Abstract
Accumulating evidences indicated that UCA1 expression was up-regulated in various cancers, and high UCA1 expression was correlated with metastasis and prognosis. This meta-analysis collected all eligible studies and explored the relationships between UCA1 expression and lymph node metastasis (LNM) or overall survival (OS). Literature collection was performed by using electronic databases PubMed, Cochrane Library, and Web of Science (up to June 13, 2016). According to the inclusion and exclusion criteria, twelve studies were included in the meta-analysis. The result showed that high UCA1 expression was correlated with more LNM (OR=2.50, 95 %CI: 1.58-3.96, p<0.0001) in a random-effects model (I2=45 %, p=0.08) and could predict poor OS in cancer patients, with pooled hazard ratio (HR) of 1.65 [95% confidence interval (CI) 1.44-1.88, p<0.00001] indicated by a fixed-effects model (I2=35%, p=0.11). In conclusion, the present meta-analysis demonstrated that high expression of UCA1 might serve as a common molecular marker for predicting lymph node metastasis and prognosis in various cancers.
Keywords: UCA1, lncRNA, prognosis, survival, lymph node metastasis
INTRODUCTION
With increasing incidence and mortality in each year, cancer is becoming the leading cause of death worldwide and is a major public health problem. In the United States, there will be 1,685,210 new cancer cases and 595,690 cancer deaths in 2016 [1]. For patients in early stage of cancers without lymph node metastasis or distant metastasis, some treatments are available, such as surgery, chemotherapy and radiation therapy. However, in patients with lymph node metastasis or distant metastasis, there are no available therapies. Therefore, finding a common molecular cancer marker for predicting lymph node metastasis and prognosis is indispensable for observing the progression of cancers.
With the rapid development of second-generation sequencing technology, lots of long noncoding RNAs (lncRNAs) have been found to be dysregulated in expression and involved in the development of various human diseases, particularly in cancers [2]. Accumulating evidences reveal that lncRNAs play vital regulatory roles in diverse cellular processes, such as regulation of gene expression, posttranslational processing and tumorigenesis [3]. Urothelial Carcinoma Associated 1 (UCA1), a 2314-bp lncRNA encoded on human chromosome 19, was firstly identified in bladder cancer [4]. A large amount of studies have demonstrated that UCA1 expression was up-regulated in many cancers, such as hepatocellular cancer, colorectal cancer, gastric cancer, esophageal squamous cell carcinoma and lung cancer, and that high UCA1 expression was strongly correlated with clinicopathologic characteristics, including lymph node metastasis and overall survival [5–10]. According to these findings, we carried out this meta-analysis to explore the relationships between UCA1 expression and lymph node metastasis or overall survival, and to evaluate whether UCA1 could serve as a common molecular marker for LNM and OS.
RESULTS
Characteristics of eligible studies
As shown in the flow diagram (Figure 1), a total of 126 published articles were revealed in our initial search by using the keywords. After screening the title and abstract carefully, 111 articles were excluded. After further inspection of the full articles, 2 were excluded because there is no available information in their records. In the 13 potential candidate studies, 1 study was excluded because of inadequate data of HR with 95 % CI. Finally, according to the criteria for selection, a total of 12 studies were eligible [5–16].
Figure 1. The flow diagram of this meta-analysis.
These studies included a total of 986 patients. The mean patient sample size was 82.17 (range from 40 to 117). Among the twelve studies, one focused on esophageal squamous cell carcinoma, one on non-small cell lung cancer, one on lung cancer, one on colon cancer, one on gastric cancer, one on hepatocellular carcinoma, one on prostate cancer, two on ovarian cancer and three on colorectal cancer. All the diagnoses of lymph node metastasis were based on pathology. In all of the studies, the patients were divided into two groups: high and low expression of UCA1. All studies used qRT-PCR to detect UCA1 expression. The main characteristics of the eligible studies were summarized in Table 1 and Table 2.
Table 1. Characteristics of studies in this meta-analysis.
| Author | Year | Country | Tumor type | Sample size(n) | UCA1 expression | Method | |||
|---|---|---|---|---|---|---|---|---|---|
| High expression | High with LNM | Low expression | Low with LNM | ||||||
| Li | 2014 | China | ESCC | 90 | 41 | 22 | 49 | 12 | RT-qPCR |
| Nie | 2016 | China | NSCLC | 112 | 39 | 14 | 73 | 21 | RT-qPCR |
| Wang | 2015 | China | LC | 60 | 36 | 26 | 24 | 8 | RT-qPCR |
| Zhang | 2016 | China | OC | 110 | 57 | 26 | 53 | 12 | RT-qPCR |
| Yang | 2016 | China | OC | 53 | 27 | 13 | 26 | 5 | RT-qPCR |
| Ni | 2015 | China | CRC | 54 | 27 | 12 | 27 | 5 | RT-qPCR |
| Tao | 2015 | China | CC | 80 | 20 | 13 | 60 | 21 | RT-qPCR |
| Zheng | 2015 | China | GC | 112 | 56 | 35 | 56 | 37 | RT-qPCR |
ESCC esophageal squamous cell carcinoma, NSCLC non-small cell lung cancer, LC lung cancer, OC Ovarian cancer, CRC colorectal cancer, CC colon cancer, GC gastric cancer, qRT-PCR quantitative real-time PCR, LNM lymph node metastasis.
Table 2. Overall survival of the eligible studies in this meta-analysis.
| Study | Year | Disease | Number | MALAT1 assay | Survival analysis | Multivariate analysis | HR statistic | Hazard ratios(95% CI) | Follow-up, moths |
|---|---|---|---|---|---|---|---|---|---|
| Zheng | 2015 | GC | 112 | RT-qPCR | OS | Yes | Data in paper | 2.351(1.222-4.521) | 60 |
| Zhang | 2016 | OC | 117 | RT-qPCR | OS | Yes | Data in paper | 1.688(1.005-2.834) | >60 |
| Yang | 2016 | OC | 53 | RT-qPCR | OS | Yes | Data in paper | 6.318(1.119-35.679) | NA |
| Bian | 2016 | CRC | 90 | RT-qPCR | OS | Yes | Data in paper | 2.395 (1.044-5.495) | >75 |
| WangF | 2015 | HCC | 98 | RT-qPCR | OS | Yes | Data in paper | 1.859 (1.077-3.210) | 60 |
| Nie | 2016 | NSCLC | 112 | RT-qPCR | OS | Yes | Data in paper | 1.409(1.077-1.844) | >60 |
| Tao | 2015 | CC | 80 | RT-qPCR | OS | Yes | Data in paper | 2.002 (1.007-3.981) | 60 |
| Ni | 2015 | CRC | 54 | RT-qPCR | OS | NO | Survival curve | 3.137(1.1707-8.4056) | NA |
| Na | 2015 | PC | 40 | RT-qPCR | OS | Yes | Survival curve | 1.52(1.2309- 1.8769) | 60 |
| Han | 2014 | CRC | 80 | RT-qPCR | OS | NO | Survival curve | 0.190 (0.0372-0.9709) | NA |
| WangH | 2015 | LC | 60 | RT-qPCR | OS | Yes | Data in paper | 1.936 (1.062-3.258) | 60 |
| Li | 2014 | ESCC | 90 | RT-qPCR | OS | Yes | Data in paper | 2.627(1.416-5.874) | 60 |
ESCC esophageal squamous cell carcinoma, NSCLC non-small cell lung cancer, LC lung cancer, OC ovarian cancer, CRC colorectal cancer, CC colon cancer, GC gastric cancer, HCC hepatocellular carcinoma, PC prostate cancer, qRT-PCR quantitative real-time PCR,NA not available.
Meta-analysis results
Association between UCA1 and LNM
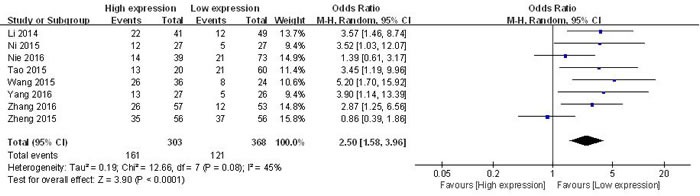
All of the eligible studies reported the LNM of 671 patients based on different UCA1 expression levels. The random-effects model was adopted because of the significant between-study heterogeneity (I2 = 45 %, p = 0.08). The odds ratios, expressed as high UCA1 expression group versus low UCA1 expression group, was 2.50 (CI 95 % 1.58-3.96, p < 0.0001 in random-effects model) (Figure 2). The result revealed that patients with high UCA1 expression in tumor tissues were more prone to LNM.
Figure 2. Forest plot for the association between UCA1 expression levels with LNM.
Association between UCA1 and OS
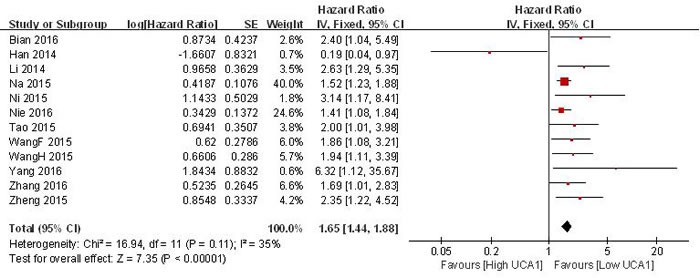
Data of pooled HRs and 95 %CI of overall survival were collected from the 12 eligible studies. The fixed-effects model was used as the small heterogeneity (I2 = 35 %, p = 0.11). Analysis showed a pooled OR of 1.65 with 95 %CI 1.44-1.88 (p < 0.00001) (Figure 3). Compared with low UCA1 expression group, high UCA1 expression group showed a statistically significant reduction in OS and a correlation with a worse survival.
Figure 3. Forest plot of the pooled HRs of elevated UCA1 expression for OS for the included studies.
Sensitivity analysis
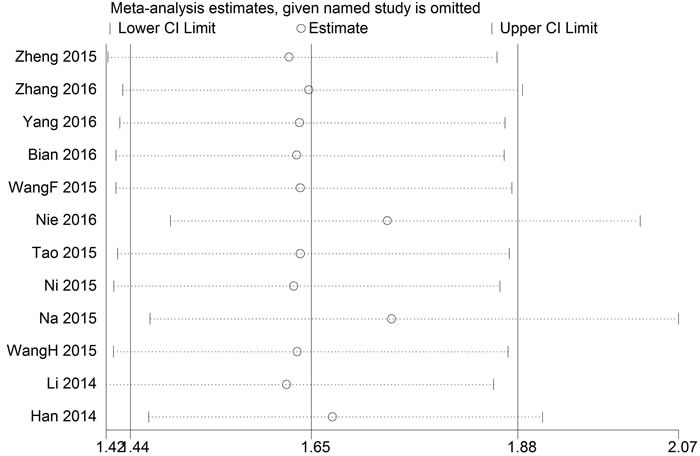
Sensitivity analysis was performed to assess the effect of a single study on the overall meta-analysis results by omitting one study at a time in total population. When each study was excluded sequentially, none of the results were significantly altered each time (Figure 4). Because of the small number of studies, the sensitivity analysis was not analyzed in LNM group.
Figure 4. Result of sensitivity analysis in OS group.
Publication bias
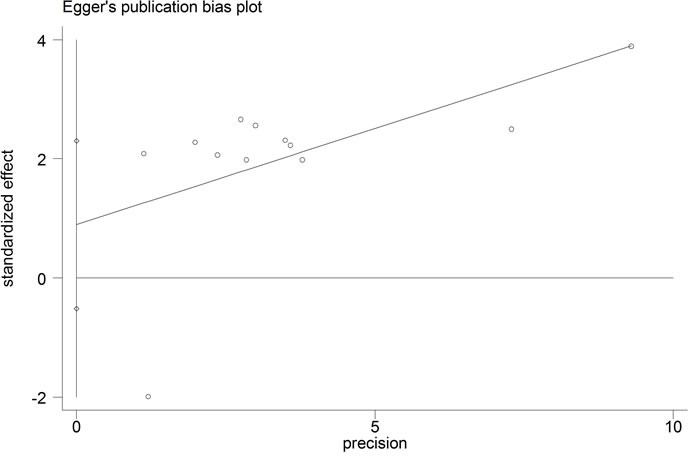
The publication bias of the present meta-analysis was evaluated by Egger's test. In the OS group, the Egger's test did not display obvious publication bias for the HR evaluations of OS (Egger's test, t = 1.42, p = 0.187) (Figure 4). As for LNM group, the publication bias was not analyzed because of the small number of studies.
DISCUSSION
UCA1 has been recently found to be up-regulated in several cancers, contributing to tumor proliferation, apoptosis, metastasis and survival [4]. UCA1 increases cell proliferation through KLF4-KRT6/13 signaling pathway and FGFR1/ERK signaling pathway, respectively, in prostate cancer [11] and hepatocellular carcinoma [5]. Upregulated UCA1 is associated with tumor progression through targeting miR-193a-3p, miR-204-5p in non-small cell lung cancer [14] and colorectal cancer [7] individually. Moreover, UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis [15].UCA1, significantly associated with LNM and OS, may function as on oncogene and a potential indicator of prognosis in some cancers, including colorectal cancer, esophageal squamous cell carcinoma, prostate cancer, hepatocellular carcinoma and ovarian cancer [5–16].
This meta-analysis explored the relationships between UCA1 expression levels and LNM, DM and OS. The results indicated that high UCA1 expression was significant correlated with LNM (OR = 2.50, 95 %CI: 1.58-3.96, p < 0.0001), and poor OS (HR = 1.65, 95 %CI: 1.44-1.88, p < 0.00001). Through the above analysis, we found that high expression of UCA1 might serve as a common molecular marker for lymph node metastasis and poor prognosis in various cancers. Nevertheless, several limitations in the present meta-analysis should be emphasized. First, only twelve studies were included in this meta-analysis; it has weakened the reliability of this meta-analysis' results. So larger-size and better design studies are needed to be conducted to confirm our results. Second, because of different types of cancers, the cutoff value of UCA1 expression was different in each eligible study. Third, patients included in this present meta-analysis were all Asians. Because of this, our results may just account for patients from Asia. Fourth, many included studies reported positive results because negative results would have little chance to be published.
In conclusion, the meta-analysis offers evidence that up-regulated UCA1 is significantly corrected with LNM and poor OS in patients with various cancers. UCA1 may serve not only as a molecular marker for LNM, but also as a prognostic factor for patients with various cancers. Nevertheless, larger-size and better design studies are to be conducted to confirm its precise role among other well-known molecular markers and prognostic factors.
Figure 5. Funnel plot analysis of potential publication bias in OS group (Egger's test).
MATERIALS AND METHODS
Literature collection
We exhaustively searched electronic databases PubMed, Cochrane Library, and Web of Science, by using “UCA1 or urothelial carcinoma associated 1” as the keyword, in order to identify potentially relevant studies. The last update of searching time was June 13, 2016.
Inclusion and exclusion criteria
Inclusion criteria are as the following: (1) evaluation of a link between UCA1 expression and prognosis of patients with any type of cancer. (2) patients were grouped according to the expression levels of UCA1.(3) related clinicopathologic parameters were described, such as lymph node metastasis.(4) reporting of outcomes, including overall survival (OS). (5) studies containing sufficient data for the computation of odds ratios (OR) and corresponding 95 % confidence intervals (CI). Exclusion criteria are as the following: (1) nonhuman research, letters, editorials, expert opinions, case reports and reviews. (2) studies without usable data. (3) duplicate publications.
Quality assessment
The Newcastle-Ottawa Scale (NOS) was applied to assess the quality of all included studies by three investigators (ABH, ZCC and DLW) independently. All eligible studies were assessed to be of high quality by using the Newcastle-Ottawa Scale (NOS).
Date extraction
Data extraction from the eligible studies were carried out independently by three authors (HAB, CZC and LXH), according to the inclusion and exclusion criteria. Disagreements were discussed with two investigators (WDL,LJF) in conference. For each eligible study, the following information was collected: first author, publication date, country of origin, tumor type, total number of patients, number of high UCA1 expression group and low UCA1 expression group, number of patients with lymph node metastasis and distant metastasis, detection method of UCA1 expression levels, follow-up period and cut-off values, HRs, and corresponding 95% CIs for OS.
Statistical methods
Data of pooled hazard ratios (HRs) were collected from the eligible studies; the log HR and standard error (SE) were used for aggregation of the survival results [17]. So as to evaluate the heterogeneity of the eligible studies, pooled HRs were executed using I2 statistics in this meta-analysis [18]. If the between-study heterogeneity was absent in the included studies (p > 0.1), we used fixed-effects model to analyze the results, while the random-effects model was applied for this meta-analysis when between-study heterogeneity was statistical (p < 0.1). The potential publication bias was assessed using the Egger's test. All the statistical analyses were carried out by using the Review Manager 5.3 and Stata 12.0. p values < 0.05 were considered statistically significant.
Acknowledgments
This research was supported by the Shenzhen Key Medical Discipline Fund, Special Support Funds of Shenzhen for Introduced High-Level Medical Team and the National Science Foundation for Young Scientists of China [81602421].
Footnotes
CONFLICTS OF INTEREST
All authors declare that there is no conflict of interests.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. British journal of cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Xue M, Chen W, Li X. Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer. Journal of cancer research and clinical oncology. 2016:1421407–1419. doi: 10.1007/s00432-015-2042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han Y, Yang YN, Yuan HH, Zhang TT, Sui H, Wei XL, Liu L, Huang P, Zhang WJ, Bai YX. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 7.Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, Zhou L, Qi X, Huang S, Hua D, Xing C, Huang Z. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Scientific reports. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni B, Yu X, Guo X, Fan X, Yang Z, Wu P, Yuan Z, Deng Y, Wang J, Chen D, Wang L. Increased urothelial cancer associated 1 is associated with tumor proliferation and metastasis and predicts poor prognosis in colorectal cancer. International journal of oncology. 2015;47:1329–1338. doi: 10.3892/ijo.2015.3109. [DOI] [PubMed] [Google Scholar]
- 9.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. International journal of clinical and experimental pathology. 2014;7:7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C, Ye H, Zhou B, Chen JJ, Chen P. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clinical & translational oncology. 2015;17:640–646. doi: 10.1007/s12094-015-1290-2. [DOI] [PubMed] [Google Scholar]
- 11.Na XY, Liu ZY, Ren PP, Yu R, Shang XS. Long non-coding RNA UCA1 contributes to the progression of prostate cancer and regulates proliferation through KLF4-KRT6/13 signaling pathway. International journal of clinical and experimental medicine. 2015;8:12609–12616. [PMC free article] [PubMed] [Google Scholar]
- 12.Tao K, Yang J, Hu Y, Sun Y, Tan Z, Duan J, Zhang F, Yan H, Deng A. Clinical significance of urothelial carcinoma associated 1 in colon cancer. International journal of clinical and experimental medicine. 2015;8:21854–21860. [PMC free article] [PubMed] [Google Scholar]
- 13.Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. International journal of clinical and experimental medicine. 2015;8:11824–11830. [PMC free article] [PubMed] [Google Scholar]
- 14.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer letters. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Jiang Y, Wan Y, Zhang L, Qiu J, Zhou S, Cheng W. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour biology. 2016 doi: 10.1007/s13277-016-4917-1. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Cao X, Zhang L, Zhang X, Sheng H, Tao K. UCA1 overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer chemotherapy and pharmacology. 2016;77:629–634. doi: 10.1007/s00280-016-2963-4. [DOI] [PubMed] [Google Scholar]
- 17.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]


