ABSTRACT
Wee1 and Cdc25 are conserved regulators of mitosis. Wee1 is a kinase that delays mitosis via inhibitory phosphorylation of Cdk1, while Cdc25 is a phosphatase that promotes mitosis by removing the inhibitory phosphorylation. Although Wee1 and Cdc25 are conserved proteins, it has remained unclear whether their functions and regulation are conserved across diverse species. Here, we analyzed regulation of Wee1 and Cdc25 in fission yeast. Both proteins undergo dramatic cell cycle-dependent changes in phosphorylation that are dependent upon PP2A associated with the regulatory subunit Pab1. The mechanisms that control Wee1 and Cdc25 in fission yeast appear to share similarities to those in budding yeast and vertebrates, which suggests that there may be common mechanisms that control mitotic entry in all eukaryotic cells.
KEYWORDS: Cdc25, Cell cycle, fission yeast, PP2APab1, Wee1
Introduction
Mitosis is a conserved feature of all eukaryotic cells. One might therefore expect the signals that control mitosis to be conserved as well. Two important conserved regulators of mitosis are the Wee1 kinase and the Cdc25 phosphatase. Wee1 delays mitosis by phosphorylating and inhibiting cyclin-dependent kinase 1 (Cdk1), the master controller of mitosis.1 Cdc25 promotes mitosis by removing the inhibitory phosphorylation.2-5 Pioneering work in fission yeast discovered that Wee1 mutants cause premature passage through mitosis at a reduced cell size.6,7 This observation led to the idea that Wee1 and Cdc25 function in a cell size checkpoint that delays mitosis until sufficient growth has occurred.8 However, there is no evidence yet that Wee1 and Cdc25 function in a cell size checkpoint in vertebrate cells. Moreover, it has proven difficult to discover signaling mechanisms that link the activity of Wee1 and Cdc25 to cell growth or size. A coherent understanding of the function and regulation of Wee1 and Cdc25 in diverse species has not yet emerged.
In budding yeast, Wee1 is initially phosphorylated by mitotic Cdk1, which activates Wee1 to bind and inhibit Cdk1.9,10 Thus, Cdk1 activates its own inhibitor. Activation of Wee1 by Cdk1 is opposed by a form of protein phosphatase 2A associated with the Cdc55 regulatory subunit (PP2ACdc55).10 The opposing activity of PP2ACdc55 is thought to restrain activation of Wee1, thereby allowing a low level of Cdk1 activity to accumulate in early mitosis to initiate early mitotic events. Later in mitosis, Wee1 undergoes additional multi-site phosphorylation events that promote inactivation of Wee1. There is extensive evidence that each of these mechanisms is conserved in vertebrates.11-15
Like Wee1, Cdc25 also undergoes cell cycle-dependent changes in phosphorylation. Cdk1 phosphorylates and activates Cdc25 in a conserved feedback loop that promotes Cdk1 activation.16-22 An additional layer of control has been observed in budding yeast.18 In this case, Cdc25 is hyperphosphorylated early in the cell cycle by an unknown kinase. Cdc25 then undergoes dephosphorylation during entry into mitosis. Genetic data suggest that hyperphosphorylation of Cdc25 early in the cell cycle is inhibitory, and that dephosphorylation is necessary for full activity. Dephosphorylation of Cdc25 during entry into mitosis is performed by PP2ACdc55.18,23 Thus far, this added layer of control has appeared to be an oddity of budding yeast, although in Xenopus there is evidence for an additional kinase that phosphorylates Cdc25 in opposition to PP2A.20,24 There is also evidence that human Cdc25C is phosphorylated on an inhibitory site during interphase by the kinases Chk1 and C-TAK1, which promotes the binding of a 14–3–3 protein.25,26
There is little evidence that the mechanisms that control Wee1 and Cdc25 in budding yeast and vertebrates are conserved in fission yeast. For example, fission yeast Wee1 has not been observed to undergo significant changes in phosphorylation during the cell cycle. Similarly, although PP2A has emerged as a critical regulator of Wee1 and Cdc25 in budding yeast and vertebrates, there is limited evidence for a similar role in fission yeast.27,28 A significant problem has been detecting the low abundance Wee1 protein by western blot in fission yeast. Here, we have analyzed regulation of fission yeast Wee1 and Cdc25 using new reagents and methods that yield improved sensitivity and preservation of phosphorylation. The results suggest that key mechanisms that control Wee1 and Cdc25 are conserved, which raises the hope that it may yet be possible to discover common mechanisms that control mitosis and cell size in all eukaryotic cells.
Results
Wee1 and Cdc25 undergo changes in phosphorylation during the cell cycle in Schizosaccharomyces pombe.
We used western blotting to analyze changes in the abundance and phosphorylation of Wee1 and Cdc25 in synchronized cells progressing through 2 rounds of cell division. Since epitope tags often disrupt function, we raised polyclonal antibodies against Wee1 and Cdc25. The antibodies recognized multiple isoforms of both Wee1 and Cdc25 (Fig. 1A, B). Phosphatase treatment caused the isoforms to collapse into a single band, which indicated that they represented phosphorylated forms of the protein (Fig. 1C).
Figure 1.
Characterization of polyclonal antibodies that recognize Wee1 and Cdc25. (A) An anti-Cdc25 antibody was used to probe samples from wild type and cdc25Δ cdc2–3w cells. (B) An anti-Wee1 antibody was used to probe samples from wild type, wee1–3xHA, and wee1Δ cells. The HA tag causes a shift in the electrophoretic mobility of bands corresponding to Wee1. (C) Western blots of log phase Cdc25 and Wee1 with or without treatment with λ-phosphatase. Asterisks indicate background bands that serve as loading controls.
To obtain a synchronous population of cells, we used centrifugal elutriation to isolate small cells in G2 phase. In addition to monitoring Wee1 and Cdc25, we used a polyclonal antibody to detect Cdc13, the fission yeast homolog of cyclin B, and a phospho-specific antibody to detect Wee1-dependent inhibitory phosphorylation of Cdk1 at tyrosine 15 (Fig. 2A). Cells were fixed and stained with DAPI and calcofluor to assay nuclear division and formation of the septum that forms in late mitosis to complete cell division (Fig. 2B). We focused primarily on events observed during the second cell cycle, since this cycle is less likely to show perturbations associated with elutriation.
Figure 2.
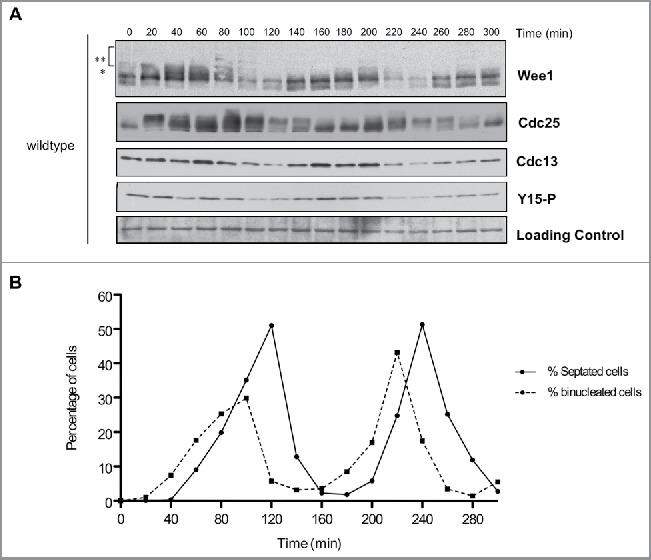
Cell cycle-dependent changes in phosphorylation of Wee1 and Cdc25. Fission yeast cells were synchronized by centrifugal elutriation and released into fresh medium at 30°C. The data in panels A and B were generated from the same time course to allow direct comparison of the timing of cell cycle events. (A) Western blots showing the behavior of Wee1, Cdc25, Cdc13, and Cdk1 inhibitory phosphorylation during the cell cycle. A background band was used as a loading control. A single asterisk is used to mark a partially phosphorylated form of Wee1; 2 asterisks mark more extensively phosphorylated forms referred to in the text as hyperphosphorylated forms. Inhibitory phosphorylation of Cdk1 was detected using a phospho-specific antibody. (B) A fluorescence microscopy assay using DAPI and calcofluor staining was used to determine the percentage of binucleated cells and cells undergoing septation.
Cdc13 was detected in cells isolated by elutriation, confirming that they were in G2 phase (Fig. 2A). Cyclin levels dropped during anaphase and then reappeared as cells entered the second mitosis. For example, in the second cell cycle cyclin levels dropped at 220 minutes when cells were in anaphase (peak of binucleate cells) and then reappeared at 240 min just after septation in early G2 and remained high during G2. Inhibitory phosphorylation of Cdk1 largely coincided with Cdc13 levels, which suggested that it could be present on at least a fraction of Cdk1 throughout much of mitosis (Fig. 2, A and B). Alternatively, it is possible that inhibitory phosphorylation of Cdk1 occurs only during mitotic entry, and the prolonged presence of inhibitory phosphorylation of Cdk1 is due to imperfect synchrony.
Multiple forms of Wee1 could be detected during the cell cycle. A partially phosphorylated form of Wee1 appeared at the end of G2 and in early mitosis (marked with an asterisk in Fig. 2A). More extensively hyperphosphorylated forms of Wee1 appeared as cells progressed through mitosis; we refer to these forms as hyperphosphorylated Wee1 (marked with 2 asterisks in Fig. 2A). For example, in the second cell cycle, Wee1 phosphorylation was initiated at the end of G2 (180 minutes) and reached a maximum level at metaphase (200 minutes), just before Cdc13 degradation in anaphase (220 minutes). Hyperphosphorylated forms of Wee1 appeared to precede a decrease in levels of Cdc13 and Cdk1-Y15 phosphorylation. A decrease in Wee1 protein levels occurred as cells progressed through anaphase and septation, which correlated with the lowest levels of Cdc13 and Cdk1-Y15 phosphorylation (time points 100–120 and 220–240 min, Fig. 2, A and B). A previous study observed similar fluctuations in Wee1 protein levels.29
We also analyzed the behavior of a version of Wee1 tagged with 3 copies of the HA epitope (Fig. S1). Wee1–3XHA showed reduced phosphorylation compared with untagged Wee1. For example, hyperphosphorylated forms of Wee1–3XHA were difficult to detect, and a higher fraction of Wee1–3XHA appeared to persist in a dephosphorylated form. Cell cycle-dependent changes in Wee1 phosphorylation that can be detected via electrophoretic mobility shifts were largely eliminated, although cell cycle-dependent changes in protein abundance were largely unaffected. Others have found that Wee1–3XHA suppresses cell size defects caused by deletion of the genes for Cdr1 or Cdr2, which are upstream regulators of Wee1 (J. Moseley, personal communication). Thus, genetic data further suggest that a 3XHA tag interferes with Wee1 function.
Cdc25 underwent large changes in phosphorylation during the cell cycle. In the first cell cycle, Cdc25 was hyperphosphorylated in G2 or early mitosis and became dephosphorylated when cyclin levels were high (Fig. 2A, compare 20 and 40 minute time points). Cdc25 then became hyperphosphorylated as cyclin levels began to decline at 80 min and remained hyperphosphorylated through the peak of septation. In the second cell cycle, Cdc25 underwent dephosphorylation as Cdc13 levels increased (Fig. 2A, compare time points 140 and 160 min). At the end of the second mitosis, Cdc25 again underwent hyperphosphorylation as mitotic cyclin levels declined. A previous study also observed that Cdc25 undergoes hyperphosphorylation as mitotic cyclin levels decline.30 However, in the previous study hyperphosphorylation of Cdc25 did not persist during interphase, which may be due to the use of sample preparation methods that did not fully preserve phosphorylation.
To investigate whether cell cycle dependent changes in phosphorylation of Cdc25 are dependent upon Cdk1, we used a mutant version of Cdc25 that lacks all minimal Cdk1 consensus sites (cdc25–13A). Previous work has shown that cdc25–13A causes increased cell size, presumably due to a failure in the positive feedback loop the promotes mitotic entry.19 The mutant eliminated electrophoretic mobility shifts caused by phosphorylation in asynchronously growing cells (Fig. 3A). It also eliminated cell cycle-dependent changes in phosphorylation of Cdc25 that can be detected by electrophoretic mobility shifts in synchronized cells (Fig. 3B and C). Thus, the phosphorylation of Cdc25 detected under the conditions of these experiments is dependent upon Cdk1 consensus sites. Previous work found that cdc25–13A becomes phosphorylated during a prolonged mitotic arrest caused by depolymerization of microtubules, which suggests that at least one kinase in addition to Cdk1 can phosphorylate Cdc2519. However, we did not detect activity of this kinase in synchronized wild type cells.
Figure 3.
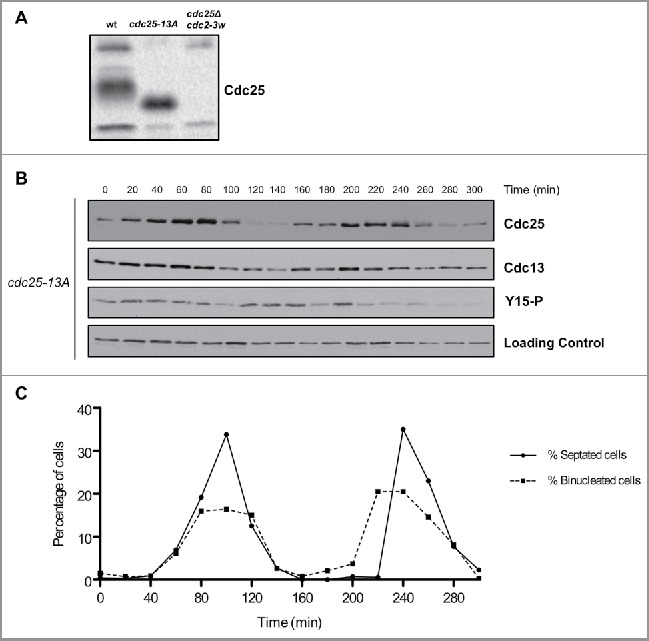
Cell cycle-dependent changes in phosphorylation of Cdc25 are dependent upon Cdk1 consensus sites. (A) Cdc25 was analyzed by western blot in asynchronous wild type, cdc25-13A and cdc25Δ cdc2-3w cells. (B) Cdc25, Cdc13, and Cdk1 inhibitory phosphorylation were analyzed by western blot in cdc25–13A cells synchronized by centrifugal elutriation. a-tubulin was used as a loading control. (C) A fluorescence microscopy assay using DAPI and calcofluor staining was used to determine the percentage of binucleated cells and cells undergoing septation.
Both wild type Cdc25 and cdc25–13A proteins underwent cell cycle-dependent changes in abundance, with the amount of Cdc25 decreasing when mitotic cyclin levels are low (Fig. 2A and Fig. 3B).
Wee1 and Cdc25 are regulated by PP2A
In budding yeast, both Wee1 and Cdc25 are controlled by PP2ACdc55.10,18,23 PP2ACdc55 opposes the initial partial hyperphosphorylation of Wee1 and is also required for dephosphorylation of Cdc25 during mitotic entry. Due to these roles, inactivation of Cdc55 causes constitutive partial phosphorylation of Wee1, as well as constitutive hyperphosphorylation of Cdc25. There is also evidence that the Cdc14 phosphatase binds to Wee1 and controls its phosphorylation.31
The phosphatases that control Wee1 and Cdc25 in budding yeast have also been implicated in fission yeast, although much less is known about their roles. For example, PP2A is thought to control mitotic events and nutrient modulation of cell size in fission yeast; however, the underlying mechanisms and targets have remained unknown.27 Pab1, the fission yeast homolog of the Cdc55 regulatory subunit, causes cell size defects and has been genetically linked to regulation of Wee1 or Cdc25.27,28,32 Clp1, the fission yeast homolog of Cdc14, causes reduced cell size and has been implicated in controlling inhibitory phosphorylation of Cdk1, although the mechanisms are poorly understood.33-35
To explore how PP2A controls mitosis in fission yeast, we tested whether pab1Δ cells show defects in phosphorylation of Wee1 or Cdc25 in cells synchronized by centrifugal elutriation. pab1Δ cells had a significantly longer cell cycle: whereas wild type cells required 120 min to complete the cell cycle (i.e. the time from the first peak of septation to the second peak), pab1Δ cells required 180 min. Wee1 failed to undergo significant changes in phosphorylation during the cell cycle in pab1Δ cells (Fig. 4). Direct comparison of key time points in wild type and pab1Δ cells indicated that pab1Δ caused Wee1 to remain in the partially phosphorylated form throughout the cell cycle (Fig. S2). Similarly, Cdc25 was hyperphosphorylated throughout the cell cycle and failed to undergo normal dephosphorylation when mitotic cyclin levels rise (Figs. 4 and S2). Thus, pab1Δ largely eliminated cell cycle-dependent changes in phosphorylation of Wee1 and Cdc25. Cell cycle-dependent changes in Wee1 and Cdc25 protein levels appeared to be unaffected by pab1Δ.
Figure 4.

PP2APab1 is required for cell-cycle dependent changes in phosphorylation of Wee1 and Cdc25. (A) Wee1, Cdc25, Cdc13, and Cdk1 inhibitory phosphorylation were analyzed by western blot in pab1Δ cells synchronized by centrifugal elutriation, as in Figure 2. A background band was used as a loading control. (B) A fluorescence microscopy assay using DAPI and calcofluor staining was used to determine the percentage of binucleated cells and cells undergoing septation.
We also tested the effects of clp1Δ on phosphorylation of Wee1 and Cdc25. The cell cycle was 20 min longer in clp1Δ cells compared with wild type cells. Cell cycle dependent changes in Wee1 phosphorylation were reduced, and a significant fraction of Wee1 persisted in a dephosphorylated form (Figs. 5A, B and S2). clp1Δ caused Cdc25 to persist in a phosphorylated form, and it largely eliminated cell cycle dependent changes in Cdc25 phosphorylation, similar to pab1Δ. However, some phosphorylation and dephosphorylation of Cdc25 could still be detected in clp1Δ cells. For example, hyperphosphorylated forms of Cdc25 could be detected at 40 and 180 min, and a dephosphorylated form was detected between 120 and 180 min.
Figure 5.

Inactivation of Clp1 and PP2APab1 does not show additive effects upon phosphorylation of Wee1 or Cdc25. (A, C) Western blots of Wee1, Cdc25, Cdc13, and Cdk1 inhibitory phosphorylation during the cell cycle in clp1Δ and pab1Δ clp1Δ cells synchronized by centrifugal elutriation. A background band was used as a loading control. (B, D) A fluorescence microscopy assay using DAPI and calcofluor was used to determine the percentage of binucleated cells and cells undergoing septation.
To determine whether Pab1 and Clp1 play overlapping roles in controlling phosphorylation of Wee1 and Cdc25, we tested pab1Δ clp1Δ double mutants to determine if loss of both phosphatases causes additive effects (Fig. 5C and D). In pab1Δ clp1Δ cells, Wee1 phosphorylation during the cell cycle most closely resembled clp1Δ single mutants, which suggests that Clp1 is required for hyperphosphorylation of Wee1 in pab1Δ cells. Phosphorylation of Cdc25 was similar in the single or double mutants, so there was no evidence for additive effects.
Discussion
Evidence for conserved mechanisms of mitotic control
These observations provide new insight into mechanisms that control Wee1 and Cdc25. A number of similarities between fission yeast and budding yeast suggest the existence of conserved mechanisms. For example, in both yeasts, Wee1 shifts to a partially phosphorylated form in early mitosis and to fully phosphorylated forms later in mitosis, followed by destruction (Fig. 2 and refs. 9, 10, 36-38). Similarly, Cdc25 in both yeasts undergoes dephosphorylation as mitotic cyclin levels rise (Fig. 2 and refs. 18, 23). Our analysis also points to a conserved role for PP2APab1/Cdc55 in coordinately regulating Wee1 and Cdc25. In both yeasts, inactivation of PP2APab1/Cdc55 causes Wee1 to accumulate in a partially phosphorylated form, while also blocking full hyperphosphorylation (Fig. 2, Fig. 3, Fig. S2 and refs. 10, 23). Similarly, inactivation of PP2APab1/Cdc55 in both yeasts causes a failure in cell cycle-dependent dephosphorylation of Cdc25 (Fig. 2, Fig. 3, Fig. S2 and refs. 18, 23).
Previous work in vertebrates, budding yeast and fission yeast suggests that Cdk1 phosphorylates and activates Cdc25 during mitotic entry in a positive feedback loop that promotes further Cdk1 activation 16-22 This model for Cdc25 activation suggests that Cdc25 should undergo Cdk1-dependent hyperphosphorylation during mitotic entry and should remain phosphorylated during mitosis, when Cdc25 must be active to maintain Cdk1 activity. However, we observed that Cdc25 becomes hyperphosphorylated only as mitotic cyclin levels decline at the end of the first mitosis. Cdc25 then remains hyperphosphorylated and undergoes dephosphorylation as cyclin levels rise during the second mitosis. Hyperphosphorylation of Cdc25 late in mitosis as cyclin levels decline was also observed in a previous study.30 These observations are not consistent with simple versions of the positive feedback model, but do not rule out the model. One possibility is that phosphorylation of a subset of sites drives positive feedback, while phosphorylation of another set of sites plays roles late in mitosis that remain to be discovered.
Dephosphorylation of Cdc25 as cyclin levels rise is also observed in budding yeast. Moreover, in both budding yeast and fission yeast, dephosphorylation of Cdc25 during the cell cycle is dependent upon PP2APab1/Cdc55. An apparent difference between budding yeast and fission yeast is that dephosphorylation of Cdc25 as mitotic cyclin levels rise in budding yeast is thought to occur on sites that are not Cdk1-consensus sites. Previous work suggested that casein kinase 1 phosphorylates Cdc25; however, more recent work suggests that the effects of casein kinase 1 are likely to be indirect (V. Thai and D. Kellogg, unpublished data). The kinase responsible for hyperphosphorylation of budding yeast Cdc25 therefore remains unknown.
Our data here suggest that dephosphorylation of fission yeast Cdc25 as cyclin levels rise occurs mainly on Cdk1 consensus sites. However, we detect some Cdc25 hyperphosphorylation and dephosphorylation in clp1Δ cells. Since Clp1 is thought to play an important role in dephosphorylation of Cdk1 sites, this observation suggests the existence of Cdc25 phosphorylation events that are not controlled by Cdk1 or Clp1. In addition, the cdc25–13A mutant eliminated phosphorylation of Cdc25 that can be detected by an electrophoretic mobility shift; however, this does not rule out the possibility that another kinase phosphorylates Cdc25 in a manner that is dependent upon prior phosphorylation of Cdc25 on Cdk1 consensus sites.
Genetic analysis of Wee1 suggested that it controls mitotic entry.6,39 However, we found that inhibitory phosphorylation of Cdk1 could be detected as long as mitotic cyclin was present (Fig. 2). Since mitotic cyclin destruction is initiated at the beginning of anaphase,40 this observation suggests that inhibitory phosphorylation is present on a fraction of Cdk1 through much of mitosis. Inhibitory phosphorylation of Cdk1 during mitosis is also detected in budding yeast, where it has been shown to control the duration of metaphase.41 There is also evidence that human Wee1 controls the duration of mitotic events that occur after mitotic entry, and that Drosophila Wee1 controls the morphology of the mitotic spindle.42,43 Thus, it is possible that Wee1 carries out conserved functions after mitotic entry.
Numerous observations suggest that the mechanisms that control Wee1 and Cdc25 in budding and fission yeast are conserved in vertebrates. For example, there is evidence in both yeast and vertebrates that Cdk1 binds and directly phosphorylates Wee1, which triggers formation of a Wee1-Cdk1 complex in which Wee1 inhibits Cdk1.9-13 Moreover, there is evidence in both yeast and vertebrates that PP2A opposes phosphorylation of Wee1 by Cdk1.10-12,18,23 The opposing activity of PP2ACdc55 is thought to set a threshold that limits activation of Wee1 by Cdk1, thereby allowing a low level of Cdk1 activity to accumulate in early mitosis.10 Another mechanism that appears to be shared between yeast and vertebrates is that full hyperphosphorylation of Wee1 leads to its inactivation.9,12,13,15 Finally, a kinase other than Cdk1 phosphorylates Cdc25 in opposition to PP2A in vertebrates, which suggests that the added layer of Cdc25 control observed in budding yeast may be conserved in vertebrates.20,24
Wee1 and Cdc25 have been under study for 4 decades, yet a coherent understanding of their function and regulation across diverse species has remained elusive. The discovery that a number of mechanisms that control Wee1 and Cdc25 may be conserved in fission yeast, budding yeast and vertebrates raises the hope that it may yet be possible to define a core set of common signaling mechanisms that control Wee1 and Cdc25 in all eukaryotic cells, leading to a unified understanding of the signals that integrate mitotic entry with cell growth.
Materials and methods
Yeast methods and strains
The strains used in this study are listed in Table S1. Cells were grown at 30°C in standard YES media with supplements (Sunrise Science 2011–500). clp1Δ pab1Δ strain was made amplifying the pab1Δ::kanR cassette from JJ1745 using the oligos: ACTGAGCTTTTGACTCGCCG and TGTATCGATTCACGGCATAC. The resulting fragment was transformed into JJ2068 and positives clones were checked by PCR and G418 resistance.
Production of polyclonal antibodies that recognize Wee1 and Cdc25
Antibodies that recognize Wee1 (anti-SpWee1) and Cdc25 (anti-SpCdc25) were generated by immunizing rabbits with fusion proteins expressed in bacteria. Plasmids that express Wee1 and Cdc25 were constructed with the Gateway cloning system. Briefly, PCR products that encode the first 200 amino acids of Wee1 or the first 300 amino acids of Cdc25 were cloned into the entry vector pDONR221 (Wee1 oligos: GAAAACCTGTACTTCCAGTCCatgagctcttcttctaataccagcag and GTACAAGAAAGCTGGGTCTCActttcgcagcttctgagcaggagt; Cdc25 oligos: GAAAACCTGTACTTCCAGTCCatggattctccgctttcttcactttc and GTACAAGAAAGCTGGGTCTCAagaattatcattgcaagcaccaaaatc. Gene-specific sequences are in lower case). The resulting donor plasmids were used to generate plasmids that express proteins fused at their N-terminus to 6XHis or GST, using expression vectors pDEST17 (6XHis) and pDEST15 (GST).
The 6XHis-Cdc25 fusion was expressed in BL21 cells and purified in the presence of 8M urea using standard procedures, yielding 4 mg from 4 Ls of bacterial culture. A milligram of the purified protein was used to immunize a rabbit. Since the yield of 6XHis-Cdc25 was low, a GST-Cdc25 fusion was purified using standard procedures, yielding 12 mg from 4 Ls of culture. The purified GST-Cdc25 fusion was linked to Affigel 10 to generate an affinity column for purification of antibody.
The 6XHis-Wee1 fusion protein was purified in the presence of 8M urea using standard procedures, yielding 60 mg from 4 Ls of culture. Some of the purified protein precipitated after dialysis into 25 mM HEPES, pH 7.6, 150 mM NaCl, 30% glycerol. Therefore, the protein was solubilized with a minimal amount of sodium dodecyl sulfate (approximately 0.1%) and coupled to Affigel 15 to generate a column for affinity purification of antibody.
Synchronization by centrifugal elutriation
To prepare cells for centrifugal elutriation, 3 L of cells were grown overnight in YES media at 30°C to an OD595nm of 0.5. Centrifugal elutriation was performed at 25°C using a Beckman J6-MI centrifuge equipped with a JE-5.0 rotor. The culture was pumped into a 40 ml chamber at 35 ml/min at a rotor speed of 2,800 rpm. After the elutriation, small G2 cells at OD595nm 0.25 were released into fresh YES media at 30°C and samples were taken at 20 min intervals. At each time point, 1.6 ml of culture were collected for western blotting, and 0.5 ml were collected and fixed with 70% ethanol for binucleated cells and septation index.
Western blotting
Samples were collected and processed for western blotting as described previously.44 Briefly, 1.6 ml of cell culture was centrifuged at 13,000 rpm for 15 s. The supernatant was removed and 250 µl of glass beads were added before freezing in liquid nitrogen. Sample buffer (65 mM Tris-HCl, pH 6.8, 3% SDS, 10% glycerol, 50 mM NaF, 100 mM ß-glycerophosphate, 5% 2-mercaptoethanol, 2 mM phenylmethylsulfonylfluoride [PMSF]) was added and samples were immediately lysed in a Multibeater-16 (Biospec Products) for 2 min. The PMSF was added from a 100 mM stock in 100% ethanol immediately before addition of sample buffer to the cells. Samples were centrifuged for 10 s, immediately placed in a boiling water bath for 5 min, and then centrifuged for 5 min at 13,000 rpm. To normalize protein levels across the time course, the amount of sample buffer added to each sample was increased proportionally to the optical density of the culture, starting with 120 µl in the first sample.
SDS-PAGE was carried out as described previously.10,45 Briefly, samples were loaded on SDS-PAGE gels containing 9% acrylamide and 0.14% bisacrylamide (Wee1, Cdc25 and Cdc13) and 15% acrylamide and 0.086% bisacrylamide for Cdk1-Y15 phosphorylation. The dimensions of the resolving gel were 145×65×1 mm. Gels were run on the constant current setting at 20 mA per gel until a 58 kD marker was at the bottom of the gel in the case of Wee1 and Cdc25, and until a 29 kD marker was at the bottom of the gel in the case of Cdc13 and Cdk1-Y15 phosphorylation. After transferring to nitrocellulose, blots were probed overnight on a rocker platform at 4°C with rabbit anti-Wee1 at 1 µg/ml, rabbit anti-Cdc25 at 3 µg/ml, rabbit anti-Cdk1 Y15P at a dilution of 1:1,000 (Cell Signaling 9111S), rabbit anti-Cdc13 at a dilution 1:5,000 (a kind gift from Paul Nurse) and α-tubulin at a dilution 1:5,000 (Sigma T5168). Antibodies were diluted in a phosphate buffered saline solution containing 3% dry milk (1X PBS, 0.25 M NaCl, 0.1% Tween 20) except for anti-Cdk1 Y15, that was diluted in 5% w/v BSA, 1X TBS, 0.1% Tween 20. All blots were probed with an HRP-conjugated donkey anti-rabbit secondary antibody (GE Healthcare NA934V) at a dilution of 1:5,000 for 45 min at room temperature. Secondary antibodies were detected via chemiluminescence with Advansta ECL reagents.
A minimum of 3 biological replicates were carried out for each time course, which gave similar results.
Lambda phosphatase treatment
1.6 ml of wildtype cells at optical density of 0.5 were pelleted at 13,000 rpm. The supernatant was removed and 250 µl of glass beads were added before freezing in liquid nitrogen. Cells in the pellet were lysed by bead-beating directly into either 300 µl of λ phosphatase buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 2 mM DTT, 0.01% Brij 35, 1 mM MnCl2, 2 mM PMSF, New England Biolabs P0753L) or into 300 µl of phosphatase inhibitor buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 2 mM DTT, 0.01% Brij 35, 1 mM MnCl2 2 mM PMSF, 50 mM NaF, 100 mM ß-glycerophosphate, 20 mM okadaic acid). Samples were bead-beaten for 1 min and then transferred to an ice water bath for 1 min and centrifuged for 1 min at 13,000 rpm. A 20 ul sample of each lysate was taken and 400 U of λ-phosphatase (New England Bioloabs P0753L) was added to the sample lysed into λ phosphatase buffer, which was then incubated at 30°C for 30 min. Reactions were stopped by adding 4X SDS-PAGE sample buffer (260 mM Tris-HCl, pH 6.8, 12% SDS, 40% glycerol, 20% 2-mercaptoethanol) followed by incubation in a boiling water bath for 5 min.
Microscopy
Fixed cells were imaged using a Delta Vision microscope (GE Healthcare) equipped with a 60X N.A. 1.40 oil-immersion objective (Olympus). Cells were stained with DAPI (Sigma D9542) and Calcofluor (Sigma F3543) as described previously 46
Supplementary Material
Abbreviations
- 3xHA
three tandem repeats of the influenza virus hemagglutinin epitope.
- Cdk1
cyclin-dependent kinase 1;
- DAPI
4',6-diamidino-2-phenylindole
- PP2A
protein phosphatase 2A;
- YES
yeast extract and supplements;
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Paul Nurse, Paul Russell, Kathy Gould, Juan Jimenez and Rafael R. Daga for strains and reagents and to Needhi Bhalla for sharing the Delta Vision microscope. We also thank James Moseley for advice and sharing unpublished data, and members of the Kellogg lab for advice and support.
Funding
This work was supported by NIH Grant GM109143.
References
- [1].Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 1989; 342:39-45; PMID:2682257; http://dx.doi.org/ 10.1038/342039a0 [DOI] [PubMed] [Google Scholar]
- [2].Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 1986; 45:145-53; PMID:3955656; http://dx.doi.org/ 10.1016/0092-8674(86)90546-5 [DOI] [PubMed] [Google Scholar]
- [3].Kumagai A, Dunphy WG. The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell 1991; 64:903-14; PMID:1825803; http://dx.doi.org/ 10.1016/0092-8674(91)90315-P [DOI] [PubMed] [Google Scholar]
- [4].Gautier J, Solomon MJ, Booher RN, Bazan JF, Kirschner MW. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell 1991; 67:197-211; PMID:1913817; http://dx.doi.org/ 10.1016/0092-8674(91)90583-K [DOI] [PubMed] [Google Scholar]
- [5].Strausfeld U, Labbé JC, Fesquet D, Cavadore JC, Picard A, Sadhu K, Russell P, Dorée M. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human CDC25 protein. Nature 1991; 351:242-5; PMID:1828290; http://dx.doi.org/ 10.1038/351242a0 [DOI] [PubMed] [Google Scholar]
- [6].Nurse P. Genetic control of cell size at cell division in yeast. Nature 1975; 256:547-51; PMID:1165770; http://dx.doi.org/ 10.1038/256547a0 [DOI] [PubMed] [Google Scholar]
- [7].Thuriaux P, Nurse P, Carter B. Mutants altered in the control co-ordinating cell division with cell growth in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet 1978; 161:215-20; PMID:672898 [DOI] [PubMed] [Google Scholar]
- [8].Fantes P, Nurse P. Control of cell size in fission yeast by a growth modulated size control over nuclear division. Exp Cell Res 1977; 107:377-86; PMID:872891; http://dx.doi.org/ 10.1016/0014-4827(77)90359-7 [DOI] [PubMed] [Google Scholar]
- [9].Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell 2005; 122:407-20; PMID:16096060; http://dx.doi.org/ 10.1016/j.cell.2005.05.029 [DOI] [PubMed] [Google Scholar]
- [10].Harvey SL, Enciso G, Dephoure N, Gygi SP, Gunawardena J, Kellogg DR. A phosphatase threshold sets the level of Cdk1 activity in early mitosis in budding yeast. Mol Biol Cell 2011; 22:3595-608; PMID:21849476; http://dx.doi.org/ 10.1091/mbc.E11-04-0340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tang Z, Coleman TR, Dunphy WG. Two distinct mechanisms for negative regulation of the Wee1 protein kinase. Embo J 1993; 12:3427-36; PMID:7504624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell 1995; 6:119-34; PMID:7749193; http://dx.doi.org/ 10.1091/mbc.6.1.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Deibler RW, Kirschner MW. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol Cell 2010; 37:753-67; PMID:20347419; http://dx.doi.org/ 10.1016/j.molcel.2010.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim SY, Song EJ, Lee KJ, Ferrell JE Jr. Multisite M-phase phosphorylation of Xenopus Wee1A. Mol Cell Biol 2005; 25:10580-90; PMID:16287869; http://dx.doi.org/ 10.1128/MCB.25.23.10580-10590.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Watanabe N, Broome M, Hunter T. Regulation of the human Wee1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J 1995; 14:1878-91; PMID:7743995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hoffman I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self amplification of MPF at mitosis. EMBO J 1993; 12:53-63; PMID:8428594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Izumi T, Maller JL. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol Biol Cell 1993; 12:1337-50; http://dx.doi.org/ 10.1091/mbc.4.12.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pal G, Paraz MT, Kellogg DR. Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J Cell Biol 2008; 180:931-45; PMID:18316413; http://dx.doi.org/ 10.1083/jcb.200711014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lu LX, Domingo-Sananes MR, Huzarska M, Novak B, Gould KL. Multisite phosphoregulation of Cdc25 activity refines the mitotic entrance and exit switches. Proc Natl Acad Sci U S A 2012; 109:9899-904; PMID:22665807; http://dx.doi.org/ 10.1073/pnas.1201366109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kumagai A, Dunphy WG. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell 1992; 70:139-51; PMID:1623517; http://dx.doi.org/ 10.1016/0092-8674(92)90540-S [DOI] [PubMed] [Google Scholar]
- [21].Kovelman R, Russell P. Stockpiling of Cdc25 during a DNA replication checkpoint in Schizosaccharomyces pombe. Molec Cell Biol 1996; 16:86-93; PMID:8524332; http://dx.doi.org/ 10.1128/MCB.16.1.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Trunnell NB, Poon AC, Kim SY, Ferrell JE Jr. Ultrasensitivity in the Regulation of Cdc25C by Cdk1. Mol Cell 2011; 41:263-74; PMID:21292159; http://dx.doi.org/ 10.1016/j.molcel.2011.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wicky S, Tjandra H, Schieltz D, Yates J 3rd, Kellogg DR. The Zds proteins control entry into mitosis and target protein phosphatase 2A to the Cdc25 phosphatase. Mol Biol Cell 2011; 22:20-32; PMID:21119008; http://dx.doi.org/ 10.1091/mbc.E10-06-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Izumi T, Maller JL. Phosphorylation and activation of the Xenopus Cdc25 phosphatase in the absence of Cdc2 and Cdk2 kinase activity. Mol Biol Cell 1995; 6:215-6; PMID:7787247; http://dx.doi.org/ 10.1091/mbc.6.2.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 1997; 277:1501-5; PMID:9278512; http://dx.doi.org/ 10.1126/science.277.5331.1501 [DOI] [PubMed] [Google Scholar]
- [26].Kumagai A, Yakowec PS, Dunphy WG. 14-3-3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol Biol Cell 1998; 9:345-54; PMID:9450960; http://dx.doi.org/ 10.1091/mbc.9.2.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kinoshita N, Yamano H, Niwa H, Yoshida T, Yangida M. Negative regulation of mitosis by the fission yeast protein phosphatase ppa2. Genes Dev 1993; 7:1059-71; PMID:8389306; http://dx.doi.org/ 10.1101/gad.7.6.1059 [DOI] [PubMed] [Google Scholar]
- [28].Chica N, Rozalen AE, Perez-Hidalgo L, Rubio A, Novak B, Moreno S. Nutritional Control of Cell Size by the Greatwall-Endosulfine-PP2A.B55 Pathway. Curr Biol 2016; 26:319-30; PMID:26776736; http://dx.doi.org/ 10.1016/j.cub.2015.12.035 [DOI] [PubMed] [Google Scholar]
- [29].Aligue R, Wu L, Russell P. Regulation of Schizosaccharomyces pombe Wee1 tyrosine kinase. J Biol Chem 1997; 272:13320-5; PMID:9148953; http://dx.doi.org/ 10.1074/jbc.272.20.13320 [DOI] [PubMed] [Google Scholar]
- [30].Creanor J, Mitchison JM. The kinetics of the B cyclin p56cdc13 and the phosphatase p80cdc25 during the cell cycle of the fission yeast Schizosaccharomyces pombe. J Cell Sci 1996; 109(Pt 6):1647-53; PMID:8799851 [DOI] [PubMed] [Google Scholar]
- [31].Raspelli E, Cassani C, Chiroli E, Fraschini R. Budding yeast Swe1 is involved in the control of mitotic spindle elongation and is regulated by Cdc14 phosphatase during mitosis. The J Biol Chem 2015; 290:6006; PMID:25750263; http://dx.doi.org/ 10.1074/jbc.A114.590984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lahoz A, Alcaide-Gavilan M, Daga RR, Jimenez J. Antagonistic roles of PP2A-Pab1 and Etd1 in the control of cytokinesis in fission yeast. Genetics 2010; 186:1261-70; PMID:20876564; http://dx.doi.org/ 10.1534/genetics.110.121368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Trautmann S, Wolfe BA, Jorgensen P, Tyers M, Gould KL, McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr Biol 2001; 11:931-40; PMID:11448769; http://dx.doi.org/ 10.1016/S0960-9822(01)00268-8 [DOI] [PubMed] [Google Scholar]
- [34].Wolfe BA, Gould KL. Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation. Embo J 2004; 23:919-29; PMID:14765109; http://dx.doi.org/ 10.1038/sj.emboj.7600103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Esteban V, Blanco M, Cueille N, Simanis V, Moreno S, Bueno A. A role for the Cdc14-family phosphatase Flp1p at the end of the cell cycle in controlling the rapid degradation of the mitotic inducer Cdc25p in fission yeast. J Cell Sci 2004; 117:2461-8; PMID:15128870; http://dx.doi.org/ 10.1242/jcs.01107 [DOI] [PubMed] [Google Scholar]
- [36].Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol 1999; 19:7123-37; PMID:10490648; http://dx.doi.org/ 10.1128/MCB.19.10.7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sreenivasan A, Kellogg D. The Elm1 kinase functions in a mitotic signaling network in budding yeast. Mol Cell Biol 1999; 19:7983-94; PMID:10567524; http://dx.doi.org/ 10.1128/MCB.19.12.7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sia RAL, Bardes ESG, Lew DJ. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J 1998; 17:6678-88; PMID:9822611; http://dx.doi.org/ 10.1093/emboj/17.22.6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nurse P, Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics 1980; 96:627-37; PMID:7262540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell 1989; 58:361-72; PMID:2665944; http://dx.doi.org/ 10.1016/0092-8674(89)90850-7 [DOI] [PubMed] [Google Scholar]
- [41].Lianga N, Williams EC, Kennedy EK, Dore C, Pilon S, Girard SL, Deneault JS, Rudner AD. A Wee1 checkpoint inhibits anaphase onset. J Cell Biol 2013; 201:843-62; PMID:23751495; http://dx.doi.org/ 10.1083/jcb.201212038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Toledo CM, Ding Y, Hoellerbauer P, Davis RJ, Basom R, Girard EJ, Lee E, Corrin P, Hart T, Bolouri H, et al.. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Rep 2015; 13:2425-39; PMID:26673326; http://dx.doi.org/ 10.1016/j.celrep.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Stumpff J, Kellogg DR, Krohne KA, Su TT. Drosophila Wee1 interacts with members of the gammaTURC and is required for proper mitotic-spindle morphogenesis and positioning. Curr Biol 2005; 15:1525-34; PMID:16139207; http://dx.doi.org/ 10.1016/j.cub.2005.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lucena R, Dephoure N, Gygi SP, Kellogg DR, Tallada VA, Daga RR, Jimenez J. Nucleocytoplasmic transport in the midzone membrane domain controls yeast mitotic spindle disassembly. J Cell Biol 2015; 209:387-402; PMID:25963819; http://dx.doi.org/ 10.1083/jcb.201412144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Anderson CW, Baum PR, Gesteland RF. Processing of adenovirus 2-induced proteins. J Virol 1973; 12:241-52; PMID:4747985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 1991; 194:795-823 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



