Abstract
Nontuberculous mycobacteria (NTM) are ubiquitous and have been isolated from a variety of environmental sources, including water. Various NTM were isolated from biofilms in drinking water distribution systems in two urban and two semiurban areas in South Africa. Most of the isolates belonged to opportunistic pathogenic species of the NTM group, but none belonged to the Mycobacterium avium complex.
The genus Mycobacterium comprises both the strictly pathogenic species that are transmitted by human or animal reservoirs only (M. tuberculosis, M. leprae) and the so-called nontuberculous mycobacteria (NTM) (3, 13, 14). The NTM have generally been associated with soil and water, and while many of them are considered to be nonpathogenic, an increasing number are being reported as opportunistic pathogens (3, 5, 16). This growing number of atypical pathogenic mycobacteria includes M. abscessus (8), M. chelonae (10), M. fortuitum (16, 27), M. gordonae (19), M. mageritense (9), and M. xenopi (5). Several NTM constitute a risk not only to immunosuppressed persons but also to otherwise healthy persons (10). They can cause pulmonary and cutaneous diseases, lymphadenitis, and other infections (14).
M. abscessus, M. gilvum, M. gordonae, and M. mageritense have been associated with municipal water supplies (7, 9, 12). In a recent report, furunculosis, caused by M. mageritense, was also linked to the water supply of a salon where two women received footbaths (9). In another report, cervical lymphadenitis in children below 2 years of age has been linked with mycobacteria in the United States, the United Kingdom, and Australia (21). These infections were linked to the prevalence of M. avium and M. scrofulaceum in water (21). There are also reports that NTM can be present in aerosols, such as at swimming pools and spas, of water that may contain mycobacteria and that individuals exposed to the aerosols for extended periods are more at risk of contracting an infection (6). In the United States alone, over a million workers are exposed to aerosols generated by metal grinding, and exposure to such aerosols can lead to hypersensitivity, pneumonitis, and chronic obstructive pulmonary disease (6, 21).
NTM are tolerant to a much wider pH and temperature range than are most other bacterial pathogens detected in municipal water supplies. They are also generally tolerant to chlorine, making them potentially more difficult to eliminate (12, 13). Adding to this is their ability to form biofilms on surfaces in drinking water distribution systems (12, 24). The growth of NTM in biofilms may lead to dissemination into the bulk water, constituting a risk to consumers both by drinking and by inhalation of aerosols though showering and swimming.
NTM can form biofilms under low-nutrient conditions, making surfaces of drinking water distribution systems an environment for their growth and possible dissemination (12). The aim of this study was to determine the presence and diversity of NTM in biofilms in drinking water distribution systems by analyzing samples from various points in urban and semiurban areas.
Seventy-eight samples were collected from two well-serviced urban areas (Pretoria [31 samples] and Pietermaritzburg [20 samples]), a semiurban developing community (Botshabelo [5 samples]), and other towns with small distribution networks (22 samples) in South Africa. Water from the two urban areas was chloraminated, while the rest was chlorinated. Samples were collected during the period from September 2001 to August 2002. Prior to collection, water was allowed to run to waste at a uniform rate for 2 to 3 min. Water samples were collected in sterile bottles containing sodium thiosulfate to a final concentration of 0.01% (wt/vol) to neutralize any free or combined residual chlorine (2). Following removal of the tap, biofilm samples were taken from the inner surface of the service pipe using a sterile cotton-tipped swab moistened in 1 ml of sterile one-quarter-strength Ringer's solution (Merck). The area of biofilm removed in this way was assumed to be approximately 1 cm2. The swab was then returned to the Ringer's solution tube. All of the samples were transported to the laboratory on ice and analyzed within 12 to 18 h. Water from the systems was tested for the presence of fecal coliforms by filtering duplicate 100-ml volumes through 0.45-μm-pore-size nitrocellulose filters (Millipore) and incubating them on mFC agar (Merck) at 44.5°C for 22 to 24 h. Biofilm samples were dispersed by vigorous vortexing in sterile diluent. The heterotrophic culturable count was determined for both the water and biofilm phases of the samples by plating serial dilutions (10−1 to 10−5 in one-quarter-strength Ringer's solution) onto R2A agar (Oxoid) and incubating them at 28°C for 5 days.
NTM in biofilm samples were enumerated by a three-tube most probable number technique. Cetylpyridinium chloride (Sigma) was added to triplicate decimal dilutions of biofilm suspensions to a final concentration of 0.005% (wt/vol). The tubes were incubated at room temperature (25 ± 2°C) for 30 min before 100-μl aliquots were plated onto Middlebrook 7H10 agar (Difco) supplemented with 0.5% glycerol and 10% oleic acid-albumin. Plates were incubated for 21 days before being scored as negative (7). Isolates obtained were restreaked onto Middlebrook agar, and their DNA was extracted with a DNeasy tissue kit (QIAGEN).
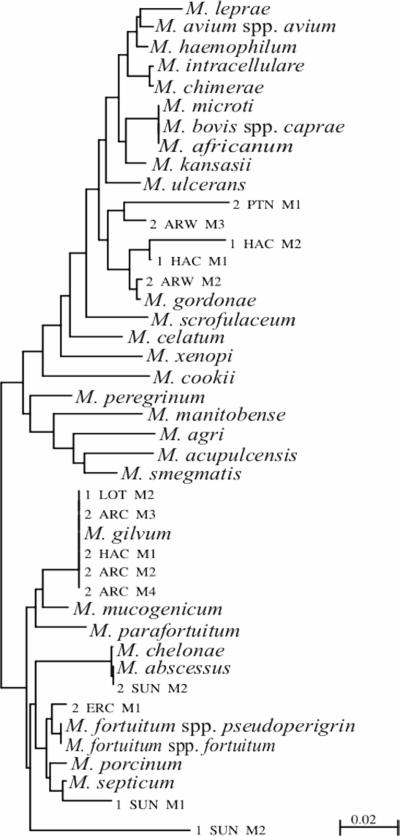
The 16S rRNA genes of isolates were amplified by PCR using both universal primer sets 63f and 1387r (17) and fD1 and rP2 (26). PCR products were purified with a QIAquick PCR purification kit (QIAGEN). The nucleotide sequences were determined by sequencing on an ABI PRISM 377 or ABI Prism 3100 sequencer (Perkin Elmer) using the BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems). Sequences were edited by Sequence Navigator (Applied Biosystems), and the relationship to known bacteria was determined by searching known sequences in GenBank using a basic BLAST search of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences reduced to approximately 400-bp-long fragments corresponding to positions 170 to 566 of the Escherichia coli 16S rRNA genes (1, 11) were used to draw a phylogram with Clustal X (version 1.81) (25) and viewed with TreeView (Win 32) and NJPlot (Win 95) (20).
The heterotrophic plate counts for the analysis of the water samples ranged from 1.0 × 101 to 3 × 108 CFU · ml−1 (Table 1), indicating a wide range of water quality. Plate counts of biofilm samples ranged from 3 × 101 to 2 × 109 CFU · cm−2 (Table 1), indicating that biofilms were present in all of the drinking water systems tested. Fourteen out of the 78 sites sampled, 11 of which were from Pretoria, tested positive for NTM with Middlebrook 7H10 agar. These numbers could have been even higher if a different decontamination method had been used or if samples had been incubated for a longer period (18). Fourteen NTM isolates were identified by 16S rRNA gene sequencing and were found to be M. gordonae (five isolates), M. gilvum (five isolates), M. abscessus (one isolate), M. fortuitum (one isolate), M. septicum (one isolate), and one isolate that did not cluster with known species (Fig. 1).
TABLE 1.
Bacterial water quality data for samples taken from of the distribution networks of urban and semiurban areas
| Bacteria | Measurement | Value for:
|
|||
|---|---|---|---|---|---|
| Pretoria (31)a | Pietermaritzburg (20) | Botshabelo (5) | Towns (22) | ||
| Heterotrophic plate counts | |||||
| In water (CFU · ml−1) | Minimum | 10 | NDb | ND | ND |
| Maximum | 3 × 108 | 3 × 108 | 3 × 108 | 3 × 108 | |
| Geometric mean | 1.2 × 104 | 1.7 × 104 | 4 × 106 | 1.8 × 104 | |
| In biofilm (CFU · cm−2) | Minimum | 30 | 30 | 3 × 108 | 7,400 |
| Maximum | 3 × 108 | 2 × 109 | 3 × 108 | 6 × 107 | |
| Geometric mean | 3.2 × 105 | 1.4 × 105 | 3 × 108 | 8.6 × 105 | |
| NTM (CFU · cm−2) | Minimum | ND | ND | ND | ND |
| Maximum | 2,300 | 9,300 | ND | 4.6 × 105 | |
| Geometric mean | 3 | 0 | ND | 0 | |
| Fecal coliforms (CFU · 100 ml−1) | Minimum | ND | ND | ND | ND |
| Maximum | ND | 163 | ND | 3 | |
| Geometric mean | ND | 0 | ND | 0 | |
Number of sites sampled is given in parentheses.
ND, not detected.
FIG. 1.
Rooted phylogram after phylogenetic analysis of a 400-bp-long fragment of the 16S rRNA gene sequences. Sequences from databases are indicated by their full taxonomic name, while sequences from isolates in this study are indicated by a code.
No isolates appeared to belong to the M. avium complex, but most of the NTM isolated did belong to species with documented pathogenicity. M. gordonae may cause nosocomial pulmonary and systemic infections in the elderly (22) and the immunosuppressed, as well as skin lesions (15). It has been reported from diverse environments, including ice machines (19). M. fortuitum caused intractable pulmonary disease following repeated exposure to aerosols from a contaminated hot tub, as well as osteomyelitis, cellulitis, and wound infections (16, 18). M. abscessus has been the cause of abscesses due to injection with medication (8), and M. septicum has been associated with central line sepsis in immunosuppressed and immunocompetent individuals (23). M. gilvum, also detected in this study, has, however, not been linked to any clinical infection to date.
Fecal coliforms (Table 1) were detected in 6 out of 78 (7.7%) samples tested, occurring mainly in the small towns fed by boreholes (four samples) and from Pietermaritzburg (two samples). Neither fecal coliforms nor high heterotrophic counts were associated with high levels of NTM, showing that normal microbial water quality parameters do not provide any indication of the possible presence of NTM in distribution networks and the potential risk to consumers.
The findings of this study are in agreement with those conducted by Dailloux et al. (4), Falkinham et al. (7), and Le Dantec et al. (13, 14), who also found that NTM occur in biofilms in drinking water distribution systems. We would thus recommend that water authorities include sporadic testing for this group of bacteria, especially in countries or areas with a high prevalence of immunocompromised persons.
Nucleotide sequence accession numbers.
Accession numbers for the partial 16S rRNA genes of the new isolates are AY624031 to AY624044.
Acknowledgments
We thank P. Jagals and I. Bailey for supplying biofilm and drinking water samples from Botshabelo and Pietermaritzburg, respectively, and F. A. Els for processing some of the samples from Pretoria and the small towns.
Footnotes
Journal series publication 3434 from the South Dakota Agricultural Experiment Station.
REFERENCES
- 1.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1999. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, D.C.
- 3.Collins, C. H., J. M. Grange, and M. D. Yates. 1984. Mycobacteria in water. J. Appl. Bacteriol. 57:193-211. [DOI] [PubMed] [Google Scholar]
- 4.Dailloux, M., M. Albert, C. Laurain, S. Andolfatto, A. Lozniewski, P. Hartemann, and L. Mathieu. 2003. Mycobacterium xenopi and drinking water biofilms. Appl. Environ. Microbiol. 69:6946-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emmerson, A. M. 2001. Emerging waterborne infections in health-care settings. Emerg. Infect. Dis. 7:272-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falkinham, J. O., III. 2003. Mycobacterial aerosols and respiratory disease. Emerg. Infect. Dis. 9:763-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falkinham, J. O., III, C. D. Norton, and M. W. LeChevalier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galil, K., L. A. Miller, M. A. Yakrus, R. J. Wallace, D. G. Mosley, B. England, G. Huitt, M. M. McNeil, and B. A. Perkins. 1999. Abscesses due to Mycobacterium abscessus linked to injection of unapproved alternative medicine. Emerg. Infect. Dis. 5:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gira, A. K., A. H. Reisenauer, L. Hammock, U. Nadiminti, J. T. Macy, A. Reeves, C. Burnett, M. A. Yakrus, S. Toney, B. J. Jensen, H. M. Blumberg, S. W. Caughman, and F. S. Nolte. 2004. Furunculosis due to Mycobacterium mageritense associated with footbaths at a nail salon. J. Clin. Microbiol. 42:1813-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, D. R. 2002. Hot tub-associated mycobacterial infections in immunosuppressed persons. Emerg. Infect. Dis. 8:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutell, R. R., N. Larson, and C. R. Woese. 1994. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 58:10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall-Stoodley, L., C. W. Keevil, and H. M. Lappin-Scott. 1999. Mycobacterium fortuitum and Mycobacterium chelonae biofilm formation under high and low nutrient conditions. J. Appl. Microbiol. 85:60S-69S. [DOI] [PubMed] [Google Scholar]
- 13.Le Dantec, C., J.-P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Appl. Environ. Microbiol. 68:1025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Dantec, C., J.-P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl. Environ. Microbiol. 68:5318-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, X. J., Q. X. Wu, and X. S. Zeng. 2003. Nontuberculous mycobacterial cutaneous infection confirmed by biochemical tests, polymerase chain reaction-restriction fragment length polymorphism analysis and sequencing of hsp65 gene. Br. J. Dermatol. 149:642-646. [DOI] [PubMed] [Google Scholar]
- 16.Mangione, E. J., G. Huitt, D. Lenaway, J. Beebe, A. Bailey, M. Figoski, M. P. Rau, K. D. Albrecht, and M. A. Yakrus. 2001. Nontuberculous mycobacterial disease following hot tub exposure. Emerg. Infect. Dis. 7:1039-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchesi, J. R., N. J. Russell, G. F. White, and W. A. House. 1998. Effects of surfactant adsorption and biodegradability on the distribution of bacteria between sediments and water in a freshwater microcosm. Appl. Environ. Microbiol. 57:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metchock, B. G., F. S. Nolte, and R. J. Wallace, Jr. 1999. Mycobacterium, p. 399-458. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 19.Panwalker, A. P., and E. Fuhse. 1986. Nosocomial Mycobacterium gordonae pseudo infection from contaminated ice machines. Infect. Control 7:67-70. [DOI] [PubMed] [Google Scholar]
- 20.Perrière, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 21.Primm, T. P., C. A. Lucero, and J. O. Falkinham III. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 17:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeki, S., H. Matsuse, T. Shimoda, Y. Soejima, H. Ohno, and S. Kohno. 2004. A case of pulmonary Mycobacterium gordonae infection with pleural effusion. Nihon Kokyuki. Gakkai Zasshi 42:103-107. [PubMed] [Google Scholar]
- 23.Schinsky, M. F., M. M. McNeil, A. M. Whitney, A. G. Steigerwalt, B. A. Lasker, M. M. Floyd, G. G. Hogg, D. J. Brenner, and J. M. Brown. 2000. Mycobacterium septicum sp. nov., a new rapidly growing species associated with catheter-related bacteraemia. Int. J. Syst. Evol. Microbiol. 50:575-581. [DOI] [PubMed] [Google Scholar]
- 24.Szewzyk, U., R. Szewzyk, W. Manz, and K.-H. Schleifer. 2000. Microbiological safety of drinking water. Annu. Rev. Microbiol. 54:81-127. [DOI] [PubMed] [Google Scholar]
- 25.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winthrop, K. L., M. Abrams, M. Yakrus, I. Schwartz, J. Ely, D. Gillies, and D. J. Vugia. 2002. An outbreak of mycobacterial furunculosis associated with footbaths at a nail salon. N. Engl. J. Med. 346:1366-1371. [DOI] [PubMed] [Google Scholar]