Aneuploid cells either are missing or carry extra chromosomes.1 Such imbalances usually result from unequal chromosome segregation during cell division. In humans, aneuploidy often causes spontaneous abortions or neurological impairments. Aneuploidy also characterizes the vast majority of cancers.1 Hence, understanding the causes and consequences of aneuploidy is of considerable biomedical interest.
Many researchers have turned to the yeast Saccharomyces cerevisiae as a tractable model system to analyze the molecular and cellular mechanisms of aneuploidy. An extra chromosome leads to a parallel increase in mRNA and protein expression of the genes located on the duplicated chromosome.2 This indicates that yeast do not have a general dosage compensation mechanism to correct for gene expression imbalances resulting from additional chromosomes. Although aneuploidy generally leads to reduces cell fitness, it can provide a rapid response to various cellular stresses, including DNA damage, gene loss, and nutrient restrictions.3 This is thought to work by increasing the dosage of a gene or genes that help to neutralize the stress. Therefore, the duplication of entire chromosomes can yield a selective growth advantage under specific stresses.
In a recent study,4 we discovered that yeast lacking the SUMO (small ubiquitin-related modifier) protease Ulp2 rapidly develop a specific double disomy involving chromosome I (ChrI) and ChrXII. To our knowledge, this is the first example of a specific aneuploidy involving multiple chromosomes that occurs in response to deletion of a single gene.
In the SUMO system, substrate proteins are covalently linked to SUMO or SUMO polymers through a series of enzymatic steps.5 Ulp2 is one of 2 yeast proteases that specifically cleave SUMO from substrates.6 A major role for Ulp2 is in depolymerizing SUMO chains. It is involved in chromosome segregation and contributes to the maintenance of centromere cohesion,6 but there is as yet no evidence that loss of these functions is required for the chromosome duplications of ulp2 cells. Ulp2 catalytic activity, however, is necessary to prevent aneuploidy.4
To identify genes on ChrI whose increased dosage might account for the disomy of ChrI in ulp2 mutants, we screened a high-copy, genomic ChrI DNA library for suppression of ChrI duplication.4 Overexpression of either of 2 genes, CLN3, encoding a G1 cyclin, or CCR4, encoding a catalytic deadenylase subunit of the Ccr4–Not complex, suppressed the ChrI disomy (Fig. 1). Notably, loss of Whi5, a transcriptional repressor negatively regulated by the Cdc28–Cln3 kinase, also partially suppressed the ChrI disomy in ulp2Δ cells. These results suggest that restoration of normal cell-cycle progression by increased levels of Cln3 or deletion of its target Whi5 prevents development of ChrI disomy in cells lacking ULP2.
Figure 1.
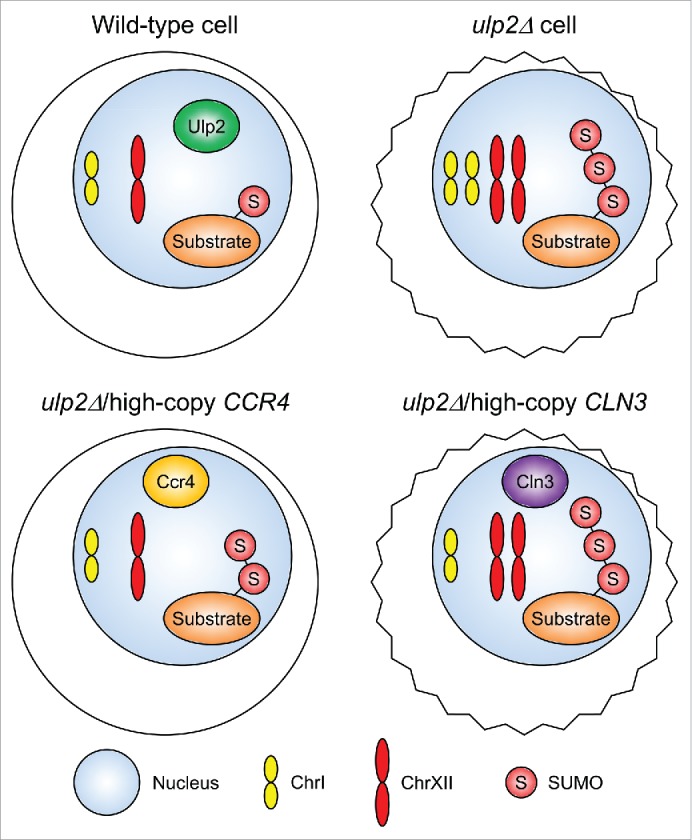
Overproduced Ccr4 or Cln3 suppresses aneuploidy triggered by loss of Ulp2. Loss of Ulp2 (top right) leads to accumulation of polySUMO-conjugated substrates, duplication of Chromosome I (ChrI) and XII, and reduced cell fitness (irregular cell outline). Increased dosage of CCR4 (bottom left) suppresses the aneuploidy and growth defects and partially reduces the aberrantly high SUMO-conjugate levels of ulp2Δ cells (Ref.4 and unpublished data). Overexpressing CLN3 (bottom right) suppresses the ChrI, but not ChrXII, disomy, likely through (partial) normalization of cell-cycle progression.
High-copy CCR4 also suppressed the ChrXII duplication, suggesting that the Ccr4–Not complex is at the heart of the aneuploidy triggered by loss of Ulp2 (Fig. 1). The multisubunit Ccr4–Not complex is evolutionarily conserved from yeast to humans. It modulates gene expression at multiple levels, including transcription, deadenylation and subsequent degradation of mRNA, translation, and even protein degradation.7 We found that overexpression of the NOT5 gene, encoding another component of the Ccr4–Not complex, also suppressed the ulp2Δ ChrI disomy. This suggests that the ChrI duplication triggered by ULP2 loss increases the expression of CCR4 in order to boost activity of the Ccr4–Not complex.
We propose that the development of the ChrI disomy is an essential adaptation that serves to amplify 2 genes, CLN3 and CCR4, in response to the potentially lethal effects inflicted by loss of the SUMO protease Ulp2. A specifc ChrXII gene amplification might also contribute to this response, although this has not yet been explored. Previous reports have documented how specific aneuploidies may confer a selective advantage under stressful conditions.3 Our data support an aneuploidy-mediated compensatory mechanism that overcomes the stress caused by a dysregulated SUMO system.
These observations raise several questions for further study. Since altered SUMO dynamics have been linked to tumorigenesis, do aberrant SUMO dynamics contribute to any of the aneuploidies observed in human tumors? How exactly does increased expression of the Ccr4-Not complex relieve the stress caused by loss of Ulp2? Since Ccr4 is a catalytic deadenylase subunit, it will be interesting to determine if this biochemical activity is indeed involved, and if so, determine the relevant RNA targets for deadenylation. Finally, are there specific proteins that must be desumoylated by Ulp2 to avoid the deficiency that leads to aneuploidy in ulp2 cells?
Recent genetic studies in our lab implicate Ulp2 in the regulation of histone modifications and transcriptional activation (unpublished data), and we are analyzing the Ulp2-dependent links between these processes. In addition, we have observed that both the aneuploidy and growth defects of ulp2Δ cells can be suppressed when mutant cultures are allowed to evolve over many generations (without reintroduction of ULP2). Therefore, we are examining these evolved ulp2Δ strains for potential adaptive changes such as histone modifications, novel gene mutations, and/or altered gene expression profiles. These studies in yeast could provide useful models for uncovering mechanistic links between SUMO pathway dysregulation and the pervasive aneuploidies that characterize human cancers.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet 2012; 13:189-203; PMID:22269907; https://dx.doi.org/ 10.1038/nrg3123 [DOI] [PubMed] [Google Scholar]
- [2].Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 2010; 468:321-325; PMID:20962780; https://dx.doi.org/ 10.1038/nature09529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mulla W, Zhu J, Li R. Yeast: a simple model system to study complex phenomena of aneuploidy. FEMS Microbiol Rev 2014; 38:201-212; PMID:24118136; https://dx.doi.org/ 10.1111/1574-6976.12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ryu HY, Wilson NR, Mehta S, Hwang SS, Hochstrasser M. Loss of the SUMO protease Ulp2 triggers a specific multichromosome aneuploidy. Genes Dev 2016; 30:1881-1894; PMID:27585592; https://dx.doi.org/ 10.1101/gad.282194.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 2012; 13:755-766; PMID:23175280; https://dx.doi.org/ 10.1038/nrm3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Miller JE, Reese JC. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 2012; 47:315-333; PMID:22416820; https://dx.doi.org/ 10.3109/10409238.2012.667214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].D'Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 2004; 117:455-469; PMID:15137939; http://dx.doi.org/ 10.1016/S0092-8674(04)00413-1 [DOI] [PubMed] [Google Scholar]


