (wt/vol) Bacto Peptone (GWP-18) (Becton Dickinson). Also, modified cellobiose-polymyxin-colistin (mCPC) agar and thiosulfate-citrate-bile-sucrose (TCBS) agar were used to obtain viable plate counts for the three vibrio species (2).
Genomic DNA extraction.
Genomic DNA was extracted from exponential cultures (1 ml) of the organisms by centrifugation (9,000 × g) followed by resuspension of the cell pellet in 567 μl of TE buffer (10 mM Tris-Cl, 1.0 mM EDTA [pH 8]). Alkaline lysis was performed with 30 μl of 10% (wt/vol) sodium dodecyl sulfate (SDS) and 3 μl of proteinase K (20 mg/ml; Sigma, St. Louis, Mo.) by using a procedure adapted from that of Ausubel et al. (3). After 1 h of incubation, 100 μl of 5 M NaCl was added along with 80 μl of cetyltrimethylammonium bromide (CTAB)-NaCl solution to complex with polysaccharides. DNA was purified from proteins and other cellular constituents with an equal volume (780 μl) of chloroform-isoamyl alcohol (24:1) and was centrifuged at 10,000 × g for 5 min. The supernatant was transferred to a new tube with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). The resulting supernatant was transferred to a new tube with 300 μl of isopropanol and was centrifuged at 10,000 × g for 5 min to pellet DNA. The pellet was washed once with 1 ml of cold 70% (vol/vol) ethanol before being dried under vacuum. Purified DNA was resuspended in 50 μl of TE buffer.
Oligonucleotide primers and probes.
The targeted genes, oligonucleotide primer sets, and DNA probes used for the detection of each of the three pathogens are listed in Table 1. The oligonucleotide probes were designed based on the nucleotide sequences internal to the amplified segments of the respective targeted genes. All oligonucleotide probes were selected by using Primer Quest software and custom synthesized along with the primers at Integrated DNA Technology, Inc. (Coralville, Idaho). The oligonucleotides synthesized at Integrated DNA Technology were deprotected and desalted. Potential probe sequences were analyzed for specificity by comparison with known gene sequences in GenBank by using the BLAST N search program provided by the National Center for Biotechnology Information.
TABLE 1.
Oligonucleotide primers and probes used for multiplex PCR and DNA array hybridization
| Pathogen | Target gene | Primer and probe sequencesa | Oligonucleotide length | % GC content | Tmb (°C) | Amplicon size (kbp) | Reference |
|---|---|---|---|---|---|---|---|
| V. vulnificus | vvh | F-VVH: 5′-TTCCAACTTCAAACCGAACTATGAC-3′ | 25 | 40.0 | 61.3 | 0.205 | 6 |
| R-VVH: 5′B-ATTCCAGTCGATGCGAATACGTTG-3′ | 24 | 45.8 | 62.8 | ||||
| P-VVH: 5′-GAAGCGCCCGTGTCTGAAACTGGCGTAACG-3′ | 30 | 60.0 | 72.3 | ||||
| viuB | F-VIUB1728: 5′-GGTTGGGCACTAAAGGCAGATATA-3′ | 24 | 45.8 | 62.8 | 0.504 | 34 | |
| R-VIUB2231: 5′B-CGGCAGTGGACTAATACGCAGC-3′ | 22 | 59.9 | 66.4 | ||||
| P-VIUB: 5′-GAGAAAGCGACCTCGCTGTCTGCTTTTATC-3′ | 30 | 50.0 | 68.7 | ||||
| V. parahaemolyticus | tlh | F-TLH: 5′-AAAGCGGATTATGCAGAAGCACTG-3′ | 24 | 45.8 | 62.8 | 0.45 | 4 |
| R-TLH: 5′B-GCTACTTTCTAGCATTTTCTCTGC-3′ | 24 | 41.6 | 61.1 | ||||
| P-TLH: 5′-ACGGACGCAGGTGCGAAGAACTTCATGTTG-3′ | 30 | 53.3 | 70.1 | ||||
| P-TLH2: 5′-TTGCGCTCTGAGTGTGCAGCGTCTGGTGCT-3′ | 30 | 60.0 | 72.8 | ||||
| tdh | F-TDH: 5′-GTAAAGGTCTCTGACTTTTGGAC-3′ | 23 | 43.5 | 57.7 | 0.269 | 4 | |
| R-TDH: 5′B-TGGAATAGAACCTTCATCTTCACC-3′ | 24 | 41.7 | 61.1 | ||||
| P-TDH: 5′-GCGGTGTCTGGCTATAAGAGCGGTCATTCT-3′ | 30 | 53.3 | 70.0 | ||||
| trh | F-TRH: 5′-TTGGCTTCGATATTTTCAGTATCT-3′ | 24 | 33.3 | 57.7 | 0.5 | 4 | |
| R-TRH: 5′B-CATAACAAACATATGCCCATTTCCG-3′ | 25 | 40.0 | 61.3 | ||||
| P-TRH: 5′-AAAACTGAATCACCAGTTAACGCAATCGTT-3′ | 30 | 36.6 | 63.2 | ||||
| P-TRH2: 5′-ACAAGCCTAAATCAGAACTATTCTTCTGTTAG-3′ | 32 | 34.3 | 63.3 | ||||
| P-TRH3: 5′-TTGACCTACCATCCATACCTTTTCCTTCTCCA-3′ | 32 | 43.7 | 67.2 | ||||
| ORF8 | F-O3MM824: 5′-AGGACGCAGTTACGCTTGATG-3′ | 21 | 52.4 | 62.5 | 0.369 | 32 | |
| R-O3MM1192: 5′B-CTAACGCATTGTCCCTTTGTAG-3′ | 22 | 45.4 | 60.8 | ||||
| P-ORF8: 5′-CTCACTCCTGCTGTACTTTTAGCTCGGTTA-3′ | 30 | 46.6 | 67.3 | ||||
| V. cholerae | ompU | F-OMPU: 5′-ACGCTGACGGAATCAACCAAAG-3′ | 22 | 50.0 | 62.6 | 0.869 | 37 |
| R-OMPU: 5′B-GCGGAAGTTTGGCTTGAAGTAG-3′ | 22 | 50.0 | 62.6 | ||||
| P-OMPU: 5′-TCACTGACTTCACCGATATCATGTCTTACC-3′ | 30 | 43.3 | 66.0 | ||||
| toxR | F-TOXR: 5′-CCTTCGATCCCCTAAGCAATAC-3′ | 22 | 50.0 | 62.6 | 0.779 | 37 | |
| R-TOXR: 5′B-AGGGTTAGCAACGATGCGTAAG-3′ | 22 | 50.0 | 62.6 | ||||
| P-TOXR: 5′-GAATCAAGCAGTGTGCCTTCATCATCCACT-3′ | 30 | 46.6 | 67.3 | ||||
| tcpI | F-TCPI: 5′-TAGCCTTAGTTCTCAGCAGGCA-3′ | 22 | 50.0 | 62.6 | 0.862 | 37 | |
| R-TCPI: 5′B-GGCAATAGTGTCGAGCTCGTTA-3′ | 22 | 50.0 | 62.6 | ||||
| P-TCPI: 5′-ATCTGATCTCACTCACAGTACCTTAATTGG-3′ | 30 | 40.0 | 64.6 | ||||
| Classical or El Tor | hlyA | F-HLYA: 5′-GGCAAACAGCGAAACAAATACC-3′ | 22 | 45.4 | 60.8 | 0.738 or 0.727 | 37 |
| R-HLYA: 5′B-CTCAGCGGGCTAATACGGTTTA-3′ | 22 | 50.0 | 62.6 | ||||
| P-HLYA: 5′-AGCCAAACTAGAGGCGAGAGCAAGTTATAC-3′ | 30 | 46.7 | 67.4 |
F, forward primer; R, reverse primer (biotinylated [B] at the 5′ end); P, oligonucleotide probe. The oligonucleotide probes for the respective genes were selected for this study.
Tm, melting temperature.
Multiplex PCR and nick translation.
Each multiplex PCR amplification was performed with a 50-μl reaction volume consisting of 10 ng of purified genomic DNA, 200 μM deoxynucleoside triphosphates (Sigma), 3 U of AmpliTaq DNA polymerase (Promega Corporation, Madison, Wis.), and PCR buffer F [60 mM Tris-Cl (pH 9.0), 15 mM (NH4)2SO4, 2 mM MgCl2] or buffer C [50 mM Tris-Cl (pH 8.9), 50 mM KCl, 2.5 mM MgCl2]. Amplification of the targeted genes for V. cholerae and V. parahaemolyticus was carried out with 1 μM each oligonucleotide primer. However, for the detection of V. vulnificus, 1 μM each of oligonucleotide primers F-vvh and R-vvh and 2 μM each of oligonucleotide primers F-viuB and R-viuB were used in order to achieve optimum multiplex PCR amplification of vvh and viuB, respectively. All PCR amplifications were performed by using a GeneAmp 2400 thermocycler (Perkin-Elmer, Shelton, Conn.) with parameters specific for the gene sets to be amplified. Reaction parameters included initial denaturation at 94°C for 3 min; 30 cycles of amplification with denaturation at 94°C for 1 min and primer annealing for 1 min at 55°C for V. parahaemolyticus, 65°C for V. vulnificus, or 60°C for V. cholerae; primer extension at 72°C for 1 min; and final extension of the incompletely synthesized DNA at 72°C for 5 min. Amplified products (5 μl) were analyzed by agarose gel electrophoresis (2% [wt/vol] Nusieve or 1% [wt/vol] SeaKem agarose [FMC Bioproducts, Rockland, Maine]) or SDS-polyacrylamide gel electrophoresis (5% [wt/vol] polyacrylamide). Gels were stained with ethidium bromide (0.005% [wt/vol]) and visualized under a Photoprep I UV transilluminator (Fotodyne, Inc., Hartland, Wis.).
Multiplex PCR mixtures (50 μl) were subjected to nick translation by using a nick translation kit (Amersham Biosciences Corp., Piscataway, N.J.). Following the incorporation of biotinylated dCTP, the reactions were terminated with 5 μl of 0.2 M disodium EDTA, and the reaction mixtures were denatured at 95°C for 5 min. The nick translation-PCR products were dried by using a Speed Vac DNA 110 instrument (Savant, Sunnyvale, Calif.) and were stored at −80°C until used for hybridization with DNA arrays.
DNA array preparation and use.
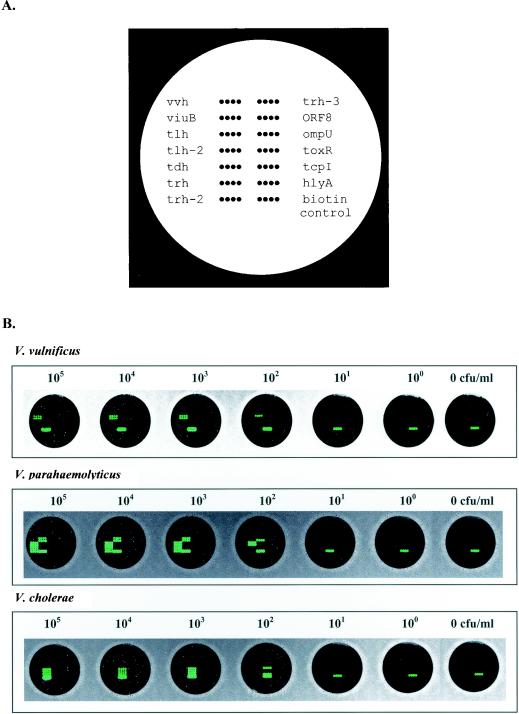
Teflon-masked, 12-well microscope slides (Erie Scientific, Porthsmouth, N.H.) were cleaned, acid washed, and derivatized with epoxysilane (3-glycidoxypropyltrimethoxysilane; Sigma-Aldrich, Milwaukee, Wis.) as described by Call et al. (9). Oligonucleotide probes were diluted in spotting buffer (0.1 M NaH2PO4, 0.2 M NaCl, 0.01% SDS) to a final concentration of 60 μM and transferred to a 96-well polystyrene plate (Robbins Scientific Corp., Sunnyvale, Calif.) for spotting. Twelve independent DNA arrays (1 per masked well) were spotted onto each slide by using a MicroGrid II arrayer (BioRobotics Inc., Woburn, Mass.). Each probe was spotted as four replicates on each array. Every array included an arbitrary oligonucleotide probe (25-mer) conjugated with biotin. These biotin pseudoprobes served as positive controls for the detection chemistry and as orientation points for image processing. A schematic diagram of the positions of the probes spotted on the array is shown in Fig. 1A. After spotting was completed, slides were baked for 1 h at 130°C and stored at room temperature until used.
FIG. 1.
(A) Schematic diagram showing the positions of the immobilized probes spotted in quadruplicate within a single well of the 12-well Teflon-masked glass slides. (B) Sensitivity of detection of genomic DNAs extracted from 10-fold serially diluted pure cultures of V. vulnificus, V. parahaemolyticus, and V. cholerae prior to enrichment. The range of dilutions used for these cultures extended from 105 CFU/ml to extinction. The results shown here are representative of experiments carried out in duplicate.
Biotinylated targets (biotin conjugated to primers or incorporated by nick translation; 10 μl) were diluted to a total volume of 70 μl in hybridization buffer (4× SSC [1× SSC is 15 mM NaCl plus 0.15 mM sodium citrate] [pH 7.0], 5× Denhardt's solution [0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin]), heat denatured at 98°C for 3 min, and hybridized overnight at 50°C to replicate arrays on the same slide. Slide preblocking and detection protocols were carried out as described by Call et al. (7). Briefly, after hybridization, slides were washed three times for 1 min each time with 100 mM Tris-HCl (pH 7.5)-150 mM NaCl-0.05% Tween 20 buffer with agitation. A tyramide signal amplification biotin system (Perkin-Elmer, Boston, Mass.) was used in conjunction with Alexa Fluor 546 (Molecular Probes, Eugene, Oreg.) to detect target-probe hybrids. Upon completion of detection chemistry, arrays were imaged by using an arrayWoRx scanner (Applied Precision, Issaquah, Wash.), and images were processed by using softWoRx software. The assay was repeated on replicate arrays from the same print lot.
Detection of Vibrio spp. in unenriched pure cultures.
The Vibrio spp. were grown separately in 10 ml of GWP-18 (autoclaved at 121°C and 15 lb/in2 for 20 min) to an optical density at 600 nm of 0.2. The cultures were 10-fold serially diluted, and viable plate counts were determined. Aliquots (1 ml) of each of the undiluted cultures were mixed together and 10-fold serially diluted to extinction in 10 ml of GWP-18. Aliquots (1 ml) were centrifuged at 12,000 × g for 10 min, and the cell pellets were treated with 200 μl of Instagene (Bio-Rad Laboratories, Hercules, Calif.) matrix (35). The samples were incubated at 56°C for 10 min, boiled for 20 min, cooled to room temperature, and centrifuged at 3,000 × g for 3 min to collect DNA. A 3-μl aliquot of each sample was used for PCR amplification.
PCR inhibition study with oysters.
Live oysters were purchased from a local seafood store and processed by standard methods (1). The inhibition control experiments were performed with unseeded oyster tissue homogenates that were enriched for 5 h at 37°C in a rotary shaker incubator (Innova 4000; New Brunswick Scientific Co. Inc., Edison, N.J.). Following enrichment, 5-ml aliquots were centrifuged and treated with Instagene matrix as described above. Next, 10-fold serial dilutions of purified genomic DNA ranging from 1.0 μg to 0.1 pg of V. cholerae 154 serovar O:1 Inaba Tox (ATCC 14100), V. vulnificus MO6-24 (O), V. parahaemolyticus F113-A, and V. parahaemolyticus BAC 98-03372 were added to 3-μl aliquots of the Instagene matrix-treated unseeded oyster tissue homogenates and used for PCR. The cycling and reaction parameters used for PCR were as stated above. The sensitivity of detection obtained with multiplex PCR of oyster tissue homogenates was compared with the sensitivity of detection obtained with purified DNA serially diluted in MilliQ water.
Detection of seeded microbial pathogens in oysters.
Three aliquots (1 g each) of market-oyster tissue homogenates were 10-fold serially diluted in GWP-18 and plated on mCPC agar for V. vulnificus and TCBS agar for V. cholerae and V. parahaemolyticus to determine viable plate counts. Individual yellow cellobiose-positive colonies on mCPC agar plates for V. vulnificus or yellow or blue colonies on TCBS agar for V. cholerae or V. parahaemolyticus, respectively, were PCR amplified with species-specific primers and the parameters stated above to confirm the actual numbers of viable vibrio cells in the samples. Cells of each of the microbial strains were grown separately in 10 ml of GWP-18 to an optical density at 600 nm of 0.2, and viable plate counts were determined. Equal samples of each of the cultures were mixed together and 10-fold serially diluted to extinction in 50 ml of GWP-18 with 1 g of oyster tissue homogenate. The seeded oyster tissue homogenates were enriched for 5 h at 37°C in a rotary shaker incubator (Innova 4000). Following enrichment, 5-ml aliquots were centrifuged at 12,000 × g for 10 min, and the supernatants were discarded. The cell pellets were treated with 200 μl of Instagene matrix as described above, and DNA was released (35). A 3-μl aliquot of each sample was used for PCR amplification.
Detection of Vibrio spp. in natural oysters.
Oysters were collected from the Gulf of Mexico near Bayou LaBatre, Ala., from June through August 2003. The temperature was recorded, and the salinity was determined by using a salinity refractometer (Reichert Scientific Instruments, Buffalo, N.Y.) during collection. Each collection included three samples, and each of these samples consisted of 10 oysters. The oysters from each sample were chilled immediately after collection, the shell surfaces were cleaned and shucked, and the oysters were homogenized for 2 min in a Waring blender (Fisher Scientific) according to the procedure outlined by the American Public Health Association (1). One gram of oyster tissue homogenate from each sample was added to 50 ml of GWP-18 and enriched for 5 h as described above. For testing of the samples, 5-ml aliquots were centrifuged at 12,000 × g for 10 min, and the cell pellets were treated with 200 μl of Instagene matrix as described above to release DNA (35). A 3-μl aliquot of each sample was used for PCR amplification. All reactions were performed in triplicate.
RESULTS
Multiplex PCR.
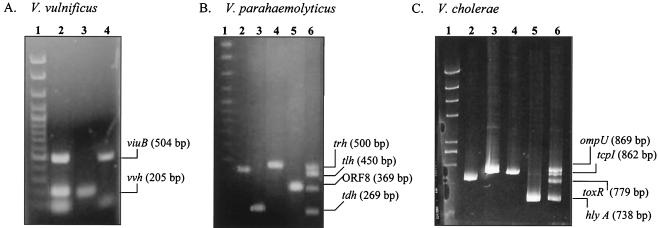
The oligonucleotide primers selected for PCR amplification of the three Vibrio spp. were specific for their respective targeted gene segments, as shown in previous studies (4, 32, 34, 35, 37, 38). At an annealing temperature of 65°C and with 2.5 mM MgCl2, PCR amplification of purified genomic DNA from V. vulnificus MO6-24 yielded DNA fragments of 205 and 504 bp for vvh (total V. vulnificus) and viuB (pathogenic strains), respectively (Fig. 2A). Using the PCR conditions described above, we successfully multiplexed four targeted genes for V. parahaemolyticus strains (Fig. 2B): tlh (450 bp) for total V. parahaemolyticus and tdh (269 bp), trh (500 bp), and open reading frame 8 (ORF8) (369 bp), representing pathogenic strains. Also, targeted genes for total and pathogenic V. cholerae strains were amplified at an optimum primer annealing temperature of 60°C and with 2 mM MgCl2 (Fig. 2C): hlyA (738 bp) (toxigenic and nontoxigenic V. cholerae O1 and O139 and V. cholerae non-O1 and non-O139), tcpI (862 bp) (toxigenic and nontoxigenic V. cholerae O1 and non-O139), toxR (779 bp) (all V. cholerae serogroups), and ompU (869 bp) (V. cholerae O1 and O139 and environmental V. cholerae non-O1 and non-O139). These targeted genes were selected based on differences in distributions in various V. cholerae serogroups as described by Rivera et al. (37). Multiplex PCR targeting the simultaneous amplification of 10 targeted gene segments from the three Vibrio spp. in a single reaction tube produced inconsistent results (data not shown).
FIG. 2.
Agarose gel electrophoresis showing the results of multiplex PCR amplification of three Vibrio spp. (A) V. vulnificus: lane 1, DNA size standard (Clone-sizer; Norgen Biotek); lane 2, multiplex PCR of both vvh and viuB targets; lane 3, 205-bp vvh amplicon; lane 4, 504-bp viuB amplicon. (B) V. parahaemolyticus: lane 1, DNA size standard; lane 2, 450-bp tlh amplicon; lane 3, 269-bp tdh amplicon; lane 4, 500-bp trh amplicon; lane 5, 369-bp ORF8 amplicon; lane 6, multiplex PCR amplification of all four targeted gene segments. (C) V. cholerae: lane 1, DNA size standard; lane 2, 779-bp toxR amplicon; lane 3, 869-bp ompU amplicon; lane 4, 862-bp tcpI amplicon; lane 5, 738-bp hlyA amplicon; lane 6, multiplex PCR amplification of all four targeted gene segments.
Specificity of oligonucleotide probes used in DNA array hybridization.
PCR products were labeled with biotinylated PCR primers or by nick translation. Both individual and multiplexed targets from the three Vibrio spp. hybridized to their complementary probes (100% specificity) at a hybridization temperature of 50°C (Fig. 1B). For V. parahaemolyticus, three out of the four targets (tlh, tdh, and ORF8) could not be detected unless the products were nick translated prior to hybridization. This was true for both individual and multiplexed PCR products. Similar results were observed with the ompU target in V. cholerae. Both vvh and viuB gene segments in V. vulnificus hybridized to their respective probes with or without labeling by nick translation.
Sensitivity of detection in pure cultures.
Unenriched cultures of Vibrio spp. grown in GWP-18 exhibited a detection level of 103 CFU/ml for all targets in a multiplex PCR followed by DNA array hybridization (Fig. 1B). However, certain targets, such as vvh, tlh, ORF8, ompU, and hlyA, were detected individually at levels as low as 102 CFU/ml in the multiplex PCR.
PCR inhibition control study.
The results from the inhibition control experiments indicated that an oyster tissue matrix spiked with purified DNA had an inhibitory effect on the multiplex PCR, resulting in a level of detection that was 10-fold lower than that seen with purified DNA (data not shown). The level of detection for vvh and viuB was 10 pg for DNA mixed with oyster tissue, whereas it was 1 pg for purified DNA. For V. parahaemolyticus and V. cholerae, consistent amplification of all targets was observed at 100 pg for oyster tissue spiked with DNA. This result could have been due to inefficient amplification of four targeted genes with higher molecular weights in the presence of the oyster matrix resulting in a lower sensitivity of detection.
Sensitivity of detection in oysters following enrichment.
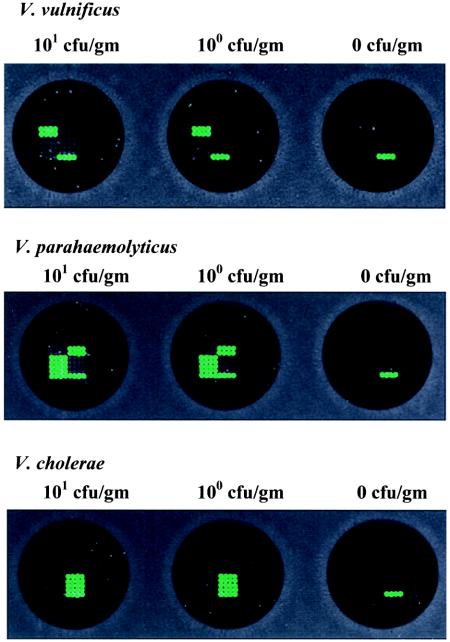
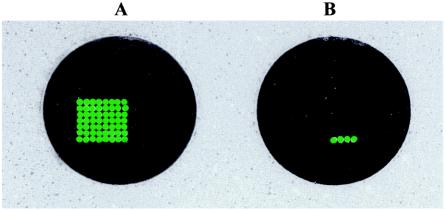
The Instagene matrix used for DNA extraction from seeded oyster tissue homogenates resulted in sufficient DNA for PCR detection followed by DNA array hybridization of all targeted genes at an initial cell count of 1 CFU/g for each Vibrio sp. (Fig. 3). In addition, simultaneous hybridization of 10 PCR-amplified targeted genes from 5-h-enriched Vibrio spp. at an initial cell count of 1 CFU/g resulted in the detection of all targeted genes on a single DNA array (Fig. 4). These results indicated that a single DNA array hybridization reaction can be used for a comprehensive, reliable, and sensitive analysis of PCR-amplified genes from all three Vibrio spp. and potentially pathogenic genes in shellfish with a level of sensitivity that is in compliance with the Interstate Shellfish Sanitation Conference (ISSC) guideline (24).
FIG. 3.
Sensitivity of detection of V. vulnificus, V. parahaemolyticus, and V. cholerae in seeded oyster tissue homogenates following 5 h of enrichment. The arrays shown here represent detection in samples at dilutions ranging from an initial cell count of 101 CFU/g to extinction. The results shown here are representative of experiments carried out in duplicate.
FIG. 4.
DNA array showing simultaneous hybridization of all 10 PCR-amplified targets of V. vulnificus, V. parahaemolyticus, and V. cholerae in seeded oyster tissue homogenates following 5 h of enrichment. The results shown here are representative of experiments carried out in duplicate. (A) 100 CFU of V. vulnificus, V. parahaemolyticus, and V. cholerae seeded in 1 g of oyster tissue homogenates. (B) Unseeded oyster tissue homogenates.
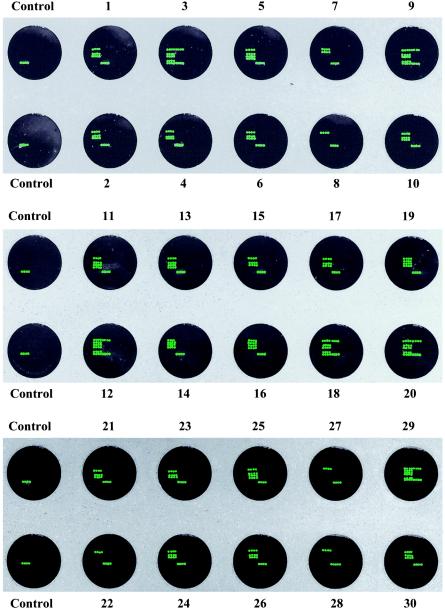
Detection of Vibrio spp. in natural oysters.
The specificity of PCR-amplified targeted genes from natural oyster samples enriched for 5 h was confirmed by DNA array hybridization (Table 2 and Fig. 5). The water temperature ranged from 24 to 26°C, and the salinity was recorded at 16 to 22 ppt. All 30 samples were found to be positive for vvh, whereas only 20% were found to be positive for viuB (Table 2). V. parahaemolyticus was also present in most samples (83.3%); 13 to 20% of the strains were pathogenic strains possessing either tdh, trh, or both (Table 2). ORF8 was not detected in any of the oyster samples that were found to be positive for V. parahaemolyticus. The distribution of pathogenic strains observed in this experiment for V. vulnificus and V. parahaemolyticus concurs with the results of previously published reports (21, 40). None of the samples was found to be positive for the ompU, hylA, toxR, and tcpI targeted genes, suggesting that V. cholerae is not prevalent in gulf water.
TABLE 2.
Results of multiplex PCR and DNA array hybridization of natural oyster samples
| Sample | Resulta for:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
V. vulnificus
|
V. parahaemolyticus
|
V. cholerae
|
||||||||
| vvh | viuB | tlh | tdh | trh | ORF8 | toxR | ompU | tcpI | hlyA | |
| 1 | + | − | + | − | − | − | − | − | − | − |
| 2 | + | − | + | − | − | − | − | − | − | − |
| 3 | + | − | + | − | + | − | − | − | − | − |
| 4 | + | − | + | − | − | − | − | − | − | − |
| 5 | + | − | + | + | − | − | − | − | − | − |
| 6 | + | − | + | − | − | − | − | − | − | − |
| 7 | + | + | − | − | − | − | − | − | − | − |
| 8 | + | − | − | − | − | − | − | − | − | − |
| 9 | + | − | + | − | + | − | − | − | − | − |
| 10 | + | − | + | − | − | − | − | − | − | − |
| 11 | + | − | + | + | − | − | − | − | − | − |
| 12 | + | + | + | − | + | − | − | − | − | − |
| 13 | + | − | + | + | − | − | − | − | − | − |
| 14 | + | + | + | − | − | − | − | − | − | − |
| 15 | + | − | + | − | − | − | − | − | − | − |
| 16 | + | + | + | − | − | − | − | − | − | − |
| 17 | + | − | + | − | − | − | − | − | − | − |
| 18 | + | − | + | − | + | − | − | − | − | − |
| 19 | + | + | + | − | − | − | − | − | − | − |
| 20 | + | − | + | − | + | − | − | − | − | − |
| 21 | + | − | + | − | − | − | − | − | − | − |
| 22 | + | − | − | − | − | − | − | − | − | − |
| 23 | + | − | + | − | − | − | − | − | − | − |
| 24 | + | − | + | − | − | − | − | − | − | − |
| 25 | + | − | + | + | − | − | − | − | − | − |
| 26 | + | − | + | − | − | − | − | − | − | − |
| 27 | + | − | − | − | − | − | − | − | − | − |
| 28 | + | − | − | − | − | − | − | − | − | − |
| 29 | + | + | + | − | + | − | − | − | − | − |
| 30 | + | − | + | − | − | − | − | − | − | − |
| % Positive samples | 100 | 20 | 83.3 | 13.3 | 20 | 0 | 0 | 0 | 0 | 0 |
+, positive; −, negative.
FIG. 5.
Application of the DNA array-based detection of PCR-amplified gene segments from V. vulnificus, V. parahaemolyticus, and V. cholerae in natural oyster samples collected from gulf water. Arrays 1 to 30 represent DNA array hybridization of PCR-amplified targets from 30 oyster samples collected from June through August 2003. The control was a PCR-negative control that was also hybridized to an individual array. The results shown here are representative of experiments carried out in duplicate.
DISCUSSION
The seafood industry, especially the Gulf Coast oyster industry, continues to suffer from elevated V. vulnificus levels in uncooked oysters and associated fatal illnesses (22). Based on the severity of this problem, the ISSC has proposed that illnesses caused by V. vulnificus due to the consumption of raw or poorly cooked shellfish must be reduced at least 60% by the year 2007 (24). Besides V. vulnificus, the presence of high levels of pathogenic V. parahaemolyticus in shellfish during the summer months causes sporadic as well as extensive outbreaks of gastroenteritis in humans (18). Although V. cholerae-related incidents are rare in the United States, routine monitoring of V. cholerae in shellfish is essential, as this species has been identified as a category B organism in the list of pathogens released by the U.S. government (www.niaid.nih.gov/biodefense/bandcpriority.htm). Moreover, frequent outbreaks of cholera in Central America, South America, Africa, and Southeast Asia could cause accidental transfer of this pathogen into coastal waters of the United States through planktonic attachment, ship ballast water, or unauthorized import of seafood products not kept at appropriate temperatures (10).
Previous studies showed that in order to identify a potentially pathogenic strain, it is necessary to target multiple genes for PCR amplification. In this study, species-specific tlh, pathogenic strain-specific tdh and trh, and serotype O3:K6-specific ORF8 were selected for the detection of V. parahaemolyticus in shellfish by multiplex PCR (4, 32). Similarly, species-specific vvh (35) and clinical strain-specific viuB (34) were selected for the detection of V. vulnificus in this study. For a comprehensive detection of various serogroups of V. cholerae, targeted genes were selected based on a previous study by Rivera et al. (37). In their study, ompU, tcpI, hlyA, and toxR were found to be present in V. cholerae strains including O1, non-O1, nontoxigenic O1, O139, and non-O139 serovars. Although there are differences in the nucleotide sequences of hlyA in V. cholerae classical and El Tor biotypes, the oligonucleotide probe used in this study was selected from the segment of the gene containing the consensus sequence, enabling the detection of both biotypes. In addition, PCR amplification of multiple genes instead of a single gene from each of the Vibrio spp. would help to reduce false-negative identification during testing of a complex matrix such as shellfish tissue. However, multigene PCR amplification is limited to individual Vibrio spp.
In this study, we established a DNA array hybridization assay that enabled us to confirm the post-PCR detection of these naturally occurring pathogenic vibrios and selected serogroups in shellfish that have a history of causing disease outbreaks, often with fatalities. This single DNA array hybridization assay would save valuable time in the detection of these pathogens, thus helping to protect consumer health by reducing shellfish-related disease outbreaks.
During this study, we were able to detect PCR products that were biotinylated via a 5′ conjugated primer. In some instances, however, terminally labeled products were detected only when they were also nick translated. The difference in detection likely was due to secondary structures on the target DNA (28). Secondary structures can interfere with microarray hybridization, whereas the nick translation process fragments the DNA and thereby disrupts interfering secondary structures. An alternative labeling approach would be to incorporate labeled nucleotides during PCR followed by sonication of amplicons to disrupt any secondary structures. While this latter approach includes a more expensive PCR, we expect some time savings from the elimination of the need for nick translation before hybridization (1 to 2 h).
These pathogens were detected in a mixed culture without enrichment at 102 to 103 CFU/ml, a level which was not sufficient to meet the recommended minimum detection level needed to declare a food product safe for human consumption (24). Therefore, in order to achieve a higher level of detection, it was necessary to enrich bacterial cells in shellfish tissue homogenates. In previous studies, enrichment of oyster tissue homogenates was used to achieve the detection of single cells of V. vulnificus and V. parahaemolyticus (5, 35). A 5-h enrichment step followed by a relatively simple DNA purification step of boiling in Instagene matrix enabled us to achieve reliable detection of a single pathogen in shellfish tissue homogenate samples. Although PCR inhibition by the oyster matrix was evident in this study, the inclusion of a 5-h enrichment step resulted in a sufficient number of cells to achieve detection of the targeted pathogens.
Agarose gel electrophoresis analysis of amplified products was found to be consistent with the positive hybridization signals of gene-specific probes on the DNA array. In addition, the use of tyramide signal amplification with DNA array hybridization enabled us to attain high colorimetric signals from samples containing 1 CFU of each of the targeted pathogens. The level of detection achieved in this study was sufficient to meet the required ISSC guideline for the detection of these pathogens in shellfish samples. The robustness of the assay was further confirmed by the detection of these vibrios with high degrees of specificity and sensitivity in natural oyster samples.
To our knowledge, this is the first report describing a comprehensive DNA array-based analysis of multiplex PCR-amplified gene segments of Vibrio spp. that are present in shellfish samples and that are considered to be the primary etiologic agents of shellfish-related bacterial illnesses in humans. The estimated cost for the assay described here is almost half that of Taqman real-time PCR or other commercially available conventional microbiological culture and biochemical methods followed by confirmatory tests, such as DNA-DNA colony hybridization. The combination, developed in this study, of DNA array hybridization with multiplex PCR of targeted genes from pathogenic vibrios in shellfish can be further expanded in the future as needed for the detection of enteric viruses, protozoans, and coliform bacteria as indicators of fecal contamination of shellfish and shellfish harvest sites. Rapid, reliable, and comprehensive detection of multigene amplicons from pathogenic vibrios and other etiologic agents in a single DNA array hybridization assay with required specificity and sensitivity is important for a steady supply of microbiologically safe seafood products in order to guard consumer health and protect the shellfish industry from financial loses.
Acknowledgments
This work was supported in part by funding from the Mississippi Alabama SeaGrant Consortium; the University of Alabama at Birmingham under research grant R/SP-8; and the Agricultural Animal Health Program, College of Veterinary Medicine, Pullman, Wash.
We thank Angelo Depaola for providing some of the bacterial strains used in this study and for helpful suggestions.
REFERENCES
- 1.American Public Health Association. 1970. Recommended procedures for the examination of seawater and shellfish, 4th ed. American Public Health Association, Washington, D.C.
- 2.Atlas, R. M. 1993. Handbook of microbiological media. CRC Press, Inc., Boca Raton, Fla.
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Smith, J. G. Sideman, and K. Struhl (ed.). 1987. Current protocols in molecular biology, p. 2.10-2.11. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bej, A. K., D. P. Patterson, C. W. Brasher, M. C. Vickery, D. D. Jones, and C. A. Kaysner. 1999. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36:215-225. [DOI] [PubMed] [Google Scholar]
- 5.Blackstone, G. M., J. L. Nordstrom, M. C. L. Vickery, M. D. Bowen, R. F. Meyer, and A. DePaola. 2003. Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. J. Microbiol. Methods 53:149-155. [DOI] [PubMed] [Google Scholar]
- 6.Brasher, C. W., A. DePaola, D. D. Jones, and A. K. Bej. 1998. Detection of microbial pathogens in shellfish with multiplex PCR. Curr. Microbiol. 37:101-107. [DOI] [PubMed] [Google Scholar]
- 7.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Call, D. R., F. J. Brockman, and D. P. Chandler. 2001. Detecting and genotyping Escherichia coli 157:H7 using multiplexed PCR and nucleic acid microarrays. Int. J. Food Microbiol. 67:71-80. [DOI] [PubMed] [Google Scholar]
- 9.Call, D. R., D. P. Chandler, and F. Brockman. 2001. Fabrication of DNA microarrays using unmodified oligonucleotide probes. BioTechniques 30:368-379. [DOI] [PubMed] [Google Scholar]
- 10.Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration. 1992. Vibrio cholerae O1 serogroup, p. 78-89. In Foodborne pathogenic microorganisms and natural toxins handbook. U.S. Food and Drug Administration, Washington, D.C.
- 11.Centers for Disease Control and Prevention. 1982. Epidemiologic notes and reports on non-O1 Vibrio cholerae gastroenteritis—New Hampshire. Morb. Mortal. Wkly. Rep. 31:538-539. [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1993. Imported cholera associated with a newly described toxigenic Vibrio cholerae O139 strain—California 1993. Morb. Mortal. Wkly. Rep. 42:501-503. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1993. Isolation of Vibrio cholerae O1 from oysters—Mobile Bay, 1991-1992. Morb. Mortal. Wkly. Rep. 42:91-93. [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 1999. Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters—Pacific Northwest, 1997. Morb. Mortal. Wkly. Rep. 47:457-462. [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2003. Preliminary FoodNet data on the incidence of foodborne illnesses—selected sites, United States, 2002. Morb. Mortal. Wkly. Rep. 52:340-343. [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. 1995. Update: Vibrio cholerae O1—Western Hemisphere, 1991-1994, and V. cholerae O139—Asia, 1994. JAMA 273:1169. [PubMed] [Google Scholar]
- 17.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croci, L., P. Serratore, L. Cozzi, A. Stacchini, S. Milandri, E. Suffredini, and L. Toti. 2001. Detection of Vibrionaceae in mussels and in their seawater growing area. Lett. Appl. Microbiol. 32:57-61. [DOI] [PubMed] [Google Scholar]
- 19.Daniels, N. A., B. Ray, A. Easton, N. Marano, E. Kahn, A. L. McShan II, L. Del Rosario, T. Baldwin, M. A. Kingsley, N. D. Puhr, J. G. Wells, and F. J. Angulo. 2000. Emergence of a new Vibrio parahaemolyticus serotype in raw oysters: a prevention quandary. JAMA 284:1541-1545. [DOI] [PubMed] [Google Scholar]
- 20.DePaola, A., C. A. Kaysner, J. Bowers, and D. W. Cook. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66:4649-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePaola, A., J. L. Nordstrom, J. C. Bowers, J. G. Wells, and D. W. Cook. 2003. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69:1521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glatzer, M. 2001. Vibrio vulnificus shellfish cases file from 1989-2000. U.S. Food and Drug Administration, Washington, D.C.
- 23.Hoshino, K., S. Yamasaki, A. K. Mukhopadhyay, S. Chakraborty, A. Basu, S. K. Bhattacharya, G. B. Nair, T. Shimada, and Y. Takeda. 1998. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol. Med. Microbiol. 20:201-207. [DOI] [PubMed] [Google Scholar]
- 24.Interstate Shellfish Sanitation Conference. 2002. Issues relating to a Vibrio vulnificus risk management plan for oysters. Interstate Shellfish Sanitation Conference, Columbia, S.C.
- 25.Kaysner, C. A., and A. DePaola, Jr. 2001. Vibrio, p. 405-420. In F. P. Downes and K. Ito (ed.), Compendium of methods for the microbiological examination of food. American Public Health Association, Washington, D.C.
- 26.Keramas, G., D. D. Bang, M. Lund, M. Madsen, S. E. Rasmussen, H. Bunkenborg, P. Telleman, and C. B. Christensen. 2003. Development of a sensitive DNA microarray suitable for rapid detection of Campylobacter spp. Mol. Cell. Probes 17:187-196. [DOI] [PubMed] [Google Scholar]
- 27.Kumamoto, K. S., and D. J. Vukich. 1998. Clinical infections of Vibrio vulnificus: a case report and review of the literature. J. Emerg. Med. 16:61-66. [DOI] [PubMed] [Google Scholar]
- 28.Lane, S., J. Evermann, F. Loge, and D. R. Call. 2004. Secondary structure prevents target hybridization to oligonucleotide microarrays. Biosens. Bioelectron. 20:728-735. [DOI] [PubMed] [Google Scholar]
- 29.Lozano-Leon, A., J. Torres, C. R. Osorio, and J. Martinez-Urtaza. 2003. Identification of tdh-positive Vibrio parahaemolyticus from an outbreak associated with raw oyster consumption in Spain. FEMS Microbiol. Lett. 226:281-284. [DOI] [PubMed] [Google Scholar]
- 30.McCarthy, S. A., R. M. McPhearson, A. M. Guarino, and J. L. Gaines. 1992. Toxigenic Vibrio cholerae O1 and cargo ships entering Gulf of Mexico. Lancet 339:624-625. [DOI] [PubMed] [Google Scholar]
- 31.Mead, P. S., L. Slutsker, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States reply to Dr. Hedberg. Emerg. Infect. Dis. 5:841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers, M. L., G. Panicker, and A. K. Bej. 2003. PCR detection of a newly emerged pandemic Vibrio parahaemolyticus O3:K6 pathogen in pure cultures and seeded waters from the Gulf of Mexico. Appl. Environ. Microbiol. 69:2194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan, T. M., T. K. Wang, C. L. Lee, S. W. Chien, and C. B. Horng. 1997. Food-borne disease outbreaks due to bacteria in Taiwan, 1986 to 1995. J. Clin. Microbiol. 35:1260-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panicker, G., M. C. L. Vickery, and A. K. Bej. Multiplex PCR detection of clinical and environmental strains of Vibrio vulnificus in shellfish. Can. J. Microbiol., in press. [DOI] [PubMed]
- 35.Panicker, G., M. L. Myers, and A. K. Bej. 2004. Rapid detection of Vibrio vulnificus in shellfish and Gulf of Mexico water by real-time PCR. Appl. Environ. Microbiol. 70:498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potasman, I., A. Paz, and M. Odeh. 2002. Infectious outbreaks associated with bivalve shellfish consumption: a worldwide perspective. Clin. Infect. Dis. 35:921-928. [DOI] [PubMed] [Google Scholar]
- 37.Rivera, I. N., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivera, I. N., E. K. Lipp, A. Gil, N. Choopun, A. Huq, and R. R. Colwell. 2003. Method of DNA extraction and application of multiplex polymerase chain reaction to detect toxigenic Vibrio cholerae O1 and O139 from aquatic ecosystems. Environ. Microbiol. 5:599-606. [DOI] [PubMed] [Google Scholar]
- 39.Sack, D. A., R. B. Sack, G. B. Nair, and A. K. Siddique. 2004. Cholera. Lancet 363:223-233. [DOI] [PubMed] [Google Scholar]
- 40.Shapiro, R. L., S. Altekruse, L. Hutwagner, R. Bishop, R. Hammond, S. Wilson, B. Ray, S. Thompson, R. V. Tauxe, P. M. Griffin, et al. 1998. The role of Gulf Coast oysters harvested in warmer months in Vibrio vulnificus infections in the United States, 1988-1996. J. Infect. Dis. 178:752-759. [DOI] [PubMed] [Google Scholar]
- 41.Stroube, R. B. 2002. Vibrio infections, Virginia, 1992-2002. Va. Epidemiol. Bull. 102:1-4. [Google Scholar]
- 42.van Ijperen, C., P. Kuhnert, J. Frey, and J. P. Clewley. 2002. Virulence typing of Escherichia coli using microarrays. Mol. Cell. Probes 16:371-378. [DOI] [PubMed] [Google Scholar]
- 43.Volokhov, D., A. Rasooly, K. Chumakov, and V. Chizhikov. 2002. Identification of Listeria species by microarray-based assay. J. Clin. Microbiol. 40:4720-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warsen, A., M. J. Krug, S. LaFrentz, D. Stanek, F. J. Loge, and D. R. Call. 2004. Simultaneous discrimination between 15 fish pathogens by using 16S ribosomal DNA PCR and DNA microarrays. Appl. Environ. Microbiol. 70:4216-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber, J. T., E. D. Mintz, R. Canizares, A. Semiglia, I. Gomez, R. Sempertegui, A. Davila, K. D. Greene, N. D. Puhr, and D. N. Cameron. 1994. Epidemic cholera in Ecuador: multidrug-resistance and transmission by water and seafood. Epidemiol. Infect. 112:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, W. J., C. L. Strout, T. Z. DeSantis, J. L. Stilwell, A. V. Carrano, and G. L. Andersen. 2002. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes 16:119-127. [DOI] [PubMed] [Google Scholar]