Abstract
A flow cytometry method was developed for rapid screening and recovery of cloned DNA containing common sequence motifs. This approach, termed fluorescence-activated cell sorting-assisted cloning, was used to recover sequences affiliated with a unique lineage within the Bacteroidetes not abundant in a clone library of environmental 16S rRNA genes.
Retrieval and sequence analysis of phylogenetically informative genes has become a standard cultivation-independent technique to investigate microbial diversity in nature (7, 18). Genes encoding the 16S rRNA, because of the relative ease of their selective amplification, have been most frequently employed for general diversity surveys (16). Environmental studies have also focused on specific subpopulations affiliated with a phylogenetic group or identified by genes encoding specific metabolic functions (e.g., ammonia oxidation, sulfate respiration, and nitrate reduction) (8, 15, 20). However, specific populations may be of low abundance (1, 23), or the genes encoding specific metabolic functions may be insufficiently conserved to provide priming sites for general PCR amplification.
Three general approaches have been used to obtain 16S rRNA sequence information from low-abundance populations: screening hundreds to thousands of clones in a general 16S rRNA gene library (21), flow cytometric sorting of a subpopulation of environmentally derived cells labeled by fluorescent in situ hybridization (FISH) (27), or selective PCR amplification using primers specific for the subpopulation (2, 23). While the first approach is simply time-consuming and tedious, the second has been restricted to fairly large and strongly fluorescent cells from aquatic samples (5, 27). The third approach often generates fragments of only a few hundred bases due to the limited number of specific priming sites. Partial sequence information often degrades analysis, obscuring or distorting the phylogenetic placement of the new sequences (11, 20).
A more robust characterization of environmental microbiota would be enabled by a method to retrieve near-full-length copies of selected genes from rare populations, not restricted to the analysis of genes encoding the rRNAs. We recently demonstrated the use of FISH of 16S rRNA gene clones (clone-FISH) to screen an environmental clone library using microscopy (24). A similar protocol has been more recently published by Ouverney et al. (17). In this study, we evaluated the combination of clone-FISH and flow cytometry to screen and sort a 16S rRNA gene clone library (termed fluorescence-activated cell sorting-assisted cloning [FACS-cloning]). A probe targeting most of the Cytophaga-Flavobacterium cluster of the phylum Bacteroidetes was used to recover near-full-length 16S rRNA gene sequences affiliated with this group not abundant in the original clone library.
Flow cytometric clone sorting.
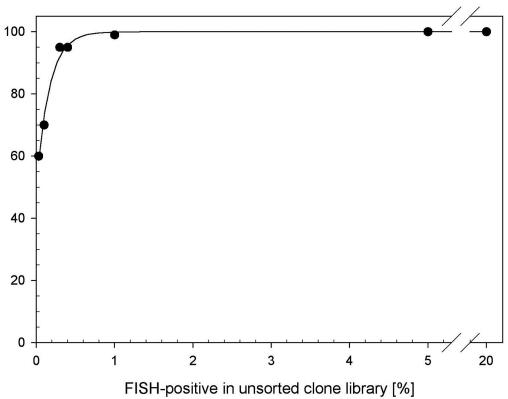
Sorting efficiency was initially evaluated with defined mixtures of plasmid-containing Escherichia coli (with and without a cloned 16S rRNA sequence). The T7 RNA polymerase promoter-containing vector pET-23(+) (Novagen, Madison, Wis.) and E. coli NovaBlue (DE3) were used to clone a 16S rRNA gene sequence from Paenibacillus polymyxa (24). A clone containing the insert in reverse orientation, yielding antisense rRNA with T7-driven transcription, was labeled (clone-FISH) as previously described (24) and sorted by flow cytometry by established methods (4). In brief, 16 μl of paraformaldehyde-fixed clones (concentration of approximately 106 μl−1) was hybridized for 2 h at 46°C in 320 μl of standard FISH buffer (19) containing 35% formamide and 2.5 ng of Cy3-labeled probe NON338 μl−1 (QIAGEN Operon, Alameda, Calif.) (13); this probe targets the in vivo-transcribed antisense rRNA of the P. polymyxa clones. Clones were pelleted by centrifugation (4,000 × g, 4 min), resuspended in 400 μl of prewarmed (46°C) hybridization buffer without probe, washed for 20 min, diluted with 800 μl of phosphate-buffered saline (pH 7.4), and placed immediately on ice. After counterstaining for 15 min with 4′,6′-diamidino-2-phenylindole (DAPI; final concentration, 1.5 μg ml−1), clones were sorted with a custom-built, high-speed cell sorter (Institute for Systems Biology, Seattle, Wash.). A UV laser line (40 mW) in combination with emission filters LP418 and BP440/80 was used for the detection of DAPI-stained cells, while a 514-nm laser line (850 mW) in combination with emission filter BP575/20 was used for Cy3-labeled cells and forward scatter. The sheath fluid was NaCl (0.9%). Sorting was calibrated with 1-μm-diameter multicolor beads (Polysciences, Warrington, Pa.), and the sorting speed was 10,000 events s−1. Sorted cells were collected in PCR tubes, an aliquot was transferred onto a microscopic slide, and FISH-positive cells (i.e., sorted target clones) were enumerated relative to all (DAPI stained) cells with an epifluorescence microscope (LSM5; Carl Zeiss, Jena, Germany). Sorting efficiency was evaluated by counting a minimum of 1,000 cells in 10 randomly chosen microscopic fields. Almost pure sorts (≥95%) were obtained with an initial target clone concentration of 0.5% or higher (Fig. 1). Lower initial concentrations yielded significantly less pure sorts. However, even very rare clones (0.01%) could be significantly enriched (ca. 60%). We anticipate that repeated sorting could achieve near pure recovery of even extremely rare clone populations, especially as cell aggregates, which frequently hamper sorting of natural populations to high purity (27), are absent in clone suspensions.
FIG. 1.
Recovery of single clones containing a 16S rRNA gene insert in the sense orientation from a defined mixture of cells containing a plasmid vector with or without the 16S rRNA gene insert. Clones were hybridized with the Cy3-labeled oligonucleotide probe NON338 and sorted by flow cytometry.
PCR amplification of sorted clones.
Sorted clones were prepared for PCR amplification as described previously (27). The sorted cells were first washed in 200 μl of sterile distilled water and recovered by centrifugation (10,000 × g, 10 min) to remove sheath fluid-derived NaCl previously shown to inhibit PCR (data not shown). After three freeze-thaw cycles, the sorted and washed cells were used directly for standard PCR reamplification of the insert with vector-specific T7 promoter and terminator primers (Novagen). A minimum of 20,000 cells was required to reproducibly obtain detectable amplification products from paraformaldehyde-fixed cells. This number is similar to those reported for reamplification of fixed bacterial cells following sorting of cells labeled by FISH of native rRNA (27). The requirement for a relatively high initial number of fixed cells has been attributed to incomplete cell lysis following paraformaldehyde fixation and general inhibitory effects associated with fixation (27). We therefore evaluated an alternative fixation using 50% ethanol, as described by Roller et al. (22). In contrast to paraformaldehyde, ethanol does not cross-link cellular components and should serve for more efficient cell lysis and recovery of unmodified DNA. Consistent with this expectation, 500 sorted ethanol-fixed cells were sufficient for reproducible PCR amplification following three freeze-thaw cycles. Ethanol fixation of clones was therefore used for all further sorting experiments. However, ethanol-fixed clones are not suitable for long-term storage and should be used within 2 to 3 weeks to avoid loss of FISH signal.
Environmental application.
FACS-cloning was then used to recover clones affiliated with the CF319a-targeted group of the Bacteroidetes from a 16S rRNA gene library developed from an estuarine sediment using PCR primers designed for general amplification of the Bacteria. Members of the Cytophaga-Flavobacterium cluster of the phylum Bacteroidetes are considered important for the degradation of high-molecular-weight dissolved organic matter in aquatic environments and are often the most abundant bacterial group in many oceanic and freshwater habitats (reviewed in reference 9). Despite a high in situ abundance suggested by 16S rRNA-targeted FISH analyses, members of the Cytophaga-Flavobacterium cluster of the phylum Bacteroidetes are commonly underrepresented in 16S rRNA gene clone libraries. Thus, several hundreds of clones often must be screened to obtain a few representative sequences of this group (9), rendering it an interesting target for flow cytometry-assisted cloning.
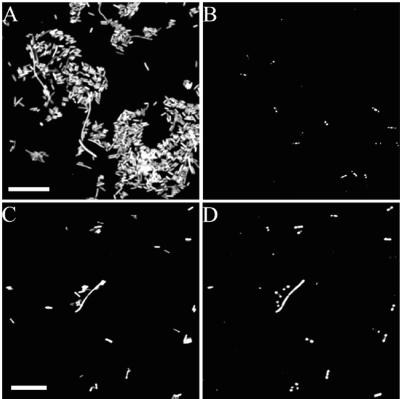
Sediment cores were collected during low tide in April 2001 from a high-salinity site (22 ppt) at the Rowley River estuary in Plum Island Sound, Mass. (6). DNA was extracted from 0.5 g of sediment (0- to 2-cm depth) by using the Fast DNA kit for soil (Bio101, Qbiogene, Carlsbad, Calif.) with 0.5 g of 0.1-mm-diameter zirconium beads instead of the bead matrix supplied with the kit. Near-full-length bacterial 16S rRNA genes were PCR amplified with the primer pair GM3 (E. coli positions 8f/1392r) (10, 14), and a clone library was constructed with the pCRII-TOPO kit and TOPO 10 One Shot competent cells (Invitrogen) following the manufacturer's instructions. A second library was constructed by ligation into vector pGEM-T (Promega), transformation into E. coli NovaBlue(DE3) (Novagen), and transcription of the plasmid insert according to the standard protocol of Schramm et al. (24). Subsequently, clones were fixed by the addition of 96% ice-cold ethanol (final concentration, 50%). The relative abundance of clones affiliated with the CF319a-targeted group of the Cytophaga-Flavobacterium cluster of the phylum Bacteroidetes in the library was determined microscopically as 1.3% ± 0.9% (mean ± standard error) by standard FISH (19) with the Cy3-labeled probe CF319a (12) (Fig. 2A). Assuming a random distribution of insert direction (i.e., only 50% of the amplified 16S rRNA genes were inserted in the forward direction and thus detectable upon in vivo transcription), the number of Cytophaga-Flavobacterium-like clones would be about twice this number. A more accurate enumeration of insert direction could be derived with sense and antisense probes to the transcripts. The library was counterstained by hybridizing with the fluorescein-labeled probe GAM42a, targeting the 23S rRNA of γ-Proteobacteria (13), serving to label all E. coli cells. Sorting for CF319a/Cy3-positive clones and reamplification of the plasmid inserts from approximately 500 sorted clones were performed as described above but with pUC/M13 primers (Promega). Microscopic examination of the sorted clones revealed that the CF319a-positive clones had been enriched to approximately 98% purity (Fig. 2B to D). Reamplified PCR products were gel purified with a commercial kit (QIAGEN Inc., Valencia, Calif.), ligated into pCRII-TOPO plasmids (Invitrogen, Carlsbad, Calif.), and transformed into TOPO 10 One Shot competent cells (Invitrogen) as recommended by the respective manufacturers. Of these clones, 96 with inserts were further examined by semispecific PCR, using probes CF319a and pUC/M13 forward (Promega) as primers. Among these clones, 78 yielded a PCR product, indicating that approximately 80% of the sorted cells contained the CF319 target site. Deviation from the microscopic observation of 98% purity may derive from lack of selectivity at one or more steps in this protocol. For example, the established conditions for in situ hybridization with CF319a probe may not resolve target from closely related nontarget sequences. As briefly discussed below, sequence analysis suggests that nontarget hybridization contributed to the recovery of sequences containing a common mismatch composition in the target region of this probe. Also, ethanol fixation, relative to paraformaldehyde fixation, may result in greater release of plasmid DNA from lysed cells. Contamination of the sheath solution in the sorted cell pool with plasmid DNA may also contribute to loss of selectivity. Whether this contamination problem is more severe with ethanol-fixed, versus paraformaldehyde-fixed, clones must be further evaluated.
FIG. 2.
Micrographs showing the results of hybridization of the GAM42a probe (A and C) and CF319a (B and D) to clones before and after sorting by flow cytometry. Identical microscopic fields are shown for FISH of the unsorted library (A and B) and the sorted clones (C and D). Scale bars = 20 μm.
Sequences of clones picked randomly from the unsorted and the sorted clone library (29 each) were determined with a MegaBACE 1000 automated sequencer (Amersham Biosciences, Piscataway, N.J.) according to the manufacturer's instructions (GenBank accession no. AY678478 to AY678530). Sequences were aligned with the Fast Aligner program in ARB (www.arb-home.de), and all alignments were checked manually, using the secondary structure to help resolve regions of ambiguity. Phylogenetic relationships were analyzed by evolutionary distance and parsimony methods with the ARB software package. Evolutionary distances were calculated with the Kimura two-parameter distance correction. Regions of ambiguous alignment were not included in the analysis. Confidence estimates of branching order were determined in PAUP* 4.0 (25) by randomly resampling the sequences 100 times (bootstrapping) (3). Short sequences from the unsorted clone library were added to the final tree with the parsimony tool in ARB.
Sequence analysis of the FACS-clone library revealed that 12 of 29 clones originated from cells associated with the Cytophaga-Flavobacterium group of the Bacteroidetes. All sequences affiliated with the Cytophaga-Flavobacterium group had a perfect match to the CF319a probe and clustered in two groups within the Bacteroidetes (Fig. 3). None of these sequences was recovered in the screen of the unsorted library, supporting the utility of this method to recover low-abundance clone types. The remaining 17 clones shared a common sequence variant in the CF319 probe target sequence, containing mismatches at positions 2 (T/G) and 15 (A/C), and were distributed phylogenetically between two groups of closely related sequences (99.5% intragroup similarity). One group was affiliated with the δ-Proteobacteria (10 clones), and the other was affiliated with the β-Proteobacteria (7 clones).
FIG. 3.
Phylogenetic relationships of 16S rRNA sequences from the clone library sorted by the CF319a probe (underlined and bold with CF319a prefix) and the unsorted clone library (bold with R8C prefix). The tree was inferred by the neighbor-joining algorithm and is based on 969 nucleotide positions. Aquifex pyrophilus was used as the outgroup (not shown). Nodes supported by bootstrap values greater than 90% by neighbor-joining (above the node) and parsimony (below the node) analyses are indicated by a filled circle. Open circles indicate nodes supported by greater than 50%. The scale bar represents 0.10 substitution per nucleotide position. Dotted lines indicate short sequences added by the parsimony tool in ARB. Sequences that have perfect matches to the CF319a probe sequence are denoted by asterisks.
The unsorted clones revealed a high sequence diversity in the original sample, with 4 out of 29 sequences affiliated with the Cytophaga-Flavobacterium group of the Bacteroidetes (Fig. 3). One of 29 sequences (ca. 3%) in the unsorted library had a perfect match to the CF319a probe but was not identified in our limited survey of the sorted library. Three had either one or three mismatches. None of these mismatch variants was identified by FACS-cloning. Only the double-mismatch sequence type was encountered in the sorted population. High selectivity for this mismatch variant strongly suggested that a stable duplex could be formed between this target sequence and the CF319a probe.
The validation of probes designed for FISH generally includes an empirical demonstration of discrimination between target and nontarget mismatch variants. However, although it is well recognized that mismatch discrimination is determined by both type and position of mismatch, it is not feasible to evaluate all permutations (26). The CF319a probe apparently failed to discriminate between the two-mismatch variant and the complementary target. Although we used hybridization conditions slightly less stringent than those reported by Manz et al. (12), 46 versus 48°C, it is likely that more comprehensive analyses of hybridization to nontarget sequences would reveal considerable variation in discrimination among different population types. Thus, we anticipate that FACS-cloning could also serve to confirm analyses of complex microbial communities based on FISH.
Conclusions.
We have demonstrated the utility of flow cytometry-assisted clone sorting after FISH (FACS-cloning) to retrieve rare clones (1% abundance) from an environmental 16S rRNA gene clone library. With a sorting speed of 40,000 events s−1 and continued optimization of hybridization stringency and cell fixation, we anticipate that the technique will allow for the “fishing” of extremely rare genes from environmental libraries. FACS-cloning should also be applicable for screening shotgun libraries and searching for functional genes, thus avoiding the limitations and biases associated with conventional, PCR-based methods.
Acknowledgments
We thank Paul Framson and Tim Petersen for technical assistance with flow cytometry.
Funding for this study was obtained by NSF grants (DEB-997897 and DEB0213186) to D.A.S., the Lawrence Berkeley National Laboratory Genomes to Life Project sponsored by the office of Biological and Environmental Research of the U.S. Department of Energy Office of Science (DOE DE-AC03-76SF00098), and an NSF postdoctoral fellowship to A.E.B. A.S. was financially supported by an Otto-Hahn-Award of the Max Planck Society, Germany, and J.L.N. was supported by the Danish-American Fullbright Commission and the Danish Research Council.
REFERENCES
- 1.Altmann, D., P. Stief, R. Amann, D. de Beer, and A. Schramm. 2003. In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ. Microbiol. 5:798-803. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs, B. M., M. V. Zubkov, K. Sahm, P. H. Burkill, and R. Amann. 2000. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2:191-201. [DOI] [PubMed] [Google Scholar]
- 6.Hopkinson, C. S. J., A. E. Giblin, J. Tucker, and R. H. Garritt. 1999. Benthic metabolism and nutrient cycling along an estuarine salinity gradient. Estuaries 22:863-881. [Google Scholar]
- 7.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joulian, C., N. B. Ramsing, and K. Ingvorsen. 2001. Congruent phylogenies of most common small-subunit rRNA and dissimilatory sulfite reductase gene sequences retrieved from estuarine sediments. Appl. Environ. Microbiol. 67:3314-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 10.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 11.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K.-H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 12.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 13.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 14.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 15.Nogales, B., K. N. Timmis, D. B. Nedwell, and A. M. Osborn. 2002. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 17.Ouverney, C. C., G. C. Armitage, and D. A. Relman. 2003. Single-cell enumeration of an uncultivated TM7 subgroup in the human subgingival crevice. Appl. Environ. Microbiol. 69:6294-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 19.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 20.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmidt, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 23.Scheid, D., and S. Stubner. 2001. Structure and diversity of Gram-negative sulfate-reducing bacteria on rice roots. FEMS Microbiol. Ecol. 36:175-183. [DOI] [PubMed] [Google Scholar]
- 24.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 25.Swofford, D. L. 2002. PAUP: phylogenetic analysis using parsimony and other methods, 4th ed. Sinauer Associates, Sunderland, Mass.
- 26.Urakawa, H. S., S. El Fantroussi, E. H. Tribou, H. Smidt, J. C. Smoot, J. J. Kelly, P. A. Noble, and D. A. Stahl. 2003. Optimization of single-base-pair mismatch discrimination on oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallner, G., B. Fuchs, S. Spring, W. Beisker, and R. Amann. 1997. Flow sorting of microorganisms for molecular analysis. Appl. Environ. Microbiol. 63:4223-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]