Abstract
Cowden syndrome (CS) is an autosomal dominant disorder that predisposes to breast, thyroid, and other epithelial cancers. Differentiated thyroid carcinoma (DTC), as one of the major component cancers of CS, is the fastest rising incident cancer in the USA, and the most familial of all solid tumours. To identify additional candidate genes of CS and potentially DTC, we analysed a multi-generation CS-like family with papillary thyroid cancer (PTC), applying a combined linkage-based and whole-genome sequencing strategy and identified an in-frame germline compound heterozygous deletion, p.[Gln1478del];[Gln1476-Gln1478del] in USF3 (previously known as KIAA2018). Among 90 unrelated CS/CS-like individuals, 29% were found to have p.[Gln1478del];[Gln1476-Gln1478del]. Of 497 TCGA PTC individuals, 138 (27%) were found to carry this germline compound deletion, with somatically decreased tumour USF3 expression. We demonstrate an increased migration phenotype along with enhanced epithelial-to-mesenchymal transition (EMT) signature after USF3 knockdown or USF3 p.[Gln1478del];[Gln1476-Gln1478del] overexpression, which sensitizes cells to the endoplasmic reticulum (ER) stress. Loss of USF3 function induced cell necrosis-like features and impaired respiratory capacity while providing a glutamine-dependent cell survival advantage, strongly suggests a metabolic survival and migration-favouring microenvironment for carcinogenesis. Therefore, USF3 may be involved in the predisposition of thyroid cancer. Importantly, the results that glutamine-dependent survival and sensitivity to ER stress in USF3-deficient cells provide avenues for therapeutic and adjunct preventive interventions for both sporadic cancer as well as cancer predisposition syndromes with similar mechanisms.
Introduction
Cowden syndrome (CS, [MIM 158350]) is an autosomal dominant disorder that predisposes to female breast cancer, differentiated thyroid carcinoma (DTC) and other cancers (1–3). The syndrome is difficult to recognize clinically because of the protean and variable manifestations of its broad phenotype (2) and remains under-diagnosed. Similar to many cancer syndromes and relevant to clinical practice, identifying clear predisposition genes in CS with associated gene-specific cancer risks can significantly impact risk assessment, genetic counseling and gene-informed surveillance and clinical management for the patients and their families.
Germline mutations in the phosphatase and tensin homolog deleted on chromosome ten (PTEN, [MIM 601728]) tumour suppressor gene were found in up to 85% of CS cases defined by the strict International Cowden Consortium criteria in familial and exaggerated cases (4). Over the last two decades, CS and PTEN have served as useful models not only for gene-informed risk assessment and management for the inherited cancer syndrome but also for sporadic malignancies with somatic PTEN and associated signalling pathway abnormalities.
In a recent prospective series comprising over 3000 community-accrued probands, 25% of classic CS individuals were found to carry germline PTEN mutations (3). When individuals have features of CS but do not meet full criteria, they are referred to as CS-like and necessarily represent a heterogeneous group of potentially heritable cancer disorders. Only 5% of CS-like individuals have germline PTEN mutations (3). Thus, other predisposition genes must exist for PTEN mutation-negative CS/CS-like individuals and families. We reported previously that autosomal-located mitochondrial complex II genes SDHB/C/D (collectively SDHx, SDHB [MIM: 185470], SDHC [MIM: 602413], and SDHD [MIM: 602690]) variants account for up to 8% of CS/CS-like individuals without germline PTEN mutations (5,6). Individuals with SDHx variants have significantly higher lifetime risks of developing breast cancer or thyroid cancer compared to PTEN mutation carriers (5,6). In addition to PTEN and SDHx, we also uncovered an alternative mechanism, namely germline hypermethylation (epimutation) of the tumour suppressor KLLN (encoding KILLIN, [MIM: 612105]), accounting for up to one third of PTEN mutation-negative CS/CS-like (7). Germline KLLN hypermethylation is associated with a 250-fold decrease in KLLN expression and increased risks of breast and renal cancers over those with PTEN mutations, but not thyroid cancer (7). More recently, germline gain-of-function mutations in PIK3CA (MIM: 171834) and AKT1 (MIM: 164730), which are downstream of the PTEN signalling pathway, were identified in 11% of PTEN/SDHx/KLLN mutation/alteration-negative CS/CS-like individuals (8). However, even after accounting for all potential candidate genes identified so far, still almost half of CS/CS-like individuals with clear clinical phenotype, especially cancers, do not have identified genetic risk factors. The lack of genetic aetiology confers a major challenge for molecular diagnosis, risk assessment and genetic counseling for patients and makes gene-specific predictive testing impossible. We therefore sought to identify additional genetic aetiologies for CS/CS-like disorders, focusing on DTC-predisposition, by applying a combined family-based whole-genome sequencing strategy.
Results
Germline USF3 compound heterozygous deletion in a multi-generational CS-like family with PTC
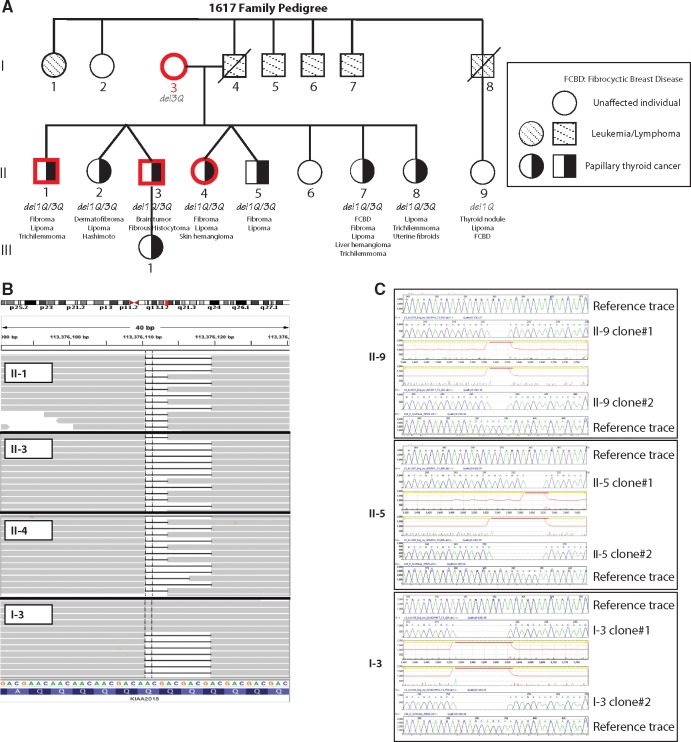
We analysed a PTEN mutation negative multi-generation CS-like family (1617) with PTC as its major malignancy (Fig. 1A). Of nine available individuals, seven are siblings and all were diagnosed with PTC in the presence of other CS features, while the mother and a paternal cousin were unaffected. The pathology report on tumour tissue from affected research participant II-8 (who was eventually found to carry the compound heterozygous USF3 deletion) confirmed bilateral PTC, with multifocal distribution, extracapsular extension in the lower right thyroid lobe, as well as metastasis to two out of three adjacent small lymph nodes. Segregation analysis together with whole-genome sequencing fine mapping revealed a 3-basepair (bp) in-frame deletion c.4416_4418delGCA/p.Gln1472del (resulting in the deletion of one glutamine, which we will hereinafter refer to as del1Q) in the USF3 found only in affected individuals (II-1, II-3 and II-4) but not in an unaffected (I-3) individual. We then confirmed the deletion through visual inspection using the IGV, and additionally found another 9-bp c.4410_4418delGCAGCAGCA/p.Gln1470_Gln1472del deletion (resulting in the deletion of three glutamine residues; del3Q) common to all four family members (Fig. 1B). We subsequently Sanger sequenced the germline DNA from all nine family members by cloning the Q deletion region, confirming the compound heterozygous deletion p.[Gln1472del];[Gln1470-Gln1472del] (del1Q/3Q) in all seven affected family members, while unaffected I-3 (mother of the affected siblings) only carries the del3Q allele, and unaffected II-9 (paternal cousin of the affected siblings) only has del1Q (Fig. 1C).
Figure 1.
USF3 germline compound heterozygous in-frame deletion in the CS-like 1617 family. (A) 1617 family pedigree. All nine available samples from family members (I-3, II-1, II-2, II-3, II-4, II-5, II-7, II-8, II-9) were analysed by genome-wide SNP array (family studies approach). Four family members (highlighted in red), three affected and one unaffected, were also analysed by whole-genome sequencing of their germline DNA. Del1Q, del3Q, and del1Q/3Q denote USF3 3bp in-frame deletion c.4410_4412delGCA/p.Gln1472del, 9bp c.4410_4418delGCAGCAGCA/p.Gln1470_Gln1472del, or compound heterozygous deletion of the two p.[Gln1472del];[Gln1470-Gln1472del]. Symbols are: square, male; circle, female; slash, deceased; black filling, papillary thyroid cancer phenotype; dashed back slash filling, leukemia or lymphoma phenotype. (B) IGV view of sequence reads. The 3bp in-frame deletion c.4410_4412delGCA/p.Gln1472del (del1Q) in gene USF3 in three of the affected family members (II-1, II-3, II-4) but not in the unaffected member I-3). The 9bp c.4410_4418delGCAGCAGCA/p.Gln1470_Gln1472del (del3Q) was observed in all four family members. The reads were label as KIAA2018 in IGV for USF3. (C) Representative cloning Sanger sequencing traces of unaffected family member II-9 only showing del1Q, affected family member II-5 showing both del1Q and del3Q, and unaffected family member I-3 showing only del3Q.
rs10606566 (c.4416_4418delGCA, p.Gln1478del) and rs112313093 (c.4410_4418delGCAGCAGCA, p.Gln1476_Gln1478del) have both been reported in the 1000 Genomes Project database without frequency information. None of the variations were noted in 6500 exomes from the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (see Web Resources) (9). rs10606566 has also been reported to exist somatically in one of 20 breast carcinoma samples in the Catalogue of Somatic Mutations in Cancer (COSMIC, see Web Resources) database (COSMIC1484461) (10). However, both variants were reported in another database including various disease-specific and population genetic studies—Exome Aggregation Consortium (ExAC, see Web Resources). In ExAC, rs10606566 and rs112313093 have minor allele frequencies as 0.4 and 0.3 respectively.
Variations called out from the mitochondrial genome were identical in all four family members with whole-genome sequencing data and regardless of disease status. The same applied to CNVs in shared genomic regions.
USF3 compound heterozygous poly-Q deletion identified in other unrelated CS individuals
Of 90 previously enrolled unrelated PTEN mutation-negative CS/CS-like individuals with CC score above 5 (score surrogate indicating CS phenotypic burden) (11), who have available exome or whole genome sequencing data, 26 (29%) harbored the USF3del1Q/3Q compound heterozygous deletion. Of 29 non-CS patients (have other disease, but irrelevant to CS), 7 (24%) also harbored this compound del1Q/3Q alteration, similar to what we can estimate based on ExAC database. In addition, by both cloning-Sanger sequencing and Light Scanner methods, we scanned 40 in-house normal controls but none of them carried del1Q/3Q. Among the 90 PTEN mutation-negative CS/CS-like individuals, the proportion of del1Q/3Q is positively correlated with an increasing CC score (p < 0.001), suggesting its association specific to CS phenotypes and with an increasing phenotypic burden in the context of CS (Supplementary Material, Table S1).
USF3 status in TCGA THCA dataset and thyroid cell lines
Because papillary thyroid cancer is the major malignant phenotype of the 1617 family, we also examined USF3 status in the TCGA thyroid cancer (THCA) dataset, in which papillary histology is the main subtype. Notably, 138 (27%) of 497 individuals were found to carry germline (ascertained from sequencing data of peripheral blood-derived DNA samples) USF3del1Q/3Q, while only 2 (0.4%) had the del1Q/del3Q exclusively in the tumours at the somatic level. Overall, thyroid tumour samples showed significantly decreased USF3 mRNA expression levels compared to normal tissues by RNA-seq analysis from the TCGA THCA sample set (Supplementary Material, Fig. S1). Samples carrying the compound heterozygous USF3del1Q/3Q also showed slight reduction in USF3 mRNA expression levels, even though not statistically significant, compared to samples with either germline USF3del1Q only or USF3del3Q only (Supplementary Material, Fig. S1, right panel).
We also examined four commonly used thyroid cancer cell lines (BCPAP, FTC236, 8505C, WRO) and three from the TCGA dataset (TT, FTC238, and SW579), with a variety of histologic subtypes (Supplementary Material, Table S2). Among these, seven thyroid cancer cell lines, two had USF3del1Q/3Q, including the only papillary line BCPAP and a squamous carcinoma line SW579. In contrast, none of the follicular cancer cell lines FTC236, FTC238, WRO, nor the medullary cancer cell line TT had the del1Q/3Q. Compared to thyroid cancer cell lines, neither the four breast cancer cell lines (BT549, MCF7, ZR-75-01, and ZR-75-30) nor the three non-malignant ‘normal’ thyroid, breast, or kidney cell lines, respectively (NThy-ori 3-1, MCF10A, HEK293T) showed the USF3del1Q/3Q.
Enhanced migration and EMT signature after USF3 knockdown
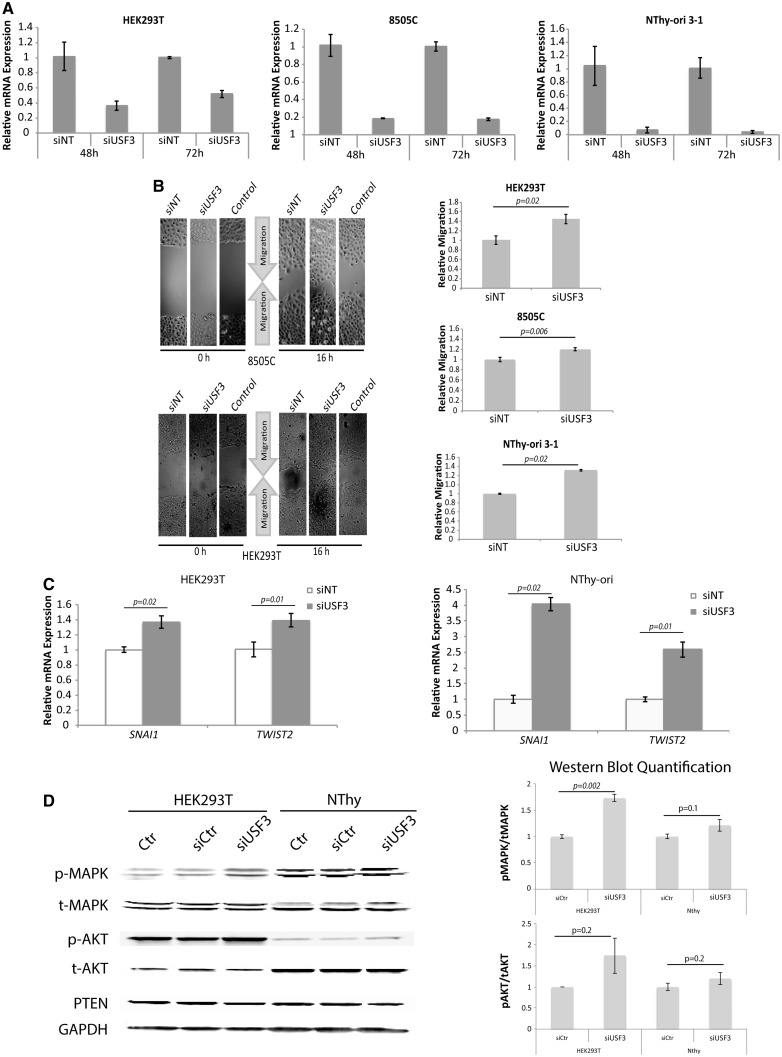
To investigate the functional impact of USF3 deletions, we first used siRNA to knock down USF3 expression in vitro to represent the loss of USF3 expression in tumour tissues observed in the TCGA THCA dataset. We selected HEK293T line and Nthy-Ori 3-1 thyroid line to represent non-malignant cells that do not have the USF3del1Q/3Q background, and 8505C thyroid cancer line as our malignant representative with USF3del3Q background. Interestingly, after USF3 knockdown (Fig. 2A), among all the cellular phenotypes we examined, including proliferation, apoptosis, migration, and clonogenicity, an enhanced migration phenotype was consistently observed in HEK293T, 8505C, and Nthy-ori 3-1 cell lines (Fig. 2B). Since increased migration without apparent proliferation can be a cellular phenotype of EMT, we next looked at the EMT marker expression profile with the hypothesis that loss-of-function of USF3 could affect EMT. Supporting our hypothesis, we observed increased mRNA expression of mesenchymal markers SNAI1 and TWIST2 after USF3 knockdown (Fig. 2C). Because PTEN-related CS results in dysfunctional downstream signalling of the PTEN pathway, we decided to look at two PTEN-downstream readouts after USF3 knockout. Interestingly, we observed consistent upregulated MAPK and slightly upregulated AKT signalling, indicating at least one shared canonical pathway (Fig. 2D). In order to test if USF3-mediated EMT is involved in RAS-MAPK-AKT-induced pathway, we checked the RAS activation status in our shUSF3 cells. However, activated RAS expression was similar between shNT and shUSF3 cells (Supplementary Material, Fig. S2), suggesting that loss-of-function of USF3 may serve as an alternative and RAS-independent activating mechanism for EMT.
Figure 2.
Enhanced migration and EMT signature after USF3 knockdown. (A) qPCR experiment showing the decrease of USF3 relative mRNA expression in siUSF3 cells compared to non-targeting controls (siNT) with GAPDH as internal loading control for normalization, in HEK293T, 8505C, and NThy-ori 3-1 cells at both 48 and 72 h after transfection. Data represent mean values ± SEM, with two independent assays performed in triplicates. (B) Phase-contrast microscopy images of wound healing assay, showing consistent increase of migration in all three cell lines after USF3 knockdown compared to both non-targeting siNT control and non-transfection control cells. The relative migration was calculated as the difference in scratch distance between time 0 and 16 h, normalized to siNT controls. For each image, distance between one side of the scratch and the other was measured and quantified using Adobe Photoshop at three different locations, in each of three different representative images (total of nine measurements) for each time point. Data represent mean values ± SEM. P value was calculated by two-sided Student’s test. (C) EMT markers SNAI1 and TWIST2 RNA expression were elevated after USF3 knockdown with qPCR quantification compared to siNT controls, normalized to GAPDH internal loading control. Data represent mean values ± SEM, with two independent assays performed in triplicates. P value was calculated by two-sided Student’s t-test. (D) Western blot showing increased pMAPK (mild increase in pAKT) after USF3 knockdown, GAPDH was used as loading control. Quantifications of pMAPK/tMAPK and pAKT/tAKT on right panel were done with ImageJ, with two independent assays for both siRNA and shRNA transfected cells. Data represent mean values ± SEM, P value was calculated by two-sided Student’s t-test.
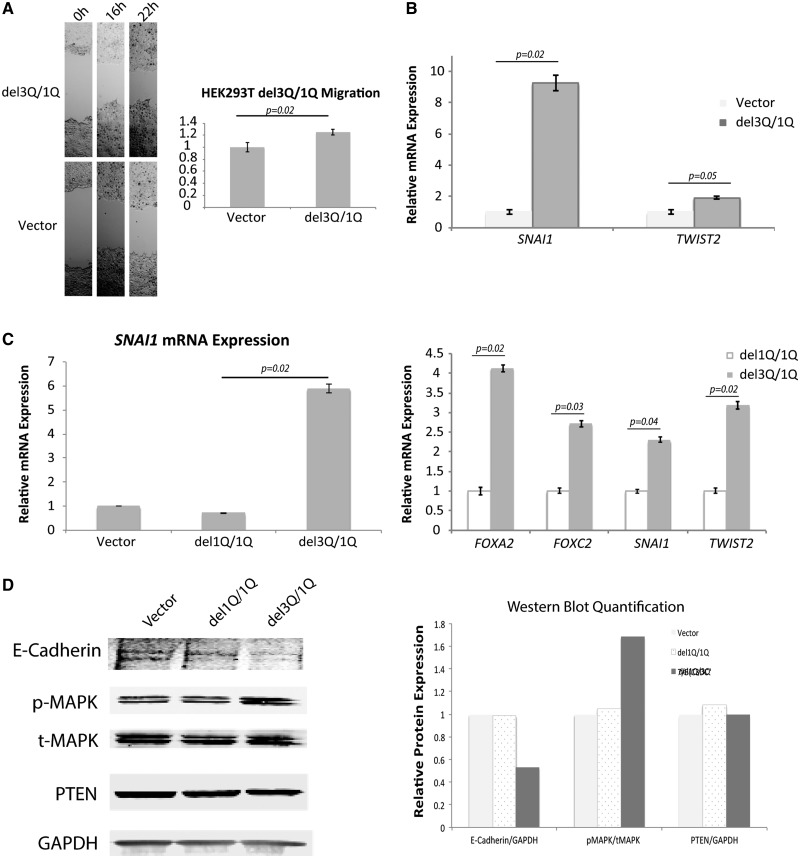
USF3del1Q/del3Q expression results in consistent increased migration and an EMT signature
We overexpressed USF3del3Q in HEK293T cells, which endogenously carry a single USF3del1Q allele (ie, WT/del1Q), to mimic the compound heterozygous deletion (del1Q/del3Q) seen in CS/CS-like individuals. Similar to our observations in USF3 knockdown cells, we observed increased migration in USF3del3Q-overexpressing del1Q cells as well as increased SNAI1 and TWIST2 EMT marker expression (Fig. 3A and B). To ensure the phenotype was not induced by overexpression of USF3 nonspecifically, but rather as a specific consequence of the USF3del1Q/3Q compound deletion, we compared USF3del3Q-overexpressing cells to USF3del1Q(only)-overexpressing cells. The activated MAPK signalling and EMT signature appear to be specific to the USF3del1Q/3Q scenario, whereas both the P-MAPK levels and EMT marker profile were unchanged in USF3del1Q(only)-overexpressing cells and empty vector transfected controls (Fig. 3C and D).
Figure 3.
Enhanced migration and EMT signature with USF3del1Q/3Q expression. (A) Phase-contrast microscopy images of wound healing assay, showing increased migration of HEK293T cells with USF3del3Q overexpression on its endogenous USF3del1Q background (mimicking the del1Q/3Q compound heterozygote state), compared to empty vector-transfected control. The relative migration was calculated as the difference in scratch distance between 0 h and 16 h, or 22 h normalized to vector controls. For each image, distance between one side of the scratch and the other was measured and quantified using Adobe Photoshop at three different locations, in each of three different representative images (total of nine measurements) for each time point. Data represent mean values ± SEM. P value was calculated by two-sided Student’s test. (B) EMT markers SNAI1 and TWIST2 RNA expression increases with USF3del3Q overexpression in USF3del1Q cells, with qRT-PCR quantification compared to vector control, normalized to GAPDH internal loading control. Data represent mean values ± SEM, with two independent assays performed in triplicates. P value was calculated by two-sided Student’s test. (C) qRT-PCR show SNAI1, FOXA2, FOXC2, and TWIST2 RNA expression increases only in USF3del3Q overexpressing cells (with the composite del1Q/del3Q genotype) but not USF3del1Q overexpressing cells. Data represent mean values ± SEM, with two independent assays performed in triplicates. P value was calculated by two-sided Student’s test. (D) Western blot showing increased p-MAPK without change in t-MAPK or PTEN, and diminished E-Cadherin with USF3del3Q overexpression. GAPDH was used as overall loading control. Quantitation of E-Cadherin/GAPDH, pMAPK/tMAPK, and PTEN/GAPDH on the right panel were performed using ImageJ for empty vector, USF3del1Q and USF3del3Q overexpressed cells.
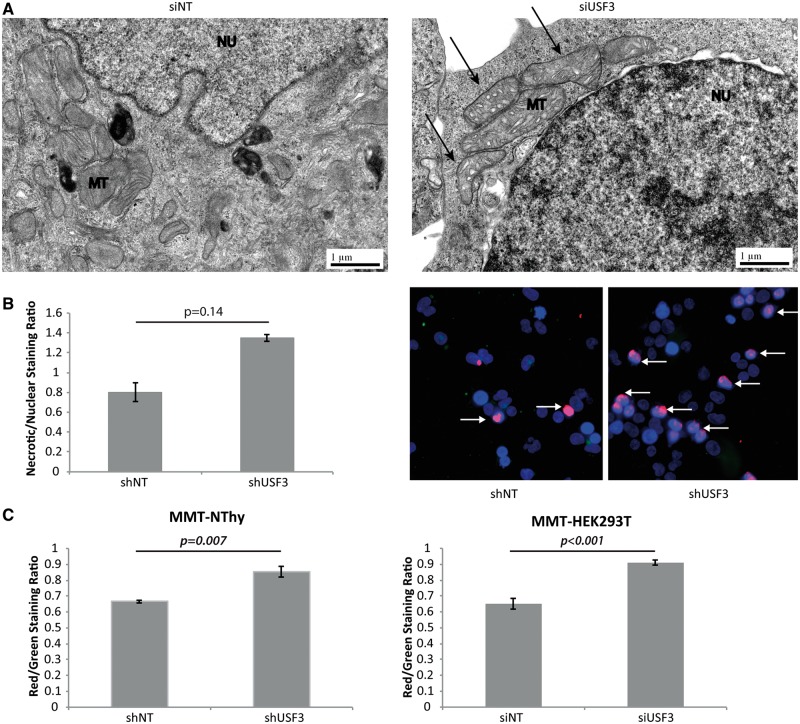
Oncotic/necrotic-like features with abnormal mitochondrial ultrastructure after USF3 knockdown
Transmission electron microscopy (TEM) can be a powerful technique to document subcellular structural changes that may result from cellular processes such as apoptosis or autophagy. TEM images of HEK293T cells with USF3 knockdown showed swelling in mitochondria or substantial loss of definition of the mitochondrial cristae (Fig. 4A, pointed by arrows), in contrast to the compact mitochondrial cristae structure in siNT HEK293T cells. As swollen mitochondria represent one of the major characteristics of necrotic cells, as opposed to cell shrinkage in apoptotic cells, we further investigated if loss of USF3 could lead to cell necrosis given that we did not observe significant changes in cell apoptosis. Indeed, by distinguishing apoptotic and necrotic cells, we observed more cells marked with the ‘necrosis probe’ in stable knock-down shUSF3 cells compared to shNT cells (Fig. 4B overlapping of red-necrosis and blue-nuclear staining, pointed by arrows), but negligible apoptotic cells in both (green staining).
Figure 4.
USF3 knock down cells have necrosis-like features. (A) Transmission electron microscopy images of non-targeting siNT control and siUSF3 transfected HEK293T cells. Arrows in the right panel point to mitochondria showing white spaces indicating swollen cristae. MT: mitochondrion; NU: nucleus. (B) Fluorescent necrosis staining. Left panel: quantification of ratio of necrosis staining to nuclear Hoechst 33342 staining, and normalized to the mean of shNT cells. Right panel: Representative fluorescent microscopy images showing more Texas Red necrosis staining in shUSF3 cells, pointed with arrows. (C) JC-1 mitochondrial membrane potential (MMP) measured in both NThy and HEK293T cells. Increased red/green fluorescence intensity indicates elevated MMP. Measurements were performed in triplicate with each triplicate repeated three times. Data represent mean values ± SEM. P-value was calculated by two-sided Student’s T-test.
Another direct measurement of mitochondrial health is the mitochondrial membrane potential (MMP). Mitochondrial depolarization is indicated by a decrease in the red/green fluorescence intensity ratio (JC-1 dye), and is dependent only on the membrane potential and not on other factors such as mitochondrial size, shape, and density that may influence single-component fluorescence signals. Compared to shNT cells, shUSF3 cells showed significantly higher JC-1 red-green ratio indicating mitochondrial hyperpolarization (Fig. 4C).
Loss of USF3 impairs mitochondrial metabolism and enhances glutamine usage for cell survival
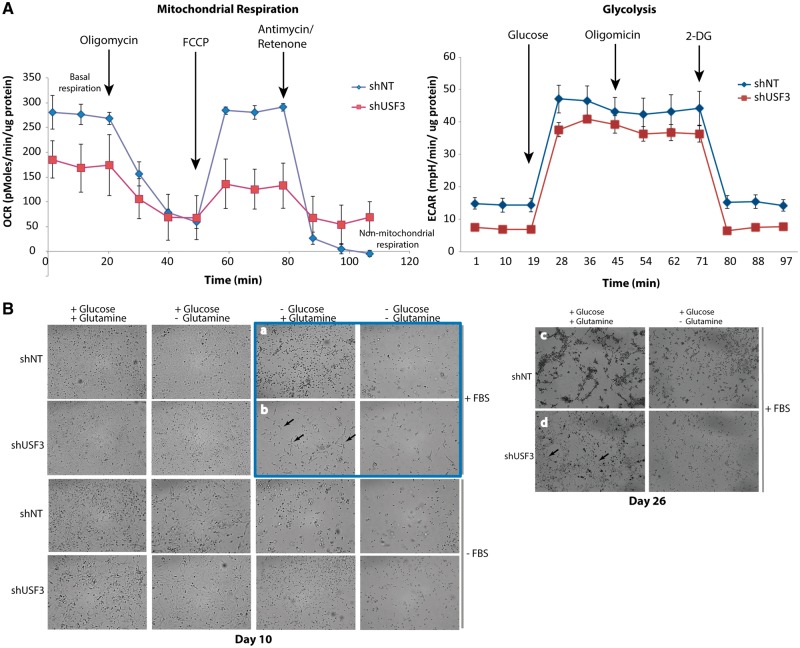
We subsequently measured cellular bioenergetics and glycolysis profile on live shNT and shUSF3 NThy-ori 3-1 cells to assess any potential impact on overall respiratory capacity and glycolytic function. shUSF3 cells showed a significant drop of basal respiration, representing decreased ATP production (Fig. 5A, left panel). Interestingly, compared to shNT cells, shUSF3 cells also showed higher non-mitochondrial respiration. However, we did not observe an obvious difference in glycolytic function between these cells (Fig. 5A, right panel). We next examined the metabolic nutrient supply that shUSF3 cells rely on. shNT and shUSF3 cells did not show any growth or survival differences under serum starvation conditions, regardless of the presence of glucose or glutamine in culture medium (Fig. 5B bottom half panel). However, under culture conditions in the presence of serum but with glucose deprivation, shUSF3 cells had a definite survival advantage compared to shNT cells at day 10. This survival advantage, however, was dependent on the supply of glutamine in the culture (Fig. 5B, highlighted in blue upper right corner group). In long-term culture (up to 26 days, Fig. 5B right panel), the shUSF3 cells still appear to be dependent on glutamine for survival even in the presence of glucose. Both shNT and shUSF3 cells did not survive in long term cultures in the absence of glucose.
Figure 5.
USF3 knock down cells had impaired mitochondrial respiration and altered glutamine usage. (A) Seahorse mitochondrial metabolic profiling. Both oxygen consumption rate (OCR, lest panel) and extracellular acidification rate (ECAR, right panel) were measured in shNT and shUSF3 NThy-ori 3-1 cells. (B) Phase-contrast microscopy images of shNT or shUSF3 NThy-ori 3-1 cells cultured in different combination of culture medium for 10 days (left panel), or extended to 26 days with FBS (right panel). As highlighted in the blue square, when cultured without glucose but with addition of glutamine, shNT cells showed mainly dotted morphology (panel a) indicating dying cells, while more shUSF3 cells (panel b) exhibited normal attached cell shape (pointed by arrows) suggesting survival. Similar observations lasted even after 26 days under the same culture conditions (panel c and d) where there were a lot more live shUSF3 cells in panel d starting to contact each other and form cellular matrix.
USF3 knockdown cells are sensitized to the endoplasmic reticulum (ER) stress
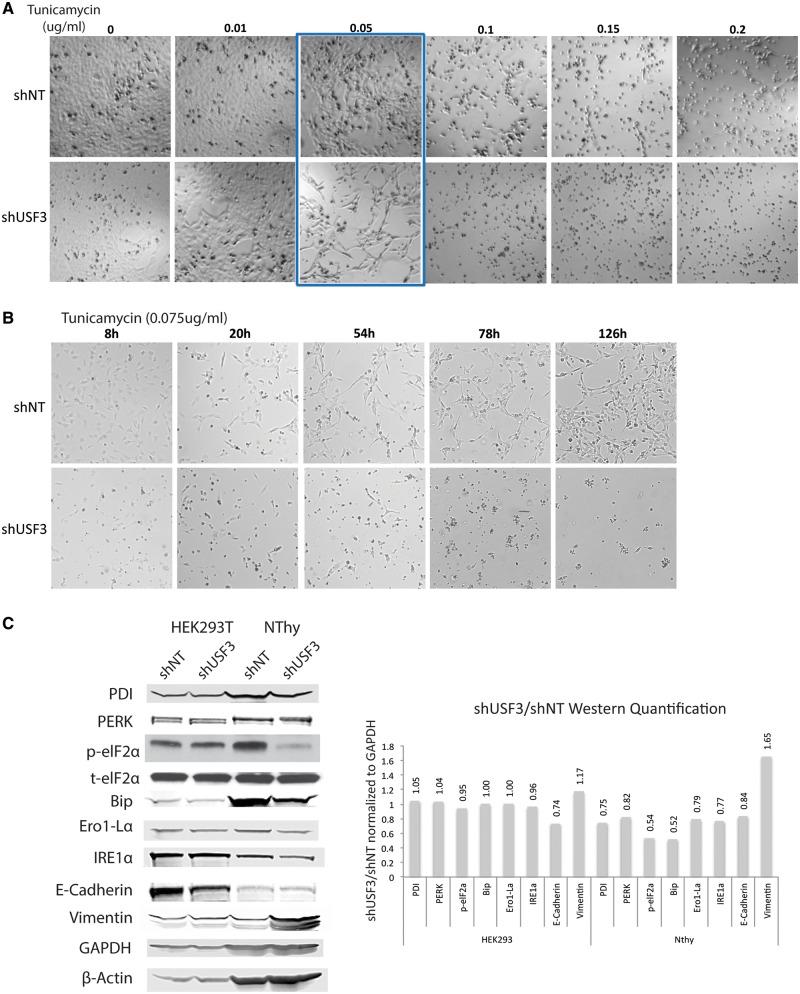
A recent study suggested EMT could sensitize cells to ER stress (12). Therefore, we sought to address our hypothesis that altered USF3 associated with EMT could be sensitive to ER stress induction. Both shNT and shUSF3 cells were treated with DMSO as a mock control or two compounds known to induce ER stress, thapsigargin (ER Ca2+ ATPase inhibitor) or tunicamycin (protein N-linked glycosylation inhibitor). We inspected cell growth and survival by examining cell morphology (attached or rounded/detached) under phase-contrast microscope and counting cells with trypan blue staining as an indication of dead cells. In HEK293T cells, both shNT and shUSF3 responded similarly to thapsigargin or tunicamycin (Supplementary Material, Fig. S3A). Conversely, in NThy-ori 3-1 ‘normal’ thyroid epithelial cells, shUSF3 resulted in cells that were more sensitive to tunicamycin treatment but not thapsigargin, with IC50shNT of 0.15 µg/ml and IC50shUSF3 of 0.067 µg/ml (Fig. 6A and Supplementary Material, Fig. S3B). Time-course experiments with a single dose (0.075 µg/ml) of tunicamycin in NThy-ori 3-1 cells showed inhibition of cell survival response in shUSF3-transfected cells as early as 8 h and lasting at least 5 days (Fig. 6B). These observations prompted us to investigate the endogenous ER stress response and unfolded protein response (UPR) in both HEK293T and NThy-ori 3-1 cells. All UPR markers (PDI, PERK, eIF2α, Bip, Ero1-Lα) showed reduction at baseline level in NThy-ori 3-1 shUSF3 cells compared to shNT cells but not in HEK293T cells (Fig. 6C). These results correspond and may explain the differential cell survival responses due to ER stress induction in HEK293T and NThy-ori 3-1 cells. Epithelial marker E-Cadherin decreased in HEK293T shUSF3 cells whereas mesenchymal marker Vimentin dramatically increased in NThy shUSF3 cells, an observation which is consistent with the observed enhanced EMT signature (above).
Figure 6.
USF3 knock down cells are sensitive to ER stress induction via tunicamycin treatment. (A) Phase-contrast microscopy images of the dose-response tunicamycin treatment in shNT or shUSF3 NThy-ori 3-1 cells. Cells were treated with 0 – 0.2 µg/ml tunicamycin for 72 h. As highlighted in the blue box, tunicamycin treatment, around the 0.05 µg/ml concentration, killed more shUSF3 cells (lower panel) compared to shNT cells (upper panel). (B) Phase-contrast microscopy images of the time-course tunicamycin treatment in shNT or shUSF3 cells. Cells were treated with a single dose of 0.075 µg/ml tunicamycin for 8–126 h. shNT cells showed normal attached morphology and even continued to proliferate after 126 h of treatment (upper panel series). However, shUSF3 cells remained rounded in shape and virtually all died after 126 h (lower panel series) (C) Western blot for UPR markers PDI, PERK, eIF2α, Bip, Ero1-Lα, EMT markers E-cadherin and Vimentin in shNT and shUSF3 stably transfected HEK293 and NThy cells. GAPDH and β-Actin were used as loading controls. Quantification of Western blots was performed with ImageJ and presented in the bar figure on the right side. Relative ratio annotated on top of each bar was calculated by protein expression level relative to GAPDH except p-eIF2α relative to t-eIF2α, and compared between shUSF3 to shNT. PDI - Protein disulfide isomerase, PERK - Protein kinase R (PKR)-like endoplasmic reticulum kinase, eIF2α - Eukaryotic translation initiation factor 2A, Bip - Binding immunoglobulin protein, Ero1-Lα - ERO1-like protein alpha, IRE1α - inositol-requiring enzyme 1.
Discussion
In this study, we discovered a compound heterozygous deletion in a rarely studied gene, USF3 with an interesting polyglutamine (polyQ) repeat in a subset of heritable CS/CS-like and sporadic PTC. The overall loss of USF3 expression was observed in sporadic thyroid tumour tissues, especially the subset of tumours with germline del1Q/3Q (TCGA data). We show that loss of USF3 leads to activation of MAPK, and possibly AKT, similar to loss-of function of one arm of canonical PTEN signalling. Of note, we also show that USF3del1Q/3Q mediates MAPK activation followed by an enhanced EMT signature and increased migration, suggesting an important role for USF3 in carcinogenesis.
It is interesting to note that the frequency of either del1Q alone or del3Q alone is common in the general and disease population, as reported in the ExAC database, yet absent in the NHLBI exome cohort and our in-house controls. It is noteworthy that the proportion of patients harboring germline del1Q/3Q increases with Cowden-phenotype disease burden. Taken together with our functional data consistent with cellular phenotypes of neoplasia, our overall observations suggest that the compound heterozygous del1Q/del3Q is associated with CS-phenotypes. Furthermore, these genetic data in the context of our glutamine and glucose data also suggest a set-up for gene-environment interactions: in the absence of a permissive environment, individuals with del1Q/del3Q may have subtle to no CS-type phenotypes (recall that CS individual features are difficult to recognize because in of themselves, can occur individually in the general population); when exposed to a certain permissive environment, del1Q/del3Q could ultimately lead to CS. The function of the upstream transcription factor family member 3 gene, USF3 (previously known as KIAA2018), has not yet been recognized. It is predicted to contain a basic helix-loop-helix (bHLH) domain (Supplementary Material, Fig. S4), a protein structural motif typical and characteristic of the transcription factor family (13,14). USF3 paralogs such as MAX (MIM: 154950) and USF1/2 (MIM: 1549523 and 600390) also have bHLH domains. USF1/2 harbor a DNA-binding E-box motif that also appears as an upstream cis-acting PTEN regulatory element that can be deleted in CS (15). It is worth noting that germline mutations in MAX, along with SDHx, have been associated with hereditary paraganglioma/pheochromocytoma (MIM: 168000, 601650, 605373, 115310, 171300), considered a metabolic neoplasia syndrome, with overlapping clinical features with CS such as a higher risk for renal cell carcinoma and PTC (16). The similarity of USF3 to MAX and USF1/2 at the sequence level may imply shared transcriptional regulatory function and warrants future investigation in that regard.
Even though the role of polyQ in the USF3 has never been elucidated before, there are at least nine inherited neurodegenerative diseases well known to be caused by an expansion (in contrast to deletion in our current report) of unstable polyQ repeats in various disease proteins, including those associated with Huntington disease (MIM: 143100), spinocerebellar ataxia (MIM: 164400, 183090,183086, 164500, 607136), spinobulbar muscular atrophy (MIM: 313700), and dentatorubropallidoluysian atrophy (MIM: 125370) (17). According to the Genotype-Tissue Expression (GTEx) database, USF3 expression levels in normal human tissues are highest in cerebellum and skeletal muscle (Supplementary Material, Fig. S5), tissues with high demand for energy, while relatively ubiquitous in other tissue types, including thyroid tissue. On the other hand, the deletion of trinucleotide repeats are not well studied but previous epidemiology studies showed correlation of the deletion of trinucleotide repeats with human ovarian and prostate cancer (18,19). Germline mutations in the same gene, with opposing functional consequences and causing completely unrelated phenotypes, have been previously demonstrated. Recent examples involve genes that are vital to energy production. For example, homozygous or compound heterozygous mutations in genes encoding mitochondrial complex II (succinate dehydrogenase, SDH) cause Leigh syndrome (MIM: 256000), an early-onset progressive neurodegenerative disorder, whereas germline heterozygous mutations in one of the 4 SDHx’s are associated with heritable pheochromocytoma/paraganglioma syndromes (20). Similarly, homozygous or compound heterozygous mutation in FH (MIM: 136850), encoding fumarate hydratase, which is the next catalytic step after SDH, results in infant-onset lethal neuropathy; in contrast, germline heterozygous FH mutations result in hereditary leiomyoma-renal cell carcinoma syndrome (MIM: 150800) (21,22). Our findings here demonstrate a different role for polyQ deletions in carcinogenesis, at least in the context of the USF3.
We observed that USF3 knockdown associates with altered mitochondrial ultrastructure and an oncotic/necrotic-like cellular phenotype, as well as compromised mitochondrial respiratory capacity. It has been shown that ATP synthase (mitochondrial complex V) inhibition by oligomycin prevents proton passage through the enzyme inducing mitochondrial membrane hyperpolarization, which also results in mitochondrial swelling (23). Furthermore, oxidative stress generated by mitochondrial dysfunction can also promote migration and stimulate MAPK-mediated necrosis (24,25), thus setting up a feedback-signalling network. We also show that the addition of glutamine confers upon cells with loss of USF3 that are also deficient in mitochondrial respiratory capacity an even greater survival advantage. Our observations corroborate accumulating evidence supporting a metabolic shift toward glutamine that promotes cancer cell survival and growth (26,27), especially the unexpected role of glutamine in cells with reduced mitochondrial function (28).
RAS-RAF activation is well documented in sporadic PTC, and is known to trigger the MAPK-SNAI-EMT signalling cascade (29,30). While we observed that USF3-knockdown was accompanied by MAPK-SNAI-EMT signalling, we also demonstrated no RAS activation. Thus, we postulate that USF3 might function in parallel with RAS in the context of inducing the EMT program (Supplementary Material, Fig. S6). However, the precise mechanism of whether this occurs through its predicted transcriptional regulation or other yet undiscovered function, will need further study. EMT involves a series of changes whereby epithelial cells disassemble their junctional structures, express mesenchymal proteins, remodel their extracellular matrix, lose polarity, and eventually become more migratory. Epithelial cancer cells use this property to enhance their invasive and metastatic potential and render them more resistant to apoptotic signals. Accordingly, cells within the invasive front of human thyroid cancers have gene expression changes consistent with EMT when compared to cells in the central part of the tumour (31). In addition, loss of E-cadherin (a hallmark of EMT), as we seen with USF3 dysfunction/loss-of-function, is a marker of progression from differentiated to undifferentiated thyroid cancers (32). In support of our observation, TCGA thyroid cancer dataset showed USF3 expression is negatively correlated with SNAI1 and SNAI3 (with no impact in SNAI2), negatively correlated with TWIST1/2. At the protein level, both phosphorylated MAPK1 and MAPK3 at amino acid T201-Y204 are up-regulated, but not pAKT1/2/3. These concordant results further strengthen our hypothesis that USF3 may be involved in the regulation of EMT signalling.
Equally relevant to potential targeted treatment, USF3 dysfunction-mediated EMT could sensitize cells to ER stress, perhaps in a tissue-specific manner. The majority of previous studies focused on its reverse signalling, ie, that ER stress induces EMT and autophagy (33); in contrast, a recent report revealed that EMT can sensitize cells to ER stress by activating the PERK-elF2α axis of the UPR (12). In our present study, this sensitization seems to be due to the loss of the endogenous ER stress response/UPR signalling although it remains unclear how the loss of/altered USF3 could lead to the loss of ER stress response/UPR.
Overall, our study uncovers a unique role of a poorly studied USF3 in cell migration regulation and associated signalling pathways. Loss of USF3 facilitates cross-talk of EMT signalling and mitochondrial dysfunction, therefore creating a microenvironment favouring tumour progression. When found in the germline, our observations suggest that USF3 dysfunction participating in carcinogenesis in the context of cancer predisposition may be amenable to gene-environment interactions, a phenomenon that is often postulated in heritable cancer but without functional hints. Importantly, we show glutamine-dependent survival and sensitivity to ER stress in USF3-deficient cells suggests an accessible therapeutic window, whether in sporadic cancer therapeutics or as a preventative adjunct in cancer predisposition syndromes dependent on similar mechanisms.
Materials and Methods
Research participants
CS or CS-like individuals were prospectively enrolled in accordance with our research protocol IRB8458-PTEN, which was approved by the Cleveland Clinic and respective Institutional Review Boards for Human Subjects Protection. All research participants provided written informed consent. To be enrolled in the IRB8458-PTEN, eligible individuals must meet the full CS diagnostic criteria established by the International Cowden Consortium (Supplementary Material, Table S3) or the relaxed criteria (full criteria minus one) according to version 2006 National Comprehensive Cancer Network (NCCN) Guidelines (34). Patients meeting the relaxed criteria are referred to as individuals with CS-like phenotypes or CS-like. In other words, CS-like was diagnosed when an individual did not fully meet the strict diagnostic criteria but had features with one or two criteria short of the operational diagnostic criteria. Matching the subjects, normal (population) controls were from Northern and Western European origin and were made anonymous prior to banking and analysis.
Genomic DNA extraction
Germline genomic DNA was extracted from peripheral blood samples from CS/CS-like and healthy individuals by the Genomic Medicine Biorepository (GMB), Genomic Medicine Institute, Cleveland Clinic (protocols are available at GMB website, see Web Resources).
Whole genome SNP array and sequencing analysis
Genome-wide SNP array was performed using Illumina HumanOmni2.5 BeadChip (Illumina, San Diego, CA) containing ∼2.5 million markers, for all nine members of 1617 family. Copy number variation (CNV) was called out by CNV partition plug-in in the Beadstudio software. SNP genotyping data were exported to PLINK (35) and non-parametric linkage analysis was done by MERLIN software package (36). Shared genomic segments among all affected individuals were collected for whole-genome sequencing analysis (Supplementary Material, Fig, S7A and B).
Germline DNA from individuals I-3, II-1, II-3 and II-4 from family 1617 were subjected to whole-genome sequencing, as previously described (37). Briefly, paired-end libraries with 100-bp sequencing were prepared and genome sequencing was performed on an Illumina HiSeq2000 instrument. Next generation sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number SRX1942054.
An in-house pipeline was used to align reads against the hg19 genome build (GRCh37) and to call single-nucleotide variants and small insertion/deletions (indels). The alignment and quality score recalibration were performed using Burrows-Wheeler Aligner (BWA) and the Genome Analysis Toolkit (GATK, see Web Resources) according to GATK best practice (38,39). Targeted regions were sequenced to an average depth of 34x, with 98% of the regions covered by 1x and 96% covered by 10x (Supplementary Material, Fig. S7C). Common polymorphisms (with reported frequency of >1%) were removed by comparison with dbSNP135, the 1000 Genomes Project database using ANNOVAR (40) (see Web Resources) and a proprietary database of exomes from in house non-CS individuals. All variants were manually inspected using the Integrative Genomics Viewer (IGV, see Web Resources) (41). The accession numbers for naming mutations in USF3 sequences reported in this paper is NM_001009899.3.
Mutation analysis by LightScanner and sanger sequencing
Genomic DNA was analysed using high-resolution melting LightScanner technology (BioFire Defense, Salt Lake City, Utah), which detects nucleic acid sequence variations, by changes in the melting curve. Primers to amplify the USF3 mutation region were designed using LightScanner Primer Design software (Supplementary Material, Table S4: primer sequence) and optimized according to the manufacturer’s instructions. Germline genomic DNA samples were amplified with LCGreen® Plus (BioFire Defense) in a final reaction volume of 10 uL with 20 uL oil overlay. The temperature cycling protocol consisted of an initial denaturation step at 95 °C for 2 min, followed by 37 cycles of denaturation at 94 °C for 30 s, optimal annealing temperature for each amplicon for 30 s, and heteroduplex formation step at 95 °C for 30 s and final hold at 25 °C. Melting curve analysis was performed on LightScanner with LightScanner software employing three steps, namely normalization, temperature shift, and generating difference plot to cluster samples. Samples with differently melting curves from positive or negative reference samples were clustered into separate genotype groups (Supplementary Material, Fig. S8).
Genomic DNA was amplified using primers flanking the USF3 poly-glutamine region and StrataCloned (Agilent, Santa Clara, CA) into E. coli competent cells. At least eight colonies were selected for each whole-cell PCR reaction and Sanger sequenced to observe allelic deletion status.
For all next-generation sequencing data available to us, the USF3 p.Gln1472del and USF3 p.Gln1470-Gln1472del were observed with the wild type (WT) allele. Please note that USF3-Gln region cloning followed by Sanger sequencing did not show any WT colonies, suggesting that bacterial cells incorporating the WT-Q region clone did not survive (which is a common gene-dependent phenomenon).
Cell lines and culture conditions
Human immortalized lymphoblastoid cell lines (LCLs) derived from peripheral blood were generated by the Genomic Medicine Biorepository, Genomic Medicine Institute, Cleveland Clinic (protocols are available at GMB website, see Web Resources). LCLs were cultured in RPMI 1640 supplemented with 20% fetal bovine serum (FBS) and 100 units/ml each of Penicillin and Streptomycin. HEK293T cell line (originally purchased from the American Type Culture Collection in 2011 and obtained in 2014 from the Cleveland Clinic Lerner Research Institute Cell Culture Core) was cultured in DMEM supplemented with 10% FBS and 100 units/ml each of Penicillin and Streptomycin. 8505C thyroid cancer cell line (#ACC219, lot # 3, purchased in 2014 from Sigma, St. Louis, MO) was cultured in EMEM (HBSS) medium supplemented with 2 mM Glutamine, 1% Non Essential Amino Acids (NEAA) and 10% FBS. The Nthy-ori 3-1 human thyroid follicular epithelium cell line (catalog number EC90011609, lot number 09C008 purchased in 2014 from Sigma, St. Louis, MO) was cultured in RPMI-1640 supplemented with 2 mM glutamine and 10% FBS. Nthy-ori 3-1 cell line was authenticated through STR PCR (AmpFLSTR SGM Plus PCR Amplification Kit, Thermo Fisher Scientific) by European Collection of Cell Cultures (ECACC, original source of the cell line, test data 14/04/2009). Cell lines used have not been listed as cells known to be misidentified according to the International Cell Line Authentication Committee (ICLAC) (42). All cell lines were maintained at 37 °C and 5% CO2 culture conditions, used within 5–16 passages from original purchase, and tested negative for mycoplasma contamination (luminescence ratios < 0.9) using the MycoAlert Mycoplasma Detection Kit (Lonza, Allendale, NJ).
For endoplasmic reticulum (ER) stress induction treatment, cells were seeded in equal starting number and allowed to grow overnight and then treated with either Thapsigargin or Tunicamycin (Sigma-Aldrich, St. Louis, MO) as noted in the text for dose-response and time course experiments. Cells were checked every day with bright field image capture. Cell number and viability were assessed by the Trypan Blue exclusion test and counting live vs dead cells.
Cell lines used to extract DNA to identify USF3 Gln-deletion status include four thyroid cancer cell lines: BCPAP, FTC236, 8505C, WRO; four breast cancer cell lines: BT549, MCF7, ZR-75-01, ZR-75-30; and three non-malignant cell lines: NThy-ori 3-1, MCF10A, HEK293T. BCPAP, FTC236 were purchased from Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (DSMZ, Germany), FTC236, 8505C, NThy-or 3-1 were purchased from Sigma, and all other lines were purchased from ATCC, utilized and frozen back within 20 passages, and authenticated by STR PCR. Additional information from three thyroid cancer cell lines (TT, FTC238, and SW579) was obtained from TCGA dataset.
Plasmid transfection
Non-targeting siRNA (siNT) and a pool of 4 USF3 siRNA (siUSF3) were commercially obtained (GE Dharmacon, Lafayette, CO), optimized and transfected into HEK293T, 8505C, and Nthy-ori 3-1 cells respectively with Lipofectamine 3000 transfection reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). USF3 mRNA expression was evaluated by qPCR to confirm knockdown efficiency at different time points. Human USF3 cDNA was cloned into pCDNATM3.1/Hygro(+) vector (Thermo Fisher Scientific) with EGFP tag at c-terminal end of open reading frame (ORF). MISSIONTM USF3 shRNA (shUSF3) and non-targeting shRNA control (shNT) was purchased from Sigma-Aldrich. USF3-del1Q-GFP, USF3-del3Q-GFP, vector-GFP, shUSF3, and shNT plasmids were transfected into HEK293T cells using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific). Fresh medium was replaced 6 h post transfection. USF3-del1Q-GFP, USF3-del3Q-GFP, vector-GFP transfected cells were observed under a fluorescent microscope to confirm the transfection efficiency, and sorted by flow cytometry for the qRT-PCR experiment. While shUSF3 and shNT transfected cells were confirmed by qPCR on USF3 mRNA expression and selected by puromycin to generate stable cell lines.
Wound-healing migration assay
Cultured cells were seeded in 12-well plates to reach a 100% confluent monolayer and scraped with a p10 pipette tip to create a straight line ‘scratch’. Debris was removed by washing the cells with 1 ml of the growth medium. First images at time 0 scratch were taken under a phase-contrast microscope and the reference position marked outside of the captured image field. Cells were incubated at 37 °C for 8–18 h and examined periodically. Post incubation images of the scratch area, located using the reference point were taken. For each image, the distance between one side of the scratch and the other was measured and quantified using Adobe Photoshop at three different locations, in each of three different representative images for each time point. The relative migration is measured by the distance of each scratch closure by comparing the images from time 0 to the last time point.
RNA extraction and quantitative reverse-transcription-PCR
Total RNA was extracted from cultured cells using the GeneJET RNA Purification Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol, and subsequently treated with Turbo DNase (Thermo Fisher Scientific). DNase-treated total RNA was reverse-transcribed into cDNA using SuperScriptIII (Thermo Fisher Scientific), as specified by the manufacturer. Quantitative PCR was performed on a LightCycler 480 system (Roche Diagnostics Corporation, Indianapolis, IN) using the TaqMan® primer-probe ready mix for USF3, and GAPDH as an endogenous loading control (Thermo Fisher Scientific).
Protein analysis
Whole-cell lysates were prepared with Mammalian Protein Extraction Reagent, M-PER (ThermoFisher Scientific) supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes (BioRad, Hercules, CA). The resulting blots were subjected to Odyssey band intensity analysis for PTEN (6H2.1, Cascade Bioscience, Portland, OR), pMAPK, tMAPK, pAKT, tAKT, PDI, PERK, p_elF2α, t_elF2α, Bip, Ero1_Lα, IRE1α, E-Cadherin, Vimentin, and GAPDH (Cell Signaling Technology, Inc, Danvers, MA) protein levels. Western blot quantification was performed with ImageJ (43).
Transmission electron microscopy
HEK293T cells were grown in 2-well plastic chamber slides (Thermo Fisher Scientific). Forty-eight h after siNT or siUSF3 knock-down, samples were fixed with 2.5% glutaraldehyde/4% paraformaldehyde in 0.2 M cacodylate buffer overnight at 4 °C. Samples were washed with sodium cacodylate buffer (0.2M, pH7.3) three times, 5 min each. Post-fixation process includes: incubate samples with 1% Osmium Tetroxide in H2O for 60 min at 4 °C; wash samples with sodium cacodylate buffer two times, 5 min each; rinse with Maleate buffer (pH 5.1) once, 5 min; change to 1% uranyl acetate in Maleate buffer, stain for 60 min; remove uranyl acetate and wash with maleate buffer three times, 5 min each. Samples then underwent a dehydration process washing serially with 30, 50, 75, 95% ethanol 5 min each, 100% ethanol three times 15 min each, and propylene oxide three times, 15 min each. Sample infiltration was conducted by adding 1:1 propylene oxide/eponate 12 medium at room temperature overnight, followed by pure eponate 12 medium incubation for 4–6 h at room temperature. Finally, the samples were polymerized for 24 h before slicing into an ultrathin section of 85 nm with diamond knife and staining with uranyl acetate and lead citrate. Sample grids were observed and representative images taken with a Tecnai G2 SpiritBT electron microscope operated at 60 kV.
JC-1 mitochondrial membrane potential measurement
Mitochondrial membrane potential can be monitored with the dual emission potentiometric dye tetrachloro‐1,1′,3,3′-tetraethylbenzimidazol-carbocyanine iodide - JC-1. JC-1 fluorescence ratio detection was carried out following the manufacturer’s protocol (Cell Technology, Fremont, CA). In brief, the cell suspension was centrifuged and resuspended in 1x JC-1 reagent, followed by 15 min incubation at 37 °C in a 5% CO2 incubator. After incubation, cell pellet was collected and washed with 1x assay buffer twice before resuspension for assay. Triplicates of each cell suspension were transferred to a black 96-well plate and measured within the red (excitation 550 nm, emission 600 nm) and green fluorescence (excitation 485 nm, emission 535 nm) regions using a fluorescence plate reader. The ratio of red to green fluorescence is decreased in dead cells and in cells undergoing apoptosis compared to healthy cells.
Necrosis and apoptosis staining
Necrotic, apoptotic, and healthy cells were stained using Apoptotic/Necrotic/Healthy Cell Detection Kit (PromoKine, Heidelberg, Germany) according to the manufacturer’s instruction. Briefly, cells were washed with 1X binding buffer followed by staining with FITC-Annexin V, Ethidium Homodimer III, and Hoechst 33342. After 15 min of incubation at room temperature, cells were washed three times and observed under fluorescent light using FITC, Texas Red, and DAPI filter sets. Green fluorescent plasma membrane staining identifies apoptotic cells, while necrotic cells are identified by a red fluorescent nuclear staining. Late apoptotic cells may show green and red staining. All cell nuclei are stained blue.
Seahorse metabolic profiling
The oxygen consumption rate (OCR, pmol/min) and extracellular acidification rate (ECAR, mpH/min) were measured using XFe24 Extracellular Flux Analyzers from Seahorse Bioscience (North Billerica, MA) following the manufacturer's protocol. Briefly, the cells were seeded at a density of 50,000 cells/well (minimum of five parallels for each treatment) in a 24-well plate and incubated overnight. Before running the assay, the cells were re-suspended in Seahorse assay medium, and the drugs from the XF Cell Mito Stress test kit (Seahorse Bioscience) were added: oligomycin (1 µM), FCCP (1 µM) and rotenone/antimycin A mix (1 µM). The data were analysed by XFe Wave software (Seahorse Bioscience).
TCGA dataset analysis
Whole genome and/or exome sequencing (WGS/WES) .bam files were obtained through the Cancer Genome Atlas (TCGA) project via download from Cancer Genomics Hub (see Web Resources). Germline data were generated by TCGA obtained from peripheral blood.
From the .bam sequence files, USF3 reads were extracted using SAMtools (39). The bam2fastq software was used to revert these aligned sequences to fastq format (see Web Resources), which were then realigned against hg19 reference genome using bowtie2 (44) and the pileup file was generated using SAMtools. From the pileup file, variants were called using bcftools and inspected through the Integrative Genome Viewer (IGV) (45).
The gene expression analysis from the TCGA dataset was based on IlluminaHiSeq_RNASeqV2 ‘rsem.gene.normalized_results’ files downloaded from TCGA’s Data Portal, in which RNA-seq reads were quantified by upper quartile normalized RSEM count estimates.
Statistical analysis
Statistical analysis was carried out using SPSS (IBM SPSS Statistics for Macintosh, Version 21. Armonk, NY: IBM Corp) with raw or adjusted (as appropriate) significance indicated at p < 0.05.
Web resources
ANNOVAR, http://www.openbioinformatics.org/annovar/
Bam2fastq software: http://www.hudsonalpha.org/gsl/information/software/bam2fastq
COSMIC, http://cancer.sanger.ac.uk/cosmic/
ExAC, http://exac.broadinstitute.org
GATK, http://www.broadinstitute.org/gatk/
GTEx, http://www.gtexportal.org/home/
Genomic Medicine Biorepository (GMB), Genomic Medicine Institute, Cleveland Clinic (http://www.lerner.ccf.org/gmi/gmb/methods.php)
Genomic Medicine Institute calculator for PTEN CC scores, http://www.lerner.ccf.org/gmi/ccscore/
IGV, https://www.broadinstitute.org/igv/home/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
TCGA Data Portal, https://tcga-data.nci.nih.gov/tcga/
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to all our patients and their families who have participated in our studies over the years. We thank Dr. Ritika Jaini for critical review of this manuscript.
Conflict of Interest statement. None declared.
Funding
This work was funded, in part, by the National Cancer Institute (NCI) grant P01CA124570 (to MDR and CE) and the Breast Cancer Research Foundation (to CE). YN is a CoGEC Scholar funded by NCI grant R25TCA094186 (to R. Elston, PhD); and LY is a Fulbright International Science and Technology Fellow and recipient of the Dr. Michael H. Fakih Predoctoral Scholarship. CE is the Sondra J. and Stephen R. Hardis Chair of Cancer Genomic Medicine at the Cleveland Clinic, and is an American Cancer Society Clinical Research Professor. Funding to pay the Open Access publication charges for this article was provided by American Cancer Society Clinical Research Professorship.
References
- 1.Eng C. (2003) PTEN: one gene, many syndromes. Hum. Mut., 22, 183–198. [DOI] [PubMed] [Google Scholar]
- 2.Zbuk K.M., Eng C. (2007) Cancer phenomics: RET and PTEN as illustrative models. Nat. Rev. Cancer, 7, 35–45. [DOI] [PubMed] [Google Scholar]
- 3.Tan M.H., Mester J.L., Ngeow J., Rybicki L.A., Orloff M.S., Eng C. (2012) Lifetime Cancer Risks in Individuals with Germline PTEN Mutations. Clin. Cancer. Res., 18, 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh D.J., Coulon V., Lunetta K.L., Rocca-Serra P., Dahia P.L., Zheng Z., Liaw D., Caron S., Duboue B., Lin A.Y., et al. (1998) Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum. Mol. Genet., 7, 507–515. [DOI] [PubMed] [Google Scholar]
- 5.Ni Y., He X., Chen J., Moline J., Mester J., Orloff M.S., Ringel M.D., Eng C. (2012) Germline SDHx variants modify breast and thyroid cancer risks in Cowden and Cowden-like syndrome via FAD/NAD-dependant destabilization of p53. Hum. Mol. Genet., 21, 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ni Y., Zbuk K.M., Sadler T., Patocs A., Lobo G., Edelman E., Platzer P., Orloff M.S., Waite K.A., Eng C. (2008) Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes. Am. J. Hum. Genet., 83, 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett K.L., Mester J., Eng C. (2010) Germline epigenetic regulation of KILLIN in Cowden and Cowden-like syndrome. JAMA, 304, 2724–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orloff M.S., He X., Peterson C., Chen F., Chen J.L., Mester J.L., Eng C. (2013) Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am. J. Hum. Genet., 92, 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu W., O'Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., et al. (2013) Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature, 493, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., et al. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res., 39, D945–9D950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan M.H., Mester J., Peterson C., Yang Y., Chen J.L., Rybicki L.A., Milas K., Pederson H., Remzi B., Orloff M.S., et al. (2011) A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am. J. Hum. Genet., 88, 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y.X., Sokol E.S., Del Vecchio C.A., Sanduja S., Claessen J.H., Proia T.A., Jin D.X., Reinhardt F., Ploegh H.L., Wang Q., et al. (2014) Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov., 4, 702–715. [DOI] [PubMed] [Google Scholar]
- 13.Murre C., Bain G., van Dijk M.A., Engel I., Furnari B.A., Massari M.E., Matthews J.R., Quong M.W., Rivera R.R., Stuiver M.H. (1994) Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta, 1218, 129–135. [DOI] [PubMed] [Google Scholar]
- 14.Massari M.E., Murre C. (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol., 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pezzolesi M.G., Zbuk K.M., Waite K.A., Eng C. (2007) Comparative genomic and functional analyses reveal a novel cis-acting PTEN regulatory element as a highly conserved functional E-box motif deleted in Cowden syndrome. Hum. Mol. Genet., 16, 1058–1071. [DOI] [PubMed] [Google Scholar]
- 16.Comino-Mendez I., Gracia-Aznarez F.J., Schiavi F., Landa I., Leandro-Garcia L.J., Leton R., Honrado E., Ramos-Medina R., Caronia D., Pita G., et al. (2011) Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat. Genet., 43, 663–667. [DOI] [PubMed] [Google Scholar]
- 17.Margulis B.A., Vigont V., Lazarev V.F., Kaznacheyeva E.V., Guzhova I.V. (2013) Pharmacological protein targets in polyglutamine diseases: mutant polypeptides and their interactors. FEBS Lett., 587, 1997–2007. [DOI] [PubMed] [Google Scholar]
- 18.Nelson K.A., Witte J.S. (2002) Androgen receptor CAG repeats and prostate cancer. Am. J. Epidemiol., 155, 883–890. [DOI] [PubMed] [Google Scholar]
- 19.Schildkraut J.M., Murphy S.K., Palmieri R.T., Iversen E., Moorman P.G., Huang Z., Halabi S., Calingaert B., Gusberg A., Marks J.R., et al. (2007) Trinucleotide repeat polymorphisms in the androgen receptor gene and risk of ovarian cancer. Cancer Epidemiol. Biomarkers Prev., 16, 473–480. [DOI] [PubMed] [Google Scholar]
- 20.Eng C., Kiuru M., Fernandez M.J., Aaltonen L.A. (2003) A role for mitochondrial enzymes in inherited neoplasia and beyond. Nat. Rev. Cancer, 3, 193–202. [DOI] [PubMed] [Google Scholar]
- 21.Alam N.A., Rowan A.J., Wortham N.C., Pollard P.J., Mitchell M., Tyrer J.P., Barclay E., Calonje E., Manek S., Adams S.J., et al. (2003) Genetic and functional analyses of FH mutations in multiple cutaneous and uterine leiomyomatosis, hereditary leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Hum. Mol. Genet., 12, 1241–1252. [DOI] [PubMed] [Google Scholar]
- 22.Mulligan L.M., Kwok J.B., Healey C.S., Elsdon M.J., Eng C., Gardner E., Love D.R., Mole S.E., Moore J.K., Papi L., et al. (1993) Germ-line mutations of the RET proto-oncogene in multiple endocrine neoplasia type 2A. Nature, 363, 458–460. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb E., Armour S.M., Harris M.H., Thompson C.B. (2003) Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ., 10, 709–717. [DOI] [PubMed] [Google Scholar]
- 24.Hung W.Y., Huang K.H., Wu C.W., Chi C.W., Kao H.L., Li A.F., Yin P.H., Lee H.C. (2012) Mitochondrial dysfunction promotes cell migration via reactive oxygen species-enhanced beta5-integrin expression in human gastric cancer SC-M1 cells. Biochim. Biophys. Acta, 1820, 1102–1110. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang S., Kinsey G.R., Yan Y., Han J., Schnellmann R.G. (2008) Extracellular signal-regulated kinase activation mediates mitochondrial dysfunction and necrosis induced by hydrogen peroxide in renal proximal tubular cells. J. Pharmacol. Exp. Ther., 325, 732–740. [DOI] [PubMed] [Google Scholar]
- 26.Yang C., Ko B., Hensley C.T., Jiang L., Wasti A.T., Kim J., Sudderth J., Calvaruso M.A., Lumata L., Mitsche M., et al. (2014) Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell, 56, 414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Moss T., Mangala L.S., Marini J., Zhao H., Wahlig S., Armaiz-Pena G., Jiang D., Achreja A., Win J., et al. (2014) Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol. Syst. Biol., 10, 728.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullen A.R., Wheaton W.W., Jin E.S., Chen P.H., Sullivan L.B., Cheng T., Yang Y., Linehan W.M., Chandel N.S., DeBerardinis R.J. (2012) Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature, 481, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiery J.P., Sleeman J.P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol., 7, 131–142. [DOI] [PubMed] [Google Scholar]
- 30.Roberts P.J., Der C.J. (2007) Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene, 26, 3291–3310. [DOI] [PubMed] [Google Scholar]
- 31.Vasko V., Espinosa A.V., Scouten W., He H., Auer H., Liyanarachchi S., Larin A., Savchenko V., Francis G.L., de la Chapelle A., et al. (2007) Gene expression and functional evidence of epithelial-to-mesenchymal transition in papillary thyroid carcinoma invasion. Proc. Natl. Acad. Sci. U. S. A, 104, 2803–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiseman S.M., Griffith O.L., Deen S., Rajput A., Masoudi H., Gilks B., Goldstein L., Gown A., Jones S.J. (2007) Identification of molecular markers altered during transformation of differentiated into anaplastic thyroid carcinoma. Arch. Surg., 142, 717–727. discussion 727-719. [DOI] [PubMed] [Google Scholar]
- 33.Moon S.Y., Kim H.S., Nho K.W., Jang Y.J., Lee S.K. (2014) Endoplasmic reticulum stress induces epithelial-mesenchymal transition through autophagy via activation of c-Src kinase. Nephron Exp. Nephrol., 126, 127–140. [DOI] [PubMed] [Google Scholar]
- 34.Eng C. (2000) Will the real Cowden syndrome please stand up: revised diagnostic criteria. J. Med. Genet., 37, 828–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. (2002) Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet., 30, 97–101. [DOI] [PubMed] [Google Scholar]
- 37.Yehia L., Niazi F., Ni Y., Ngeow J., Sankunny M., Liu Z., Wei W., Mester J.L., Keri R.A., Zhang B., et al. (2015) Germline Heterozygous Variants in SEC23B Are Associated with Cowden Syndrome and Enriched in Apparently Sporadic Thyroid Cancer. Am. J. Hum. Genet., 97, 661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K., Li M., Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res., 38, e164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capes-Davis A., Theodosopoulos G., Atkin I., Drexler H.G., Kohara A., MacLeod R.A., Masters J.R., Nakamura Y., Reid Y.A., Reddel R.R., et al. (2010) Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer, 127, 1–8. [DOI] [PubMed] [Google Scholar]
- 43.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform., 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.