Abstract
Background
Karyopherin α2 (KPNA2), a member of the Karyopherin α family, has recently been reported to play an important role in tumor progression. However, the association between KPNA2 expression and prognosis in cancer remains controversial. So we performed this meta-analysis to evaluate whether expression of KPNA2 was associated with prognosis in patients with solid tumor.
Methods/Findings
24 published eligible studies, including 6164 cases, were identified and included in this meta-analysis through searching of PubMed, EMBASE and Web of Science. We found that KPNA2 expression was an independent predictor for the prognosis of solid tumor with primary outcome (overall survival [OS]: pooled HR=1.767, 95% CI=1.503-2.077, P<0.001) and secondary outcomes (time to recurrence [TTR], recurrence free survival [RFS] and progression free survival [PFS]). However, the association between KPNA2 overexpression and disease free survival [DFS] in solid tumors was not significant (pooled HR=1.653, 95% CI=0.903-3.029, P=0.104). Furthermore, the subgroup analysis revealed that KPNA2 overexpression was associated with poor OS in East-Asian patients and European patients, as well as patients with gastric and colorectal cancer.
Conclusion
KPNA2 expression may be a useful prognostic biomarker to monitor cancer prognosis. Further prospective studies with larger sample sizes are required to confirm our findings.
Keywords: KPNA2, tumor, prognosis, overall survival
INTRODUCTION
Cancer is a main public health problem worldwide. Although, the overall mortality declined over the past two decades, cancer remains one of the main contributor of human mortality [1]. Dysfunction of cellular transport machinery is often observed in caner. The shuttling of proteins between the cytoplasm and the nucleus is mediated by karyopherins. Karyopherin α2 (KPNA2) is one of seven described members of the karyopherin α family, which is also known as importin α-1 or RAG cohort 1. KPNA2 weighs around 58 kDa and is composed of a N-terminal hydrophilicimportinb-binding domain, a central hydrophobic region, and a short acidic C-terminus [2]. KPNA2 may participate in carcinogenesis through regulating the subcellular translocation of cancer-associated cargo proteins [3]. KPNA2 overexpression was shown to promote G1/S cell cycle transition via upregulating c-Myc. KPNA2 could also enhance transcriptional activity of c-Myc, activate Akt, and suppress FOXO3a in various cancer cells. Meanwhile, downregulation of cyclin-dependent kinase (CDK) inhibitor p21 and p27, as well as upregulation of CDK regulator cyclin D1 were seen in KPNA2-over-expresssed cells [4]. Forced expression of KPNA2 could increase proliferation of breast cancer cells [5]. On the other hand, knockdown of KPNA2 was shown to inhibit proliferation of cancer cells derived from lung [6], liver [7] and prostate cancer [8]. Growth evidences have also proposed the potential role of KPNA2 in multiple cancerous behaviors, including cell proliferation, differentiation, cell-matrix adhesion, colony formation and migration [5].
Existing evidences have shown that KPNA2 was over-expressed in multiple malignancies [9–11]. Meanwhile, it has been suggest that elevated KPNA2 could be associated with poor prognosis in a variety of solid tumors, including colorectal cancer [11–13], breast cancer [14–17], gastric cancer [10, 18, 19] and hepatocellular carcinoma [20, 21]. Intriguingly, it was reported that low cytoplasmic and nuclear KPNA2 expression may also predict an adverse outcome in radiotherapy-treated head and neck squamous cell cancer [22]. The results of those individual studies were controversial. Therefore, we conducted this meta-analysis to overcome the limitation of the single study.
RESULTS
Demographic characteristics
Using the described combinations of key terms, a total of 67 articles were retrieved from a literature search of PubMed, Embase and Web of Science databases. As displayed in the search flow diagram (Figure 1) and updated Prisma checklist (Supplementary Table S1), 24 articles published from 2006-2016, which reported at least one of the mentioned outcomes, were included in this meta-analysis [4, 8, 10–31].
Figure 1. The flow chart of the selection process in our meta-analysis.
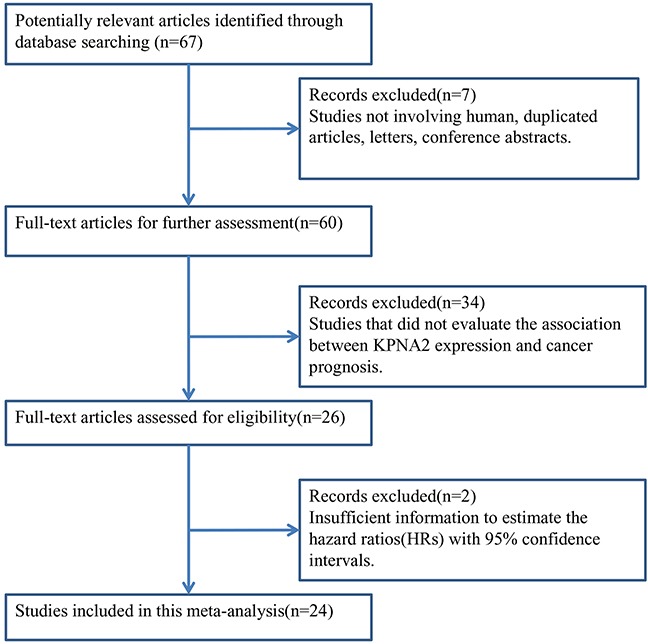
All studies were graded by Newcastle-Ottawa Scale (NOS) (Supplementary Table S2). The NOS scores ranged from 7 to 9, showing that the methodological quality was high. The main features of these eligible articles were listed in Table 1.
Table 1. Characteristics of studies included in the meta-analysis.
| First author | Year | Country | Case | Cancer type | Detection | Provided information on cutoff value | Outcome endpoints | NOS score |
|---|---|---|---|---|---|---|---|---|
| Tsai MM [18] | 2016 | Taiwan | 77 | gastric cancer | IHC | score ≥ 40 | OS | 9 |
| Zhang Y [12] | 2015 | China | 195 | colon cancer | IHC | score ≥3(range of 0-7) | OS,DFS | 8 |
| Takada T [13] | 2015 | Japan | 135 | colorectal cancer | IHC | Low = score 0-4; high = score 6, 9 | OS | 8 |
| Alshareeda AT [14] | 2015 | UK | 1494 | breast cancer | IHC | negative/low<35, positive≥35H-score(range of 0-300) | OS | 9 |
| Shi B [29] | 2015 | China | 176 | upper tract urothelial carcinoma | IHC | strong nuclear staining in at least 10% | OS,DFS | 9 |
| Erben PB [22] | 2015 | Germany | 225 | head and neck squamous cell cancer | IHC | the percentage of positive stained nuclei >15%(median) | DFS | 8 |
| Hu ZY [20] | 2014 | China | 314 | hepatocellular carcinoma | IHC | nucleus staining in more than 5% cells | OS,RFS | 7 |
| Jiang P [21] | 2014 | China | 221 | hepatocellular carcinoma | IHC | extent≥5% (range from 0 to 100%) | OS,TTR | 9 |
| Gousias K [24] | 2014 | Germany | 108 | meningiomas | IHC | the percentage of moderately or strongly immunopositive cell nuclei ≥5% (median) | PFS | 9 |
| Ikenberg K [26] | 2014 | Switzerland | 527 | endometrial cancer | IHC | strong nuclear staining in at least 10% of nuclei | OS | 9 |
| Huang L [4] | 2013 | China | 191 | epithelial ovarian carcinoma | qRT-PCR | expression level of KPNA2>3.52 | OS.RFS | 8 |
| Altan B [19] | 2013 | Japan | 179 | gastric cancer | IHC | Low = score 0-3; high = score 4, 6, 9 | OS | 9 |
| Rachidi SM [11] | 2013 | USA | 54 | colon cancer | IHC | nuclear staining intensity score > 3 | OS | 8 |
| Li C [10] | 2013 | China | 142 | gastric cancer | IHC | score ≥ 4(range of 0-9) | OS | 8 |
| He L [25] | 2012 | China | 90 | ovarian malignant germ cell tumor | IHC | score ≥ 2.5(range of 0-12) | OS,DFS | 7 |
| Gousias K (a) [23]# | 2012 | Germany | 94 | Astrocytomas | IHC | ≥5% nuclear immunoreactivity | OS,PFS | 9 |
| Gousias K (b) [23]# | 2012 | Germany | 47 | Glioblastomas | IHC | ≥10% nuclear immunoreactivity | OS,PFS | 9 |
| Mortezavi A (a) [8]# | 2011 | Switzerland | 341 | prostate cancer | IHC | Nuclear KPNA2 immunoreactivity>0% | RFS | 9 |
| Mortezavi A (b) [8]# | 2011 | Switzerland | 237 | prostate cancer | IHC | Nuclear KPNA2 immunoreactivity>0% | RFS | 9 |
| Jensen JB [27] | 2011 | Denmark | 377 | bladder cancer | IHC | nuclear staining of ≥10% of the carcinoma cells | OS,RFS | 8 |
| Zheng M [31] | 2010 | China | 102 | epithelial ovarian carcinoma | IHC | scores of (++) and (+++) were recorded as positive | OS | 9 |
| Sakai M [28] | 2010 | Japan | 116 | Esophageal Squamous Cell Carcinoma | IHC | KPNA2 LI(labeling index) ≥10.7%(range 0-44.3%) | OS | 8 |
| Gluz O [15] | 2008 | Germany | 191 | breast cancer | IHC | nuclear expression >10% of nuclei | OS | 8 |
| Dankof A [16] | 2007 | Germany | 83 | breast cancer | IHC | nuclear expression >10% of nuclei | DFS | 9 |
| Dahl E [17] | 2006 | Germany | 272 | breast cancer | IHC | nuclear expression≥10% of nuclei | OS | 9 |
| Winnepenninckx V [30] | 2006 | Belgium | 176 | melanoma | IHC | >average expression value | OS | 9 |
IHC : Immunohistochemistry; qRT-PCR:Quantitative Real Time Polymerase Chain Reaction;NOS: Newcastle-Ottawa Scale; DFS: disease free survival; TTR: time to recurrence ; RFS: recurrence free survival; PFS: progression free survival.
There were two parts of data(a and b)in each of the studies of Gousias K and Mortezavi A.
Together, the 24 eligible studies provided a sample size of 6164 patients, which were utilized to evaluate the relationship between KPNA2 expression and solid tumors' prognosis. The median sample-size was 177, with a wide range from 47 to 1494. Among all cohorts, China (n = 8) was the major source region, followed by Germany (n = 7) and Japan (n = 3). As for the cancer type, four studies evaluated breast cancer, three studies evaluated colorectal cancer, three studies evaluated gastric cancer, two studies evaluated hepatocellular carcinoma, two studies evaluated epithelial ovarian carcinoma, one study evaluated prostate cancer, one study evaluated bladder cancer, one study evaluated esophageal squamous cell carcinoma, one study evaluated endometrial cancer, one study evaluated melanoma, one study evaluated ovarian malignant germ cell tumor (OMGCT), one study evaluated upper tract urothelial carcinoma, one study evaluated meningiomas, one study evaluated anaplastic gliomas, one study evaluated astrocytomas. As for the survival outcomes, among 24 eligible studies, twenty of them focused on primary outcome (OS), thirteen studies focused on secondary outcomes (5 for DFS, 4 for RFS, 2 for PFS and 1 for TTR) (Table 1).
Evidence synthesis
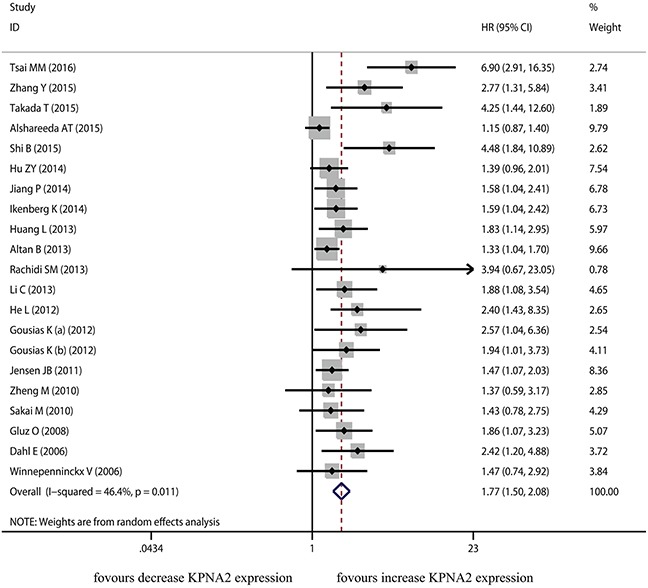
The current meta-analysis was based on primary outcome (OS) and secondary outcomes (TTR, RFS, PFS and DFS). Twenty studies were included in the meta-analysis of OS. A random-effects model was applied to calculate the pooled hazard ratio (HR) and 95% confidence interval (CI). The heterogeneity test reported the P value of 0.011 and I2 values of 46.4%. These results showed an evidence of significant association between KPNA2 overexpression and poor OS (pooled HR=1.767, 95% CI=1.503-2.077, P<0.001) (Figure 2).
Figure 2. The correlation between KPNA2 expression and overall survival in solid tumor.
A random-effects model was utilized to calculate the pooled HR and 95% CI in 5 studies which focused on DFS, as the heterogeneity test reported the P value <0.001 and I2 value of 81.0%. The pooled result showed the association between KPNA2 overexpression and DFS was not significant (pooled HR=1.653, 95% CI=0.903-3.029, P=0.104) (Figure 3).
Figure 3. The correlation between KPNA2 expression and time to tumor progression in solid tumor.
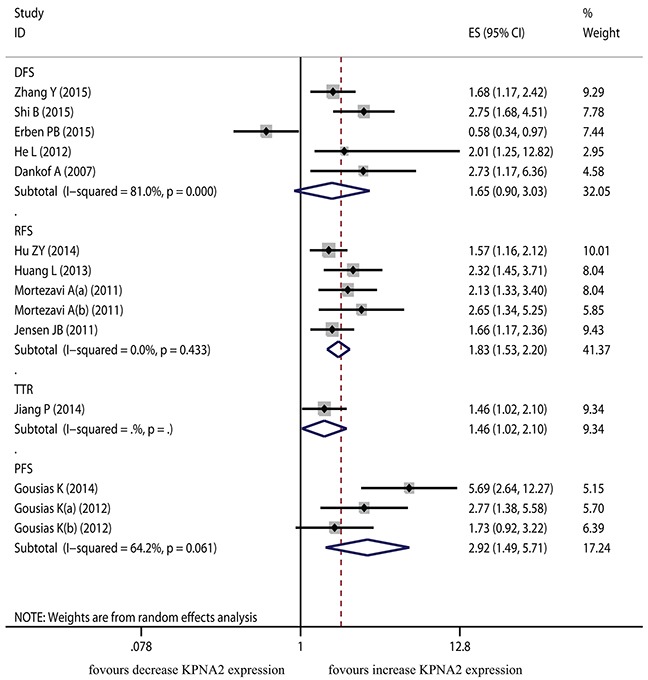
The TTR was derived from only one dataset and showed significant association with KPNA2 overexpression (HR=1.464, 95% CI=1.023-2.096, P=0.037). The pooled results from five datasets for RFS and three datasets for PFS indicated that KPNA2 overexpression was associated with poor RFS and poor PFS (HR=1.835, 95% CI=1.530-2.200, P<0.001; HR=2.921, 95% CI=1.493-5.715, P=0.002, respectively).
To explore the source of heterogeneity, subgroup analyses were conducted by origin of patients and cancer types. The results of subgroup analysis were presented in Table 2. In the subgroup stratified by origin of patients, the pooled HR was 1.962 (95% CI = 1.525-2.525, P<0.001) in East-Asian populations from 12 included studies. The pooled HR was 1.562 (95% CI = 1.407-1.734, P<0.001) for European group from the other 8 studies. Both of the two overall outcomes indicated the significant relationship between KPNA2 overexpression and poor OS. For the analysis stratified by cancer type, significant association between KPNA2 overexpression and poor OS was observed in patients with gastric cancer (HR = 2.353, 95% CI = 1.048-5.284, P = 0.038) and colorectal cancer (HR = 3.252, 95% CI = 1.82-5.811, P<0.001), but not in patients with breast cancer (HR = 1.588, 95% CI = 0.996-2.531, P = 0.052).
Table 2. Hazard ratio for the association between KPNA2 expression and solid tumor prognosis.
| Analysis | N | References | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|
| HR(95% CI) | P | I2(%) | Ph | ||||
| All Studies | |||||||
| OS | 20 | [4, 10–15, 17–21, 23, 25–31] | 1.767(1.503-2.077) | <0.001# | 46.4 | 0.011 | |
| TTR | 1 | [21] | 1.464(1.023-2.096) | 0.037 | - | - | |
| RFS | 4 | [4, 8, 20, 27] | 1.835(1.530-2.200) | <0.001# | 0.0 | 0.433 | |
| PFS | 2 | [23, 24] | 2.921(1.493-5.715) | 0.002 | 64.2 | 0.061 | |
| DFS | 5 | [12, 16, 22, 25, 29] | 1.653(0.903-3.029) | 0.104 | 81.0 | <0.001# | |
| Origin of patients | |||||||
| East-asian | OS | 12 | [4, 10, 12, 13, 18–21, 25, 28, 29, 31] | 1.962(1.525-2.525) | <0.001# | 56.7 | 0.008 |
| European | OS | 8 | [11, 14, 15, 17, 23, 26, 27, 30] | 1.562(1.407-1.734) | <0.001* | 21.6 | 0.251 |
| Cancer type | |||||||
| gastric cancer | OS | 3 | [10, 18, 19] | 2.353(1.408-5.284) | 0.038# | 85.1 | 0.001 |
| breast cancer | OS | 3 | [14, 15, 17] | 1.588(0.996-2.531) | 0.052# | 64.5 | 0.060 |
| colorectal cancer | OS | 3 | [11–13] | 3.252(1.82-5.811) | <0.001* | 0.0 | 0.796 |
N:number of studies; Ph: p value of Q-test for heterogeneity;
The pooled HR was calculated using a fixed-effects model (the Mantel–Haenszel method) according to the heterogeneity;
The pooled HR was calculated using a random-effects model (the DerSimonian and Laird method) according to the heterogeneity; Subgroup analysis was performed when there were at least three studies in each subgroup.
Publication bias and sensitivity analysis
As the amount of datasets for meta-analysis of secondary outcomes (TTR/PFS/RFS/DFS) was fewer, this meta-analysis only evaluated the publication bias for the primary outcome (OS). Begg's funnel plot and Egger's test were applied to evaluate the publication bias of the literatures. The funnel plot was asymmetrical. The P value calculated from Egger's test pointed out the presence of publication bias (P<0.001) among these studies (Figure 4A). Therefore, we performed trim and fill method to make pooled HR more reliable (Figure 4B), and the P value was less than 0.01(data not shown).
Figure 4. Begg's funnel plots for the studies involved in the meta-analysis of KPNA2 expression and the prognosis of patients with solid tumors.
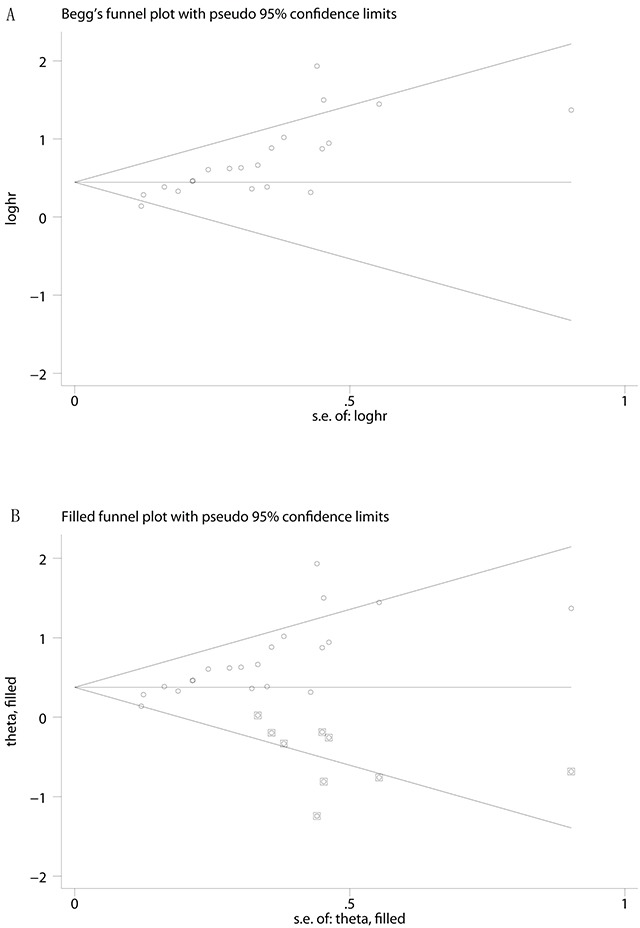
A. Publication bias influence on the overall effect was assessed by the Duval and Tweedie's trim and fill method Duval and Tweedie's trim and fill method B. Abbreviations: loghr, logarithm of hazard ratios; s.e., standard error.
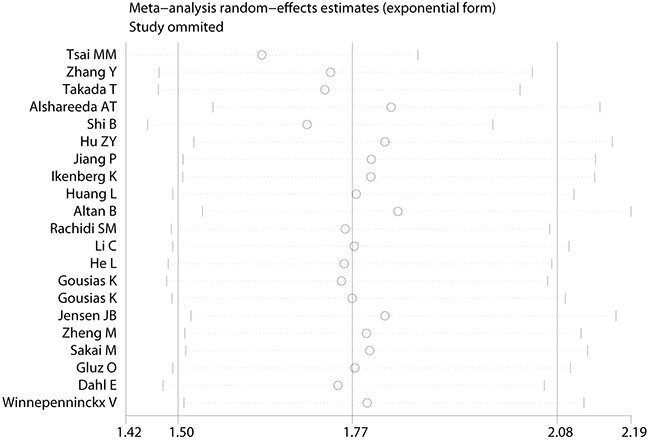
Furthermore, sensitivity analysis was conducted to assess the influence of individual study on the summary effects for the OS. None of the each single study dominated this meta-analysis, and the removal of each study had no significant effect on the overall conclusion (Figure 5). Removal of study using Quantitative Real Time Polymerase Chain Reaction (qRT-PCR) to assess the expression of KPNA2 obtained similar results of OS (HR =1.773, 95% CI = 1.495-2.102, P<0.001, I2 = 48.4%). There were 4 studies with the number of cases less than 100, elimination of these studies had no substantial impact on the outcome of OS (HR = 1.583, 95% CI = 1.372-1.826, P<0.001, I2 = 30.7%).
Figure 5. Sensitivity analysis of the meta-analysis (Overall survival).
DISCUSSION
Many studies have indicated that the aberrant expression of KPNA2 is closely associated with tumor genesis and cancer progression [8–15, 17, 19, 21, 25, 27, 29, 31]. KPNA2 is shown to participate in the translocation of cancer-associated cargo proteins, such as Chk2 [32], BRCA1 [33], NBS1 [2] and many others. In addition, clinical studies have investigated the potential prognostic value of KPNA2. Most of these studies, however, include only limited number of patients, and the results remain inconclusive. To the best of our knowledge, the current meta-analysis was the first systematic evaluation of the literatures studying tumor prognosis and KPNA2 expression.
We evaluated survival data from 6,164 solid tumor patients from 24 different studies. Our results suggest that the increased expression of KPNA2 is indeed a poor prognostic marker for solid tumors in primary outcome (OS pooled HR=1.767, 95% CI=1.503-2.077, P<0.001) and secondary outcomes (TTR/RFS/PFS).
There are several important implications from results of this meta-analysis. First, KPNA2 might serve as a reliable prognostic marker for solid tumors. In this meta-analysis, we included fifteen different cancer types, including breast cancer [14–17], colorectal cancer [11–13], gastric cancer [10, 18, 19], prostate cancer [8], hepatocellular carcinoma [20, 21], epithelial ovarian carcinoma [4, 31], bladder cancer [27], esophageal squamous cell carcinoma [28], endometrial cancer [26], melanoma [30], OMGCT [25], upper tract urothelial carcinoma [29], meningioma [24], astrocytoma [23], head and neck squamous cell cancer [22]. The overall pooled results from these cancer types indicated that elevated KPNA2 expression was associated with patients' poor OS, TTR, RFS and PFS. We therefore propose that high KPNA2 expression may have similar prognostic value for other types of tumor. Second, we demonstrated that KPNA2 overexpression correlated with poor OS in East-Asian population and European population. Different genetic background has no significant effect on the results. Finally, when data was stratified according to cancer type, the results showed the prognostic value of KPNA2 overexpression for OS was significant in gastric cancer and colorectal cancer. In breast cancer, KPNA2 overexpression was associated with poor outcome, but lack of statistical significance. The limited sample size from certain cancer types might have also been statistically insufficient to detect any small effect.
Apart from the inspiring outcomes, there are several potential limitations of this meta-analysis, which should be considered to interpret the outcomes. First, this meta-analysis only enrolled fully published studies in PubMed or EMBASE, yet conference abstracts and studies without enough data were excluded. Second, studies were more likely to be published if they have positive results than negative results. Our analysis detected some publication bias, however meta-analyses with and without the “trim and fill” method did not produce different conclusions. Third, although most of the studies detected the KPNA2 expression by IHC, the antibody concentration and the cutoff value varied across different studies, which might cause some biases in pooled analysis. Fourth, the number of patients of certain published studies, and the number of published studies of one single cancer types may not be sufficient enough for a comprehensive analysis, and our results should be extended to other specific tumor types cautiously. Therefore, our estimate of the association between increased KPNA2 and poor prognosis could possibly be overestimated.
CONCLUSION
In conclusion, our results demonstrate that overexpression of KPNA2 is associated with poor prognosis in various tumors. KPNA2 might be a promising prognostic biomarker and a potential therapeutic target for solid tumors.
MATERIALS AND METHODS
Publication search
PubMed, Embase, and Web of Science databases were searched (up to June 23, 2016) using the search terms: “KPNA2[All Fields] AND (“neoplasms”[MeSH Terms] OR “neoplasms”[All Fields] OR “cancer”[All Fields])) AND (“prognosis”[MeSH Terms] OR “prognosis”[All Fields]) OR (“mortality”[Subheading] OR “mortality”[All Fields] OR “survival”[All Fields] OR “survival”[MeSH Terms]) OR predict[All Fields] OR outcome[All Fields] OR (“life”[MeSH Terms] OR “life”[All Fields] OR “alive”[All Fields])”. All potentially eligible studies were retrieved and their bibliographies were carefully scanned to identify other eligible studies. Additional studies were identified by a hand search of the references cited in the original studies. When multiple studies of the same patient population were identified, we only included the published report with the largest sample size. Additionally, updated Prisma checklist and flow chart were used to present the search strategy.
Inclusion and exclusion criteria
Studies included in this meta-analysis had to meet all the following criteria: (a) evaluation of KPNA2 expression for predicting cancer prognosis; (b) studies reporting survival data; (c) studies provided enough data for individual HRs and 95% CIs to be extracted or calculated; and (d) studies published in English. The exclusion criteria were as follows: 1) review articles, case reports, letters to the editor, conference abstracts, experimental studies and commentary articles; 2) over-lapping or double data; 3) inadequate survival data for further quantification; and 4) the follow-up duration was shorter than 3 years.
Data extraction and methodological quality assessment
This meta-analysis of KPNA2 expression was based on following outcome endpoints: primary outcome (OS) and secondary outcomes [time to recurrence (TTR), recurrence free survival (RFS), progression free survival (PFS) and disease free survival (DFS)]. According to the inclusion and exclusion criteria above, the following items were extracted from each study: the first author's surname, year of publication, country of origin, number of cases, type of cancer, method of detection, score for KPNA2 assessment and cut-off value to determine KPNA2 positivity, Hazard ratio (HR) of KPNA2 expression for OS, TTR, RFS, PFS and DFS with the 95% CI and P-value. If only Kaplan-Meier curves were presented in the studies, we utilized Engauge Digitizer version 4.3 to obtain the survival data, and Tierney's method to calculate the HRs and 95%Cis [34]. Subgroup analysis was performed when there were at least three studies in each subgroup. Data from all eligible publications were extracted carefully and independently by two of the authors. Any disagreements between the researchers were resolved through extracting data from the original article independently by the third author, and any discrepancy was resolved by consensus review.
The methodological quality assessment of each study was performed using the Newcastle–Ottawa Scale(NOS) [35], which scored studies with 9 items including the selection of the patient population, study comparability, outcome of interest, follow-up et al. Studies with an NOS score ≥6 were considered as high-quality ones.
Statistical analysis
In order to evaluate the relationship between KPNA2 expression and solid tumor prognosis, we applied HRs with their corresponding 95% CIs from each eligible paper to calculate the pooled HR for outcome endpoints (OS, DFS, RFS, PFS and TTR). The overall HR was >1, and the 95% CI did not overlap in the forest plot, suggesting a poor prognosis in patients with high expression of KPNA2. Heterogeneity assumption among the included studies was checked using Cochran's Q test and Higgins's I2 statistic [36], P value >0.10 and I2 <50% suggested a lack of heterogeneity among studies. In absence of heterogeneity, a fixed-effects model was applied. Otherwise, the random-effects model was employed [37]. Funnel plots and the Egger's test were utilized to evaluate the possible publication bias [38]. If a publication bias did exist, its influence on the overall effect was assessed by the Duval and Tweedie's trim and fill method [39].
Sensitivity analysis was also performed by omitting each study or specific studies to find potential outliers. All statistical analyses were performed via Stata 14.0 (StataCorp, College Station, TX). All P values for comparisons were two-sided and statistical significance was defined as P<0.05, except those for heterogeneity.
SUPPLEMENTARY TABLES
Footnotes
CONFLICTS OF INTEREST
The authors have declared that there is no competing interest.
GRANT SUPPORT
This work is supported by the National Natural Science Foundation (81472786), Jiangsu Province Health Department Research Project (Z201305), Kunshan Science and Technology Program (KS1418, KS1528), the second affiliated hospital of Soochow university preponderant clinic discipline group project funding (XKQ2015005).
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Christiansen A, Dyrskjot L. The functional role of the novel biomarker karyopherin alpha 2 (KPNA2) in cancer. Cancer letters. 2013;331:18–23. doi: 10.1016/j.canlet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang CI, Chien KY, Wang CL, Liu HP, Cheng CC, Chang YS, Yu JS, Yu CJ. Quantitative proteomics reveals regulation of karyopherin subunit alpha-2 (KPNA2) and its potential novel cargo proteins in nonsmall cell lung cancer. Molecular & cellular proteomics. 2012;11:1105–1122. doi: 10.1074/mcp.M111.016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ, Yun JP, Zhang Y, Jia WH, Zheng M. KPNA2 promotes cell proliferation and tumorigenicity in epithelial ovarian carcinoma through upregulation of c-Myc and downregulation of FOXO3a. Cell death & disease. 2013;4:e745. doi: 10.1038/cddis.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noetzel E, Rose M, Bornemann J, Gajewski M, Knuchel R, Dahl E. Nuclear transport receptor karyopherin-alpha2 promotes malignant breast cancer phenotypes in vitro. Oncogene. 2012;31:2101–2114. doi: 10.1038/onc.2011.403. [DOI] [PubMed] [Google Scholar]
- 6.Wang CI, Wang CL, Wang CW, Chen CD, Wu CC, Liang Y, Tsai YH, Chang YS, Yu JS, Yu CJ. Importin subunit alpha-2 is identified as a potential biomarker for non-small cell lung cancer by integration of the cancer cell secretome and tissue transcriptome. International journal of cancer. 2011;128:2364–2372. doi: 10.1002/ijc.25568. [DOI] [PubMed] [Google Scholar]
- 7.Yoshitake K, Tanaka S, Mogushi K, Aihara A, Murakata A, Matsumura S, Mitsunori Y, Yasen M, Ban D, Noguchi N, Irie T, Kudo A, Nakamura N, Tanaka H, Arii S. Importin-alpha1 as a novel prognostic target for hepatocellular carcinoma. Annals of surgical oncology. 2011;18:2093–2103. doi: 10.1245/s10434-011-1569-7. [DOI] [PubMed] [Google Scholar]
- 8.Mortezavi A, Hermanns T, Seifert HH, Baumgartner MK, Provenzano M, Sulser T, Burger M, Montani M, Ikenberg K, Hofstadter F, Hartmann A, Jaggi R, Moch H, Kristiansen G, Wild PJ. KPNA2 expression is an independent adverse predictor of biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2011;17:1111–1121. doi: 10.1158/1078-0432.CCR-10-0081. [DOI] [PubMed] [Google Scholar]
- 9.Ma S, Zhao X. KPNA2 is a promising biomarker candidate for esophageal squamous cell carcinoma and correlates with cell proliferation. Oncology reports. 2014;32:1631–1637. doi: 10.3892/or.2014.3381. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Ji L, Ding ZY, Zhang QD, Huang GR. Overexpression of KPNA2 correlates with poor prognosis in patients with gastric adenocarcinoma. Tumour biology. 2013;34:1021–1026. doi: 10.1007/s13277-012-0641-7. [DOI] [PubMed] [Google Scholar]
- 11.Rachidi SM, Qin T, Sun S, Zheng WJ, Li Z. Molecular profiling of multiple human cancers defines an inflammatory cancer-associated molecular pattern and uncovers KPNA2 as a uniform poor prognostic cancer marker. PloS one. 2013;8:e57911. doi: 10.1371/journal.pone.0057911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Zhang M, Yu F, Lu S, Sun H, Tang H, Peng Z. Karyopherin alpha 2 is a novel prognostic marker and a potential therapeutic target for colon cancer. Journal of experimental & clinical cancer research. 2015;34:145. doi: 10.1186/s13046-015-0261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takada T, Tsutsumi S, Takahashi R, Ohsone K, Tatsuki H, Suto T, Kato T, Fujii T, Yokobori T, Kuwano H. KPNA2 over-expression is a potential marker of prognosis and therapeutic sensitivity in colorectal cancer patients. Journal of surgical oncology. 2016;113:213–217. doi: 10.1002/jso.24114. [DOI] [PubMed] [Google Scholar]
- 14.Alshareeda AT, Negm OH, Green AR, Nolan CC, Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO, Rakha EA. KPNA2 is a nuclear export protein that contributes to aberrant localisation of key proteins and poor prognosis of breast cancer. British journal of cancer. 2015;112:1929–1937. doi: 10.1038/bjc.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gluz O, Wild P, Meiler R, Diallo-Danebrock R, Ting E, Mohrmann S, Schuett G, Dahl E, Fuchs T, Herr A, Gaumann A, Frick M, Poremba C, Nitz UA, Hartmann A. Nuclear karyopherin alpha2 expression predicts poor survival in patients with advanced breast cancer irrespective of treatment intensity. International journal of cancer. 2008;123:1433–1438. doi: 10.1002/ijc.23628. [DOI] [PubMed] [Google Scholar]
- 16.Dankof A, Fritzsche FR, Dahl E, Pahl S, Wild P, Dietel M, Hartmann A, Kristiansen G. KPNA2 protein expression in invasive breast carcinoma and matched peritumoral ductal carcinoma in situ. Virchows Archiv. 2007;451:877–881. doi: 10.1007/s00428-007-0513-5. [DOI] [PubMed] [Google Scholar]
- 17.Dahl E, Kristiansen G, Gottlob K, Klaman I, Ebner E, Hinzmann B, Hermann K, Pilarsky C, Durst M, Klinkhammer-Schalke M, Blaszyk H, Knuechel R, Hartmann A, Rosenthal A, Wild PJ. Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin Cancer Res. 2006;12:3950–3960. doi: 10.1158/1078-0432.CCR-05-2090. [DOI] [PubMed] [Google Scholar]
- 18.Tsai MM, Huang HW, Wang CS, Lee KF, Tsai CY, Lu PH, Chi HC, Lin YH, Kuo LM, Lin KH. MicroRNA-26b inhibits tumor metastasis by targeting the KPNA2/c-jun pathway in human gastric cancer. Oncotarget. 2016;7:39511–39526. doi: 10.18632/oncotarget.8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altan B, Yokobori T, Mochiki E, Ohno T, Ogata K, Ogawa A, Yanai M, Kobayashi T, Luvsandagva B, Asao T, Kuwano H. Nuclear karyopherin-alpha2 expression in primary lesions and metastatic lymph nodes was associated with poor prognosis and progression in gastric cancer. Carcinogenesis. 2013;34:2314–2321. doi: 10.1093/carcin/bgt214. [DOI] [PubMed] [Google Scholar]
- 20.Hu ZY, Yuan SX, Yang Y, Zhou WP, Jiang H. Pleomorphic adenoma gene 1 mediates the role of karyopherin alpha 2 and has prognostic significance in hepatocellular carcinoma. Journal of experimental & clinical cancer research. 2014;33:61. doi: 10.1186/s13046-014-0061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang P, Tang Y, He L, Tang H, Liang M, Mai C, Hu L, Hong J. Aberrant expression of nuclear KPNA2 is correlated with early recurrence and poor prognosis in patients with small hepatocellular carcinoma after hepatectomy. Medical oncology. 2014;31:131. doi: 10.1007/s12032-014-0131-4. [DOI] [PubMed] [Google Scholar]
- 22.Erben PB, Brunner K, Hecht M, Haderlein M, Buttner-Herold M, Agaimy A, Fietkau R, Hartmann A, Distel LV. Low cytoplasmic and nuclear KPNA2 expression in radiotherapy-treated head and neck squamous cell cancer is associated with an adverse outcome. International journal of clinical and experimental pathology. 2015;8:15814–15824. [PMC free article] [PubMed] [Google Scholar]
- 23.Gousias K, Becker AJ, Simon M, Niehusmann P. Nuclear karyopherin a2: a novel biomarker for infiltrative astrocytomas. Journal of neuro-oncology. 2012;109:545–553. doi: 10.1007/s11060-012-0924-2. [DOI] [PubMed] [Google Scholar]
- 24.Gousias K, Niehusmann P, Gielen GH, Simon M. Karyopherin a2 and chromosome region maintenance protein 1 expression in meningiomas: novel biomarkers for recurrence and malignant progression. Journal of neuro-oncology. 2014;118:289–296. doi: 10.1007/s11060-014-1423-4. [DOI] [PubMed] [Google Scholar]
- 25.He L, Ding H, Wang JH, Zhou Y, Li L, Yu YH, Huang L, Jia WH, Zeng M, Yun JP, Luo RZ, Zheng M. Overexpression of karyopherin 2 in human ovarian malignant germ cell tumor correlates with poor prognosis. PloS one. 2012;7:e42992. doi: 10.1371/journal.pone.0042992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikenberg K, Valtcheva N, Brandt S, Zhong Q, Wong CE, Noske A, Rechsteiner M, Rueschoff JH, Caduff R, Dellas A, Obermann E, Fink D, Fuchs T, et al. KPNA2 is overexpressed in human and mouse endometrial cancers and promotes cellular proliferation. The Journal of pathology. 2014;234:239–252. doi: 10.1002/path.4390. [DOI] [PubMed] [Google Scholar]
- 27.Jensen JB, Munksgaard PP, Sorensen CM, Fristrup N, Birkenkamp-Demtroder K, Ulhoi BP, Jensen KM, Orntoft TF, Dyrskjot L. High expression of karyopherin-alpha2 defines poor prognosis in non-muscle-invasive bladder cancer and in patients with invasive bladder cancer undergoing radical cystectomy. European urology. 2011;59:841–848. doi: 10.1016/j.eururo.2011.01.048. [DOI] [PubMed] [Google Scholar]
- 28.Sakai M, Sohda M, Miyazaki T, Suzuki S, Sano A, Tanaka N, Inose T, Nakajima M, Kato H, Kuwano H. Significance of karyopherin-{alpha} 2 (KPNA2) expression in esophageal squamous cell carcinoma. Anticancer research. 2010;30:851–856. [PubMed] [Google Scholar]
- 29.Shi B, Su B, Fang D, Tang Y, Xiong G, Guo Z, He Q, Yang X, Zhao W, Guo Y, Li X, Zhou L. High expression of KPNA2 defines poor prognosis in patients with upper tract urothelial carcinoma treated with radical nephroureterectomy. BMC cancer. 2015;15:380. doi: 10.1186/s12885-015-1369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, Eggermont AM, Lenoir G, Sarasin A, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. Journal of the National Cancer Institute. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 31.Zheng M, Tang L, Huang L, Ding H, Liao WT, Zeng MS, Wang HY. Overexpression of karyopherin-2 in epithelial ovarian cancer and correlation with poor prognosis. Obstetrics and gynecology. 2010;116:884–891. doi: 10.1097/AOG.0b013e3181f104ce. [DOI] [PubMed] [Google Scholar]
- 32.Zannini L, Lecis D, Lisanti S, Benetti R, Buscemi G, Schneider C, Delia D. Karyopherin-alpha2 protein interacts with Chk2 and contributes to its nuclear import. The Journal of biological chemistry. 2003;278:42346–42351. doi: 10.1074/jbc.M303304200. [DOI] [PubMed] [Google Scholar]
- 33.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nature reviews Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 34.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 36.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Annals of internal medicine. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 38.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


