Abstract
The enzymatic degradation of l-methionine and subsequent formation of volatile sulfur compounds (VSCs) is believed to be essential for flavor development in cheese. l-Methionine-γ-lyase (MGL) can convert l-methionine to methanethiol (MTL), α-ketobutyrate, and ammonia. The mgl gene encoding MGL was cloned from the type strain Brevibacterium linens ATCC 9175 known to produce copious amounts of MTL and related VSCs. The disruption of the mgl gene, achieved in strain ATCC 9175, resulted in a 62% decrease in thiol-producing activity and a 97% decrease in total VSC production in the knockout strain. Our work shows that l-methionine degradation via γ-elimination is a key step in the formation of VSCs in B. linens.
Due to their low detection threshold and diversity, volatile sulfur compounds (VSCs) are of prime importance in the overall flavor of cheese and make a significant contribution to the typical aromas of different cheeses (12, 14, 33). VSCs arise primarily from the degradation of l-methionine to methanethiol (MTL) by the cheese microflora. This thiol is a common precursor for a variety of other sulfur-bearing compounds including the auto-oxidation products (11), dimethyl disulfide (DMDS), dimethyl trisulfide (DMTS), and S-methylthioesters, primarily arising from chemical reaction of MTL with acyl coenzyme A (acyl-CoA) (22). Numerous studies have therefore been done to control and/or diversify VSC synthesis during the ripening process by the use of properly selected microorganisms (4, 6, 15, 43). Many cheese microorganisms are capable of producing VSCs from l-methionine. Some of them, such as brevibacteria, especially Brevibacterium linens (17), are known to be very good VSC producers while others, such as lactic acid bacteria (LAB), can produce only limited amounts of VSCs (14).
The most direct route for MTL biosynthesis, is the l-methionine γ-elimination that directly produces MTL, α-ketobutyrate, and ammonia from l-methionine. This l-methionine γ-elimination activity is quite high in B. linens and corynebacteria (17) and is also suspected in several other cheese surface bacteria, such as Micrococcus luteus, Arthrobacter sp., and Staphylococcus equorum (8). In contrast, such activity is quite low in LAB (14). In B. linens, the methionine γ-elimination is catalyzed by a l-methionine-γ-lyase (MGL), a pyridoxal phosphate (PLP)-dependent enzyme for which l-methionine is the best substrate (16). In contrast, in LAB the reaction is catalyzed by a cystathionine β-lyase (CBL) and a cystathionine γ-lyase (CGL) which are only slightly active towards L-methionine (1, 10, 18). In LAB, another pathway for l-methionine conversion to VSCs also exists but produces limited amounts of MTL (7, 35).
Coryneform bacteria are generally found on the surface of smear cheeses and give the typical sulfur notes to cheeses such as Limburger, Tilsiter, Livarot, Epoisses, and Munster. To date, B. linens is the only food-grade bacterium from which MGL has been purified and characterized (16, 26, 31, 38, 39), but neither its protein sequence nor its gene sequence are presently available. Nevertheless, previous work (30) and a preliminary study carried out in our laboratories (unpublished data) have shown that B. linens strains could be transformed with plasmids, therefore allowing the construction of B. linens mutants. Furthermore, the recent availability of a part of the genome sequence of B. linens (U.S. Department of Energy Joint Genome Institute), together with the publication of MGL sequences of bacteria (NCBI accession no. NP 617435, NP 635109, NP 693968, NP 241665, and NP 604313), gave us new tools for picking up the gene encoding MGL in B. linens.
The objective of this study was to identify and characterize the gene responsible for MTL production from l-methionine in B. linens. For this purpose, a putative MGL gene was inactivated in B. linens and the consequences on VSC production were investigated.
MATERIALS AND METHODS
Chemicals. All chemicals were at least of analytical grade from Sigma-Aldrich (St. Quentin Fallavier, France), Bio-Rad (Marnes la Coquette, France), Boehringer (Mannheim, Germany), or Merck (Fontenay-sous-Bois, France), unless otherwise stated. Radiolabeled l-methionine was obtained from Sigma-Aldrich.
Bacterial strains, plasmids, and culture conditions.
B. linens strains were grown aerobically in 100 ml of TSYE medium (pH 7.5) at 25°C for 3 days and 150 rpm in 500-ml Erlenmeyer flasks. TSYE medium is composed of 22.7 g of tryptone peptone (Difco, Detroit, Mich.) liter−1, 4 g of papaic digest of soybean meal (Biokar Diagnostics, Beauvais, France) liter−1, 6 g of yeast extract (Labosi, Oulchy-le-Chāteau, France) liter−1, 3.33 g of glucose liter−1, 3.33 g of K2HPO4 liter−1, and 6.67 g of NaCl liter−1. Escherichia coli strains were grown aerobically in Luria-Bertani medium at 37°C with or without 1.5% agar (36). When necessary, kanamycin (5 μg ml−1 for B. linens), erythromycin (150 μg ml−1 for E. coli), or ampicillin (50 μg ml−1 for E. coli) was added to the culture medium.
DNA techniques.
All DNA manipulations were performed as described by Sambrook et al. (36). DNA restriction and modification enzymes were purchased from Gibco-BRL (Cergy Pontoise, France), Eurogentec (Seraing, Belgium), or Boehringer and used as recommended by the suppliers. E. coli electrocompetent cells were prepared and transformed by using standard techniques (24, 36).
Electrocompetent cells of B. linens were prepared as previously described (30) with a few modifications. Cells were grown aerobically in TSYE medium supplemented with 10% succinate and 1.5% glycine at 25°C. After 72 h, 0.3 μg of penicillin ml−1 was added and cell culture was continued for 5 h more. Cells were then harvested by centrifugation (10,000 × g, 10 min, 4°C), washed twice with 0.1 culture volume of cold 0.8 M saccharose, and resuspended in the same solution to a final optical density at 570 nm (OD570) of 60, and stored at −80°C until use.
Total DNA from B. linens was prepared as previously described (13). B. linens cells were harvested from 18-h-old agitated (150 rpm) liquid cultures at 25°C in 50-ml Erlenmeyer flasks containing 10 ml of TSYE medium (pH 7.5) by centrifugation (6,000 × g, 15 min, 4°C). They were washed in 10 mM Tris-HCl (pH 8) and 1 mM EDTA (TE buffer) and resuspended in 800 μl of TE buffer. Cells were treated with lysozyme (600 μg ml−1) and mutanolysine (30 U ml−1) for 2 h at 37°C, and then lysates were treated with sodium dodecyl sulfate (SDS) and proteinase K (Merck) at final concentrations of 1% (wt/vol) and 10 U (0.5 mg) ml−1, respectively, for 2 h at 55°C. NaCl and hexadecyl trimethyl ammonium bromide were added at final concentrations of 1 M and 40 mM, respectively, and the mixture was maintained for 20 min at 65°C. Then proteins were removed by sequential extraction, and DNA was precipitated and resuspended in TE buffer as previously described (13).
Plasmid DNA of E. coli was prepared with a plasmid purification kit from Qiagen, Inc. (Chatsworth, Calif.). PCR amplification was performed with a Perkin-Elmer DNA thermal cycler 2400 (Perkin-Elmer, Courtaboeuf, France) by using Taq DNA polymerase (Appligene, Illkirch, France) and the following cycling parameters. DNA denaturation was performed at 94°C for 30 s, followed by annealing at the temperature indicated in Table 1 and amplification at 72°C for 2 min. This cycle was performed 30 times before a final amplification at 72°C for 10 min. The oligonucleotide primers used in the study are listed in Table 1. They were synthesized by Invitrogen (Paisley, United Kingdom).
TABLE 1.
Oligonucleotide primers used in this study for PCR amplificationa
| Primer | Sequence (5′-3′) |
|---|---|
| MGL−148 | TTT TCC CCA CGC CCC GTG |
| MGL+148 | GCA TGA CGA AAA GTG ACA |
| MGL-F | ATG AGC ATC ACC CAG AAC |
| MGL-R | TCA TAC CGT TGC TAC AGG |
| FragMGL-F | TCG ACC TTC ATG ATG GAC ACC |
| FragMGL-R | CGA GGA TGT CGG CGA TGT CTT |
| M13F | GTA AAA CGA CGG CCA G |
| M13R | CAG GAA ACA GCT ATG AC |
The annealing temperatures were 49 and 50°C, except for primers M13F and M13R, where they were both 50°C.
Sequencing.
A 1.6-kb DNA fragment containing the putative mgl gene was amplified from the total DNA of B. linens strain ATCC 9175 by using two oligonucleotides (MGL−148 and MGL+148) obtained from the sequence of a DNA fragment of BL2 strain (Joint Genome Institute) containing a conserved sequence of mgl genes from five bacteria. The PCR fragments from three different PCR amplifications were purified with the QIAquick gel extraction kit (Qiagen, Inc.) and were used as templates for sequencing. The nucleotide sequence was determined at least twice for both strands with a model 370A automatic DNA sequencer (Applied Biosystems, Courtaboeuf, France). The samples used for sequencing were prepared with a PRISM ready reaction dye deoxy terminator cycle sequencing kit (Applied Biosystems).
The DNA and protein sequences were analyzed with the GCG program (Genetics Computer Group, Inc., Madison, Wis.). Protein homology searches were carried out with the BLAST network (2). Prosite (3) was used to search for characteristic motifs. The alignment and phylogenetic analyses were performed with ClustalW, version 1.8 (40), by using the neighbor-joining method with the blosum matrix. Phylogenetic analyses by the maximum-parsimony method were also realized by using Protpars (PHYLIP, version 3.6a3) (27).
Northern hybridization.
Total RNA was extracted from cultures of B. linens (ATCC 9175) by the method of Glatron and Rapoport (20) modified by R. Raya (unpublished data). The cell pellet from a 40-ml culture was suspended in 400 μl of ice-cold TE buffer (50 mM Tris-EDTA [pH 8.0]), and 250 μl of phenol (pH 4.7) and 250 μl of chloroform were immediately added to the suspension. Then, 0.6 g of glass beads (0.1-mm diameter; Polylabo, Strasbourg, France), 400 μl of 2% macaloid clay suspension, 15 μl of 20% SDS, and 30 μl of sodium acetate (3 M, pH 4.8) were added. The mixture of cells kept on ice for 30 min was disrupted by shaking once for 40 s at power 5 in an FP120 FastPrep cell disruptor (Savant Instruments, Inc., Holbrook, N.Y.). After centrifugation (13,000 × g for 20 min at 4°C), the RNA was extracted twice with 500 μl of phenol and chloroform and once with 1 ml of chloroform and precipitated with isopropanol and sodium acetate (3 M, pH 4.8). The pellet was resuspended in 50 μl of H2O and stored at −80°C. One hundred micrograms of total RNA from B. linens was used for electrophoresis. The DNA probe was prepared with the PCR-amplified mgl gene (obtained with oligonucleotides fragMGL-F and fragMGL-R) by using the ECL kit (Amersham, Little Chalfont, Buckinghamshire, United Kingdom). Northern hybridization was performed as described by the supplier of the ECL kit.
Gene inactivation.
The mgl mutant was constructed from B. linens ATCC 9175. To do this, a 1,042-bp internal fragment of the mgl gene was generated by PCR by using FragMGL-F and FragMGL-R primers, and this fragment was cloned into the Topo XL vector (Invitrogen) to obtain pTIL 872. This plasmid was replicated in E. coli TOP10 (Invitrogen) before being used to transform B. linens. Since the plasmid does not replicate in B. linens, it was integrated into the chromosome by single crossover, yielding TIL 872, the mgl mutant with interrupted mgl. Mutant selection was performed on TSYE agar with kanamycin (5 μg ml−1) for 4 days at 25°C.
The interruption of mgl and the integration site were verified by PCR and sequence analysis. PCR amplification with the oligonucleotides MGL-F and M13F produced a fragment of 1.3 kb, which corresponds to the expected size (1,272 bp). Sequencing of this fragment confirmed the integration of pTIL872 into the mgl gene. The plasmid profile of TIL 872 was the same as that of the wild-type strain ATCC 9175.
Preparation of cells and cell extracts.
B. linens strains were grown at 25°C with mixing (150 rpm) in TSYE medium supplemented with l-methionine (1 g liter−1). In the case of B. linens TIL 872, 5 μg of kanamycin ml of culture medium−1 were added. Cells were harvested by centrifugation (13,000 × g, 20 min, 4°C), washed twice with 1 ml of 100 mM Tris-HCl buffer (pH 8), and maintained at −20°C prior to use.
For cell extract preparation, 500 mg of B. linens cells was suspended in 1 ml of 50 mM Tris-HCl buffer. Cells were subsequently disrupted with 0.6 g of glass beads (diameter, 0.1 mm; Polylabo) by using a FP120 FastPrep cell disruptor. Three mixing sequences of 30 s (speed, 6.5 m s−1) were successively applied, each separated by a 5-min cooling period in ice. After centrifugation (20,000 × g, 5 min, 4°C), the supernatant (called CFE) was collected and enzymatic activities were determined. The protein content in CFEs was determined according to the method of Bradford (9) by using bovine serum albumin as a standard. CFEs were used for determination of thiol-producing activity (TPA).
Determination of TPA.
The TPA was determined as previously described (17) by using l-methionine as the substrate. Free thiols produced reacted with 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB; Sigma-Aldrich) to form thionitrobenzoic acid, which was then detected spectrophotometrically at 412 nm. A standard curve was obtained with different free thiol concentrations by using sodium methanethiolate (Fluka, St. Quentin Fallavier, France) as a reference. The assay mixture contained 50 mM Tris-HCl (pH 8), 0.05 mM PLP, 0.5 mM DTNB, 5 mM l-methionine as the substrate, and the CFE. Control without CFE was included. Samples were incubated at 30°C for 1 h. The total quantity of free thiols formed was determined by measuring the variation of A412 during this time interval. Thiols formed spontaneously (in the absence of CFE) were subtracted from the total quantity of thiols produced with CFE. TPA was expressed as nanomoles per gram of protein per second. We can note that DTNB not only reacts with MTL but also with all free thiols. This assay is, therefore, not specific for MGL activity.
Measurement of VSCs produced by resting cells.
The production of VSCs from l-methionine was studied by incubating B. linens resting cells in a reaction medium containing l-methionine at 30°C under agitation (150 rpm) for 20 h in 10-ml glass tubes sealed with Teflon joint caps. The reaction mixture contained 50 mM Tris-HCl (pH 8), 5 mM l-methionine, 0.05 mM PLP, and 10% (vol/vol) of the cell suspension of B. linens (OD600 = 20). Reaction mixtures were sampled before (0 h) and after (20 h) incubation for VSC analyses.
VSCs were analyzed by dynamic headspace gas chromatography-mass spectrometry (GC-MS). Five milliliters of each sample (eventually diluted) was analyzed by using a headspace analyzer (HP 7695A purge and trap concentrator; Hewlett Packard, Palo Alto, Calif.) coupled to a gas chromatograph (HP 6890; Hewlett Packard) and a mass spectrophotometer detector (HP 6890A quadrupole mass spectrometer; Hewlett Packard), as previously described by Berger et al. (4). The amount of VSCs eventually present in the samples taken initially (0 h) was subtracted from the total amount of VSCs determined in samples after a 20-h incubation period.
Catabolism of l-methionine.
The catabolism of l-methionine by resting cells of B. linens (ATCC 1975 and TIL 872) was studied by using l-[1-14C]methionine (55 mCi mmol−1) as a tracer. The reaction medium contained 100 mM Tris-HCl buffer (pH 8), 0.05 mM PLP, 2 mM unlabeled l-methionine, and 0.05 μM l-[1-14C]methionine. Fifty microliters of cell suspensions (OD480 = 150) was added to 450 μl of the reaction medium. Reaction mixtures were incubated for 24 h at 30°C and sampled after 0, 4, and 24 h. Cells were removed by centrifugation (8,000 × g for 5 min at 4°C) thereafter. The metabolites were subsequently analyzed by ion-exclusion high-performance liquid chromatography (HPLC), with both UV and radioactivity detection as previously described (44). The total radioactivity of the samples was determined under the same HPLC conditions but without separation on the column. Identification of metabolites was made by comparison of the retention times (RTs) with those of commercial standard compounds, namely α-keto-3-methylthiobutyrate, hydroxy-3-methylthiobutyrate, α-ketobutyrate, methional, 3-methylthiopropionate, propionate, hydroxybutyrate, and α-aminobutyrate. They were purchased from Sigma-Aldrich, except for 3-methylthiopropionic acid, which was prepared by acidic hydrolysis of the methyl ester of 3-methylthiopropionate (19). Identification of α-aceto-α-hydroxybutyrate was performed by GC-MS after its conversion to 2,3-pentanedione as described by Gollop et al. (21). Two milliliters of citrate buffer (1 M, pH 3.5) and 0.75 ml of FeSO4 and FeCl3 solutions (10 mM) were added to 1.5 ml of the culture supernatant. The reaction mixture was heated at 80°C for 10 min, and 2,3-pentanedione was analyzed by dynamic headspace GC-MS as described above. Commercial 2,3-pentanedione was used as a standard.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper appears in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AY622198.
RESULTS
Sequencing of the putative MGL gene from B. linens strain ATCC 9175 and Northern blot analysis of the transcript.
The putative gene encoding MGL was localized in the genome of B. linens BL2 (Joint Genome Institute) by looking for a conserved DNA sequence (5′-TN7AANCANATGAN8GG-3′, with N being any base) of the MGL gene of five bacteria. This conserved sequence was found by alignment (23) of mgl sequences of Methanosarcina acetivorans, Methanosarcina mazei, Fusobacterium nucleatum, Oceanobacillus iheyensis, and Bacillus halodurans. Such a sequence (TCGCGAAGAAGCAGATGAGCGGCTTCGG) was found only once in the DNA sequence of the BL2 genome. It was located in an open reading frame of 1,275 bp encoding a 425-amino-acid protein with a calculated molecular mass of 44,669 Da, which is in good agreement with the molecular mass of 43 kDa estimated for MGL by SDS-polyacrylamide gel electrophoresis (16). This amino acid sequence shared 35 to 39% identity with the sequences of MGL genes of O. iheyensis (35%), F. nucleatum (36%), M. acetivorans (38%), M. mazei (39%), Pseudomonas putida (37%), B. halodurans (39%), and Bacillus cereus (35%). However, the highest homologies were found with cystathionine γ-synthases (CGSs) (46 to 47% identity) and CBL or CGL (43 to 45% identity). It also exhibited a PLP attachment site of Cys/Met metabolism enzymes (Prosite accession no. PS00868) at positions 194 to 208, indicating that the gene really encodes a PLP-dependent enzyme.
The corresponding gene of the B. linens strain ATCC 9175 was amplified by PCR with two oligonucleotides designed from the sequence of the BL2 gene and was subsequently sequenced. Nucleotide sequences of both BL2 and ATCC 9175 genes shared 95% identity, and the amino acid sequences deduced from the nucleotide sequences exhibited 97% identity. A potential ribosome-binding site (AAGGAAG) was found 8 to 15 bp upstream the start codon (ATG), but no promoter or terminator was found around the open reading frame.
Northern hybridization with the mgl probe showed a stretching band around 8 kb, suggesting that mgl could be included in an operon. In our conditions, the amount of transcript was very low, since 100 μg of RNA was necessary to detect it.
Effect of inactivation of the putative MGL gene in B. linens on l-methionine conversion to VSCs.
The effect of gene inactivation on l-methionine conversion to VSCs was determined by three methods: (i) measurement of TPA with the DTNB test, (ii) analysis of VSC production by GC-MS, and (iii) HPLC analyses of metabolites produced from radiolabeled l-methionine.
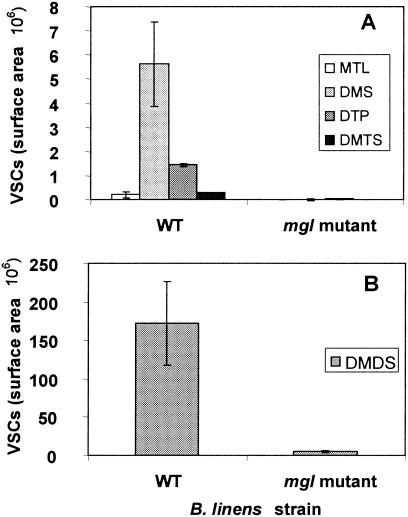
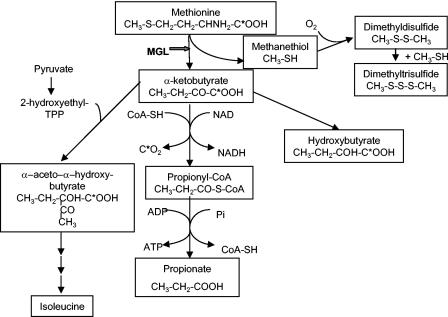
TPA (mean ± standard deviation, n = 3) was 66.0 ± 17.0 and 25.2 ± 4.8 nmol · s−1 · g−1 in the wild-type ATCC 9175 and mgl mutant TIL 872 strains, respectively. Inactivation of the putative mgl gene, therefore, led to a 62% decrease in the TPA of B. linens ATCC 9175. However, this TPA assay is not totally specific for MGL activity, since the DTNB used in the assay reacts with all free thiols and not only with MTL. Consequently, the residual 40% TPA found in the mgl mutant may be attributed to thiols other than MGL producing enzymes. The gene inactivation also reduced the total production of VSCs from l-methionine by 97% (Fig. 1). Although MTL is the reaction product of MGL on l-methionine, DMDS is, by far, the VSC produced most by the wild-type strain, representing 96% of the total VSCs produced. This is due to the fact that MTL is a highly reactive sulfur compound that quickly reacts with itself, forming the oxidized and more stable DMDS as well as, to a lesser extent, DMTS (11). HPLC analysis of metabolites produced from l-[1-14C]-methionine by the wild-type strain (ATCC 9175) (Table 2) showed one major radiolabeled compound (RT, 12.5 min) and two other compounds only detected by UV absorbance (RT, 8.1 and 11.5 min). The metabolite eluting at 8.1 min had an RT close to that of the l-methionine transamination product α-ketomethylthiobutyrate (RT, 8.3 min) but was not coeluted when α-ketomethylthiobutyrate was added as an internal standard. It corresponded to a minor radiolabeled compound that accounted for less than 1% of total initial radioactivity. This compound appeared after 4 h of incubation and did not increase thereafter. The second metabolite eluting at 11.5 min steadily accumulated with time. It was not radiolabeled and had the same RT as the propionate standard. Propionate could originate from oxidative decarboxylation of α-ketobutyrate (5), which is, together with MTL and ammonia, a reaction product of MGL on l-methionine. It was therefore expected that propionate produced from l-[1-14C]-methionine was not radiolabeled since 1-14C was eliminated through the decarboxylation step (Fig. 2). The RT of the major radiolabeled metabolite (12.5 min) did not correspond to any standard compound. This metabolite was subsequently identified as α-aceto-α-hydroxybutyrate by GC-MS analysis after its conversion to the corresponding diketone, 2,3-pentanedione. α-Aceto-α-hydroxybutyrate is also a metabolite of α-ketobutyrate produced by an anabolic acetohydroxy acid synthase, which is an isoleucine biosynthetic enzyme (42). This metabolite appeared progressively during cell incubation and accounted for 25% of initial l-[1-14C]-methionine after 24 h of incubation of l-methionine with resting cells of the wild-type strain of B. linens. No peak was detected (with UV or radioactivity detection) at the expected RT of α-ketobutyrate (7.4 min); this indicated that this metabolite did not accumulate and was readily degraded to other compounds.
FIG. 1.
Production of VSCs by the wild-type strain B. linens ATCC 9175 (WT) and the mgl mutant strain TIL 872 (mgl mutant). (A) Production of MTL, dimethyl sulfide (DMS), dithiapentane (DTP), and DMTS; (B) production of DMDS.
TABLE 2.
Peak areas of major metabolites produced from l-[1-14C]-methionine by resting cells of B. linens ATCC 9175 and mgl mutant strain TIL 872a
| RT (min) | Detection | Identification | Peak area forb:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Wild type
|
mgl mutant
|
|||||||
| 0 h | 4 h | 24 h | 0 h | 4 h | 24 h | |||
| 8.1 | UV | Not identified | 5 ± 2 | 52 ± 36 | 61 ± 12 | 0 | 0 | 0 |
| 11.5 | UV | Propionate | 4 ± 3 | 13 ± 11 | 80 ± 30 | 4 ± 1 | 5 ± 1 | 5 ± 1 |
| 12.5 | Radioactivity | α-Aceto-α-hydroxybutyrate | 322 ± 94 | 6,116 ± 777 | 13,569 ± 896 | 0 | 0 | 0 |
Cells were incubated for 0, 4, and 24 h at 30°C in the reaction medium at pH 8. Metabolites were analyzed by ion-exclusion HPLC with both UV and radioactivity detections. Data are the means of the results from three repetitions ± standard deviations.
Results for UV detection are given in millivolts, and results for radioactivity are given in disintegrations per minute.
FIG. 2.
Putative l-methionine catabolism pathways initiated by MGL. C* indicates 14C in metabolites produced from l-[1-14C]-methionine.
Inactivation of the putative mgl gene totally prevented formation of the three compounds: propionate (RT, 11.5 min), α-aceto-α-hydroxybutyrate (RT, 12.5 min), and the compound eluting at 8.1 min. No radiolabeled metabolite was detected with the mgl mutant (Table 2), even after 24 h of incubation. The absence of α-aceto-α-hydroxybutyrate formation, measured as 2,3-pentanedione formed by resting cells of the mgl mutant, was confirmed by GC-MS analysis.
DISCUSSION
In this work, the gene encoding MGL was identified in the cheese-ripening bacterium B. linens. MGL is involved in l-methionine catabolism in many bacteria (including E. coli, Proteus vulgaris, Brevibacteria, and Pseudomonas) and plays an essential role in the generation of metabolic energy from l-methionine via the production of α-ketobutyrate that can be degraded to propionyl-CoA and, subsequently, succinyl-CoA (37). Moreover, it is generally admitted that MGL is responsible for l-methionine conversion to MTL in B. linens. It is therefore believed to be highly involved in VSC biosynthesis in cheese (8, 16). MGL of B. linens has been biochemically characterized (16), but the gene location, structure, and sequence remained unknown.
Recently, the genome sequence of B. linens BL2 became available. The draft annotation did not clearly indicate the gene encoding MGL, although several genes exhibited high degrees of homology with cystathionine synthases and cystathionine lyases, which are two enzyme families closely related to MGL with respect to biochemical properties (1, 10, 16). By looking for a DNA sequence conserved in the MGL of five other bacteria in the DNA sequence of B. linens, we found a gene annotated as CGS that could be a good candidate for the MGL gene. The corresponding gene of the B. linens ATCC 9175 strain was amplified by PCR, cloned, and sequenced.
The amino acid sequence derived from the DNA sequence shared 45 to 50% identity with several CGSs and CGLs but only 35 to 39% identity with the MGL sequences (Table 3). The sequence alignments with CGLs, CGSs, and MGLs revealed that the residues unique to MGLs (absent in all other PLP enzymes), Phe41, Met81, Cys107, and Val335 (41) (amino acid numbers are based on the B. linens protein), were not conserved in the B. linens gene. The residues present at these positions were identical to those of CGLs and CGSs. Moreover, Glu335 and Arg110, which are strictly conserved in CGLs and bacterial CGSs (32), were present in the B. linens gene, whereas these residues are changed to Val335 and Ala110 or Gly110, respectively, in MGLs. However, all active site residues depicted by crystal structure of the Pseudomonas MGL (34) are conserved in the B. linens gene, even if these residues are also shared by other PLP-dependent enzymes. From the sequence alignment, we constructed a phylogenetic tree (data not shown) that showed that the B. linens protein was close to CGSs of bacteria, especially Corynebacterium glutamicum (NP601644), Mycobacterium tuberculosis (NP215595), Mycobacterium leprae (NP302550), and Bifidobacterium longum (NP696324).
TABLE 3.
Amino acid comparison of MGLs, CGSs, and CGLs by sequence alignmenta
| Gene (accession no.) | Residue at position no.b
|
Sequence of PLP-binding site (positions 194-206) | Residue at position 335 | Active site residues of MGL_Pp at positions 105, 177, 202, 368 | |||
|---|---|---|---|---|---|---|---|
| 41 | 81 | 107 | 110 | ||||
| MGL_Tv1 (CAA04124) | F | M | C | A | DVVVHSTKYING | V | Y, D, K, R |
| MGL_Tv2 (CAA04125) | F | M | C | A | DIVVHSAKYING | V | Y, D, K, R |
| MGL_Fn (AAL95612) | F | M | C | A | DIVVHSATKYLNG | V | Y, D, K, R |
| MGL_Pp1 (BAA20553) | F | M | C | A | DLVVHSATKYLSG | V | Y, D, K, R |
| MGL_Cc (NP_421962) | F | M | C | A | DLVVHSATKYLSG | V | Y, D, K, R |
| MGL_Oi (NP693968) | F | M | C | G | DFVIHSATKYISG | V | Y, D, K, R |
| MGL_Bc (NP834360) | F | M | C | G | DAVVHSATKYIGG | V | Y, D, K, R |
| MGL_Bh (BAB04518) | F | M | C | G | DFVVHSATKYIGG | V | Y, D, K, R |
| MGL_Ma (AAM05915) | F | M | C | S | DISLSSCTKYIGG | V | Y, D, K, R |
| MGL_Mm (NP635109) | F | M | C | S | DISLSSCTKYIGG | V | Y, D, K, R |
| MGL_Mb (ZP00075765) | F | M | C | S | DLSLSSCTKYIGG | Y, D, K, R | |
| MGL-B.linensc | P | T | G | R | DVVIHSTTKFING | E | Y, D, K, R |
| CGS_Mt (NP215595) | V | M | G | R | DVVLHSTTKYIGG | E | Y, D, K, R |
| CGS_Ml (NP302550) | V | M | G | R | DVVLHSTTKYIGG | E | Y, D, K, R |
| CGS_Bl (NP696324) | V | L | G | R | DVVVYSTTKYIGG | E | Y, D, K, R |
| CGS_Cg (NP601644) | P | M | G | R | HAVLHSTTKYIGG | E | Y, D, K, R |
| CGS_Cp (BAB79882) | F | L | G | R | DIVIHSATKYLGG | E | Y, D, K, R |
| CGS_Hp (P56069) | I | L | G | R | DIVAHSGTKYLGG | E | Y, D, K, R |
| CGS_Sa (BAB41649) | I | V | G | R | DIVLHSATKYLGG | E | Y, D, K, R |
| CGL_Hs (AAB24700) | A | L | G | R | DISMYSATKYMNG | E | Y, D, K, R |
| CGL_Rn (AAK52091) | A | L | G | R | DISMYSATKYMNG | E | Y, D, K, R |
| CGL_Ce (NP_495449) | N | L | G | R | DVVVHSITKYING | E | Y, D, K, R |
Multiple-sequence alignment was performed with Clustal W, version 1.8 (40). Names of organisms: Tv, Trichomonas vaginalis; Fn, Fusobacterium nucleatum; Pp, Pseudomonas putida; Cc, Caulobacter crescentus; Oi, Oceanobacillus iheyensis; Bc, Bacillus cereus; Bh, Bacillus halodurans; Ma, Methanosarcina acetivorans; Mm, Methanosarcina mazei; Mb, Methanosarcina barkeri; Mt, Mycobacterium tuberculosis; Ml, Mycobacterium leprae; Bl, Bifidobacterium longum; Cg, Corynebacterium glutamicum; Cp, Clostridium perfringens; Hp, Helicobacter pylori; Sa, Staphylococcus aureus; Hs, Homo sapiens; Rn, Rattus norvegius; Ce, Caenorhabditis elegans.
Position numbers correspond to MGL of B. linens.
The row corresponding to MGL of B. linens is in boldface type.
Although the B. linens putative mgl gene looks like a CGS gene, it really encodes MGL responsible for the degradation of l-methionine to MTL, α-ketobutyrate, and ammonia. Indeed, inactivation of the B. linens mgl gene almost totally prevented l-methionine conversion to MTL and α-ketobutyrate. With the wild-type strain of B. linens, α-ketobutyrate produced from l-methionine did not accumulate but was mainly converted to propionate and/or to α-aceto-α-hydroxybutyrate, suggesting that l-methionine catabolism is involved in the generation of energy and in isoleucine biosynthesis in B. linens.
mgl may be located in a 8-kb operon, but we were not able to identify the other genes of the operon. In P. putida, mdeA (which encodes MGL) is part of the mde operon that also contains mdeB, encoding an α-ketobutyrate dehydrogenase E1 component of an unknown α-ketoacid dehydrogenase complex (25). This complex is responsible for the α-ketobutyrate conversion to propionyl-CoA. In B. linens, a likely complex could be responsible for α-ketobutyrate conversion to propionyl-CoA, which would be readily converted to propionate by a propionate CoA transferase or a propionate kinase. In the B. linens BL2 genome, none of the genes in the flanking regions of the mgl gene shared homology with known E1 components of α-ketoacid dehydrogenase, but genes encoding such E1 components have been located elsewhere in the genome.
To our knowledge, this is the first report of the gene encoding MGL in the surface cheese-ripening bacterium B. linens. This is of considerable interest, since, until now, MGL involvement in VSC production by B. linens was rather speculative. In this study, we have demonstrated a clear-cut effect of mgl gene disruption on the ability of B. linens to produce VSCs. This new knowledge could represent a first step towards a better control over VSC biosynthesis in a wide variety of cheese types. Further studies on the regulation mechanisms of MGL in B. linens are therefore required. For instance, induction of MGL activity by l-methionine or its repression by glucose (17) could be studied at the level of gene expression. Another quite important technological property of B. linens is color development at the cheese surface. A better knowledge of MGL regulation mechanisms could therefore help in modulating VSC biosynthesis while promoting cheese coloration. Furthermore, the presence of the homologous mgl gene could be searched in other cheese-ripening surface bacteria also producing MTL from l-methionine (8).
Finally, B. linens, which is strictly aerobic and acid sensitive, only develops on the cheese surface and then during the later stages of ripening, after deacidification has been completed by yeasts such as Debaryomyces hansenii or Kluyveromyces lactis (28, 29). The use of anaerobic facultative bacteria with enhanced VSC-producing abilities could be an alternative strategy for the production of aroma compounds within the cheese matrix in the early stages of ripening. Overproduction of B. linens MGL in bacteria such as LAB could therefore be a more efficient way to increase the cheese flavor of some cheeses, among others, cheddar, than the addition of B. linens cells or extracts, which was not successful in enhancing cheese flavor (15, 43).
Acknowledgments
The sequence data of B. linens BL2 were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/).
We thank Christine Hervé, Dominique LeBars, and Nicolas Bonnaire for providing excellent technical assistance and Véronique Monnet and Henry-Eric Spinnler for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Alting, A. C., W. J. M. Engels, S. van Schalkwijk, and F. A. Exterkate. 1995. Purification and characterization of cystathionine β-lyase from Lactococcus lactis subsp. cremoris B78 and its possible role in flavor development in cheese. Appl. Environ. Microbiol. 61:4037-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch, A., P. Bucher, and K. Hofman. 1997. The Prosite database, its status in 1997. Nucleic Acids Res. 25:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, C., J. A. Khan, P. Molimard, N. Martin, and H. E. Spinnler. 1999. Production of sulfur flavors by ten strains of Geotrichum candidum. Appl. Environ. Microbiol. 65:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisswanger, H. 1981. Substrate specificity of the pyruvate dehydrogenase complex from Escherichia coli. J. Biol. Chem. 256:815-822. [PubMed] [Google Scholar]
- 6.Bloes-Breton, S., and J. Bergère. 1997. Production de composés soufrés volatils par des Micrococcaceae et des bactéries corynéformes d'origine fromagère. Lait 77:543-559. [Google Scholar]
- 7.Bonnarme, P., F. Amarita, E. Chambellon, E. Semon, H. E. Spinnler, and M. Yvon. 2004. Methylthioacetaldehyde, a possible intermediate metabolite for the production of volatile sulphur compounds from L-methionine by Lactococcus lactis. FEMS Lett. 236:85-90. [DOI] [PubMed] [Google Scholar]
- 8.Bonnarme, P., L. Psoni, and H. E. Spinnler. 2000. Diversity of l-methionine catabolism in cheese-ripening bacteria. Appl. Environ. Microbiol. 66:5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Bruinenberg, P. G., G. D. Roo, and G. K. V. Limsowtin. 1997. Purification and characterization of cystathionine γ-lyase from Lactococcus lactis subsp. cremoris SK11: possible role in flavor compound formation during cheese maturation. Appl. Environ. Microbiol. 63:561-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin, H. W., and R. C. Lindsay. 1994. Ascorbate and transition-metal mediation of methanethiol oxidation to dimethyl disulfide and dimethyl trisulfide. Food Chem. 49:387-392. [Google Scholar]
- 12.Cuer, A., G. Dauphin, A. Kergomard, S. Roger, J. P. Dumont, and J. Adda. 1979. Flavour properties of some sulphur compounds isolated from cheeses. Lebensmittelwiss. Technol. 12:258-261. [Google Scholar]
- 13.De Buyser, M. L., A. Morvan, F. Grimont, and N. el Solh. 1989. Characterization of Staphylococcus species by ribosomal RNA gene restriction patterns. J. Gen. Microbiol. 135(Pt 4):989-999. [DOI] [PubMed] [Google Scholar]
- 14.Dias, B., and B. Weimer. 1998. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl. Environ. Microbiol. 64:3320-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dias, B., and B. Weimer. 1999. Production of volatile sulfur compounds in cheddar cheese slurries. Int. Dairy J. 9:605-611. [Google Scholar]
- 16.Dias, B., and B. Weimer. 1998. Purification and characterization of l-methionine γ-lyase from Brevibacterium linens BL2. Appl. Environ. Microbiol. 64:3327-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferchichi, M., D. Hemme, M. Nardi, and N. Pamboukdjian. 1985. Production of methanethiol from methionine by Brevibacterium linens CNRZ 918. J. Gen. Microbiol. 131:715-723. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez, M., W. van Doesburg, G. A. M. Rutten, J. D. Marugg, A. C. Alting, R. van Kranenburg, and O. P. Kuipers. 2000. Molecular and functional analyses of the metC gene of Lactococcus lactis, encoding cystathionine β-lyase. Appl. Environ. Microbiol. 66:42-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fieser, L., and M. Fieser. 1956. Organic chemistry. D. C. Heath, London, United Kingdom.
- 20.Glatron, M. F., and G. Rapoport. 1972. Biosynthesis of the parasporal reclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie 54:1291-1301. [DOI] [PubMed] [Google Scholar]
- 21.Gollop, N., Z. Barak, and D. M. Chipman. 1987. A method for simultaneous determination of the two possible products of acetohydroxy acid synthase. Anal. Biochem. 160:323-331. [DOI] [PubMed] [Google Scholar]
- 22.Helinck, S., H. E. Spinnler, M. Dame-Cahagne, and P. Bonnarme. 2000. Enzymatic versus spontaneous S-methyl thioester synthesis in Geotrichum candidum. FEMS Microbiol. Lett. 193:237-241. [DOI] [PubMed] [Google Scholar]
- 23.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 24.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue, H., K. Inagaki, S. Eriguchi, T. Tamura, N. Esaki, K. Soda, and H. Tanaka. 1997. Molecular characterization of the mde operon involved in l-methionine catabolism of Pseudomonas putida. J. Bacteriol. 179:3956-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreis, W., and C. Hession. 1973. Isolation and purification of L-methionine-α-deamino-γ-mercaptomethane-lyase (L-methionase) from Clostridium sporogenes. Cancer Res. 33:1862-1865. [PubMed] [Google Scholar]
- 27.Kuhner, M. K., and J. Felsenstein. 1994. A simulation comparison of phylogeny algorithms under equal and unequal evolutionary rates. Mol. Biol. Evol. 11:459-468. [DOI] [PubMed] [Google Scholar]
- 28.Leclercq-Perlat, M. N., F. Buono, D. Lambert, E. Latrille, H. E. Spinnler, and G. Corrieu. 2004. Controlled production of Camembert-type cheeses. Part I: microbiological and physicochemical evolutions. J. Dairy Res. 71:346-354. [DOI] [PubMed] [Google Scholar]
- 29.Leclercq-Perlat, M. N., A. Oumer, J. L. Bergere, H. E. Spinnler, and G. Corrieu. 2000. Behavior of Brevibacterium linens and Debaryomyces hansenii as ripening flora in controlled production of smear soft cheese from reconstituted milk: growth and substrate consumption dairy foods. J. Dairy Sci. 83:1665-1673. [DOI] [PubMed] [Google Scholar]
- 30.Leret, V., A. Trautwetter, A. Rincé, and C. Blanco. 1998. pBLA8, from Brevibacterium linens, belongs to a Gram-positive subfamily of ColE2-related plasmids. Microbiology 144:2827-2836. [DOI] [PubMed] [Google Scholar]
- 31.Lockwood, B. C., and G. H. Coombs. 1991. Purification and characterization of methionine γ-lyase from Trichomonas vaginalis. Biochem. J. 279:675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messerschmidt, A., M. Worbs, C. Steegborn, M. C. Wahl, R. Huber, B. Laber, and T. Clausen. 2003. Determinants of enzymatic specificity in the Cys-Met-metabolism PLP-dependent enzymes family: crystal structure of cystathionine gamma-lyase from yeast and intrafamiliar structure comparison. Biol. Chem. 384:373-386. [DOI] [PubMed] [Google Scholar]
- 33.Molimard, P., and H. E. Spinnler. 1996. Compounds involved in the flavor of surface mold-ripened cheeses: origins and properties. J. Dairy Sci. 79:169-184. [Google Scholar]
- 34.Motoshima, H., K. Inagaki, T. Kumasaka, M. Furuichi, H. Inoue, T. Tamura, N. Esaki, K. Soda, N. Tanaka, M. Yamamoto, and H. Tanaka. 2000. Crystal structure of the pyridoxal 5′-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida. J. Biochem. (Tokyo) 128:349-354. [DOI] [PubMed] [Google Scholar]
- 35.Rijnen, L., M. Yvon, R. van Kranenburg, P. Courtin, A. Verheul, E. Chambellon, and G. Smit. 2003. AraT and BcaT are key enzymes for the formation of aroma compounds from amino acids in cheese. Int. Dairy J. 13:805-812. [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Soda, K., H. Tanaka, and N. Esaki. 1983. Multifunctional biocatalysis: methionine-γ-lyase. Trends Biochem. Sci. 8:214-217. [Google Scholar]
- 38.Tanaka, H., N. Esaki, and K. Soda. 1985. A versatile bacterial enzyme: l-methionine γ-lyase. Enzyme Microb. Technol. 7:530-537. [Google Scholar]
- 39.Tanaka, H., N. Esaki, T. Tammamoto, and K. Soda. 1976. Purification and properties of methionine γ-lyase from Pseudomonas ovalis. FEBS Lett. 66:307-311. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tokoro, M., T. Asai, S. Kobayashi, T. Takeuchi, and T. Nozaki. 2003. Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a key enzyme of sulfur-amino acid degradation in an anaerobic parasitic protist that lacks forward and reverse trans-sulfuration pathways. J. Biol. Chem. 278:42717-42727. [DOI] [PubMed] [Google Scholar]
- 42.Van Dyk, T. K., and R. A. LaRossa. 1987. Involvement of ack-pta operon products in alpha-ketobutyrate metabolism by Salmonella typhimurium. Mol. Gen. Genet. 207:435-440. [DOI] [PubMed] [Google Scholar]
- 43.Weimer, B., B. Dias, M. Ummadi, J. Broadbent, C. Brennand, J. Jaegi, M. Johnson, F. Milani, J. Steele, and D. V. Sisson. 1997. Influence of NaCl and pH on intracellular enzymes that influence Cheddar cheese ripening. Lait 77:383-398. [Google Scholar]
- 44.Yvon, M., S. Berthelot, and J. C. Gripon. 1998. Adding α-ketoglutarate to semi-hard cheese curd highly enhances the conversion of amino acids to aroma compounds. Int. Dairy J. 8:889-898. [Google Scholar]