Abstract
Rapid and reliable identification of bacteria directly from blood cultures is important in clinical practice to guide appropriate antibiotic therapy. In this study, the performance of the AccuProbe (Gen-Probe, Inc., San Diego, Calif.) in direct identification of Staphylococcus aureus, Streptococcus pneumoniae, enterococci, and group A and B streptococci from positive blood culture bottles was evaluated by using 6-year routine clinical laboratory blood culture material from Päijät-Häme Central Hospital, Lahti, Finland. With the enterococcal and group A and B streptococcal probes, the diagnostic performance of the test was excellent at a cutoff value of 50,000 relative light units (RLU) as recommended by the manufacturer. However, with the S. aureus probe, although the specificity was very high (99.8%), the sensitivity was low (72.4%). To improve the clinical usability of the direct AccuProbe identification, optimal cutoff values for the individual AccuProbe tests were defined by using receiver-operating characteristic analysis. Consequently, cutoff values for S. aureus and S. pneumoniae tests were adjusted to 30,000 RLU and for enterococci and to 55,000 RLU for group A and B streptococci. With these adjustments, the performance of the AccuProbe tests, especially that for S. aureus, was significantly improved.
Rapid and reliable identification of bacteria from blood culture is indisputably one of the most important functions of a clinical microbiology laboratory. Bloodstream infection is a potentially life-threatening condition that requires rapid, appropriate therapy. The increasing rate of nosocomial infections during the past decade (21), together with the threat from multiresistant infectious agents, further highlight the importance of early identification and tailored antimicrobial therapy. A rapid and accurate method for direct identification of pathogens from positive blood culture broth, combined with automated, continuously monitored blood culture systems, may greatly reduce the overall time from specimen collection to final identification. This may result in a reduction in the use of broad-spectrum antibiotics, since the diagnostic information will be available to the clinician earlier.
However, relatively few methods for direct identification of bacteria from blood culture broth have been described. Standard assays comprising the immunological, tube coagulase, and stable endonuclease methods routinely used for identification of Staphylococcus aureus after subculture have been applied directly to positive blood culture bottles, but variable sensitivities and specificities have been reported (3, 12, 20). The use of PCR for the direct identification of Streptococcus pneumoniae from blood has also been described (7, 17, 22), but overall its use for this purpose has been rather limited owing to the amplification inhibitors present in blood (6). The extreme sensitivity of the method may also be a problem (14). However, rapid identification of S. aureus from blood culture by using the LightCycler system has already been reported (18), and the use of real-time fluorescence PCR for rapid identification of bacteria from blood culture may increase in the future. Fluorescent in situ hybridization (FISH) (8, 15, 16), as well as the hybridization protection assay (2), have also been shown to be quite reasonable methods for direct identification of microorganisms from blood.
Direct identification of the most common gram-positive bacteria in blood cultures by using the commercially available DNA probe kit AccuProbe (Gen-Probe, Inc., San Diego, Calif.), which utilizes hybridization protection assay technology, has already been demonstrated (2; H. Sarkkinen, Abstr. 10th Int. Cong. Infect. Dis., p. 161, 2002). However, the test has been validated by the manufacturer only with the use of fresh growth from solid medium or from broth cultures and not with direct clinical specimens. We report here our 6-year routine clinical laboratory experience with the use of AccuProbe for direct identification of S. aureus, S. pneumoniae, enterococci, and group A and B streptococci from positive blood culture bottles. The aim of the present study was also to define the optimal conditions (cutoff values) of the individual AccuProbe tests for direct identification.
MATERIALS AND METHODS
The present study involved all blood cultures obtained between June 1997 and July 2003 in Päijät-Häme Central Hospital, Lahti, Finland. The blood cultures were carried out at the request of clinicians as part of the routine care of patients suspected of bacteremia and were processed as part of our normal clinical microbiology workflow. Blood culture bottles (BacT/Alert FAN aerobic and standard anaerobic bottles) were incubated in a BacT/Alert continuous monitoring system (Organon Technica Corp., Durham, N.C.; bioMérieux, Marcy l'Etoile, France [since July 2001]) in accordance with the manufacturer's recommendations. Organon Tecnica replaced its vented BacT/Alert aerobic blood culture bottle with a redesigned nonvent blood culture bottle in 2000, so the BacT/Alert FAN nonvent aerobic blood culture bottle was adopted in our blood culture routine in February 2001. These two types of bottles (nonvent and vented) have, however, been shown to be equivalent (10, 19).
When blood culture bottles were signaled as positive, broth from the positive bottle was Gram stained, and bottles containing gram-positive cocci were screened for S. aureus enterococcal or streptococcal rRNA. Probe selection was guided by the Gram stain pattern: when the stain revealed gram-positive cocci in clusters, the S. aureus probe was selected and, if the gram-positive cocci were clearly in pairs or chains, then Str. pneumoniae, enterococcal, and group A and B streptococcal probes were tested. All five tests were performed when the Gram stain showed gram-positive cocci without a predominance of clusters or chains. Subcultures on both chocolate and fastidious anaerobe agar plates were performed to provide isolates for conventional identification methods and susceptibility testing.
AccuProbe technology (Gen-Probe) utilizes a chemiluminescent acridinium ester labeled DNA probe directed toward the rRNA of the target organisms. The manufacturer's instructions for specimen preparation were modified to use a pellet of bacteria made directly from positive blood culture broth as previously described (2). Briefly, 1.5 ml of broth from the positive blood culture bottle was centrifuged for 6 s in a microcentrifuge at 10,000 × g to sediment the red blood cells. The supernatant was then centrifuged at 10,000 × g for 1 min to concentrate the bacteria. A 1-μl loopful of pelleted bacteria was then tested with the appropriate AccuProbe in accordance with the manufacturer's instructions by using reagents supplied by the manufacturer. The chemiluminescence of the hybridized probe was measured with a Gen-Probe Leader 50i luminometer. A positive result registered in the laboratory records was a luminometer reading above the cutoff value of 50,000 relative light units (RLU). This cutoff value has been determined by the manufacturer on the basis of readings obtained from in-house testing (sample prepared by using a colony from an agar plate).
We reviewed all consecutive signal positive blood cultures in which direct Gram stain had revealed gram-positive cocci from laboratory records. The recorded luminometer readings were tabulated with the culture results and analyzed: the culture result being considered the “gold standard.” If both bottles in the same blood culture set were positive, only the first AccuProbe result per set was included in the analysis.
During the period from June 1997 to July 2003, the total number of sets of blood cultures in our hospital was 33,695 (with one aerobic and one anaerobic bottle per set). During the same period there were 2,228 patients whose blood culture was positive. We tested 454 consecutive signal positive bottles (one per patient) directly with the AccuProbe when gram-positive cocci were seen in direct Gram stain. The number of different AccuProbe tests performed was as follows: 732 S. aureus tests (203 were culture positive and 529 were culture negative), 423 S. pneumoniae tests (142 were culture positive and 281 were culture negative), 377 enterococcus tests (55 were culture positive and 322 were culture negative), 372 group A streptococcus tests (18 were culture positive and 354 were culture negative), and 372 group B streptococcus tests (36 were culture positive and 336 were culture negative).
Receiver-operating characteristic (ROC) analysis is a method for assessing the value of a quantitative diagnostic test and deciding where to set the threshold when the test is used. In the present study, ROC analysis (StatsDirect statistical software; StatsDirect, Ltd.) was used to adjust the cutoff values (measured in RLU) for the individual AccuProbe tests.
RESULTS
There were 203 culture positive and 529 culture negative S. aureus blood culture samples in the material. A total of 147 samples were found to be positive and 528 were negative by both methods. A total of 56 samples were found to be negative with the AccuProbe test but positive in the culture. In addition, there was one culture-negative but AccuProbe-positive case, with the very high RLU value of 562,252, which was clearly negative when repeated. However, since only the first test result of each patient was included, it was considered as an AccuProbe-positive but culture-negative case in all analyses. The sensitivity and specificity values of the S. aureus AccuProbe test were 72.4% (95% confidence interval [CI] = 65.72 to 78.44%) and 99.8% (95% CI 98.95% to 100%), respectively. The predictive value of the positive test was 99.3% (95% CI = 96.29 to 99.98%), but the predictive value of the negative test was only 90.4% (95% CI = 87.73 to 92.68%). Owing to the low sensitivity of the test, 56 of 203 (27.6%) culture-positive cases were missed by the AccuProbe. On the other hand, the specificity of the test was very high, producing only one false-positive case.
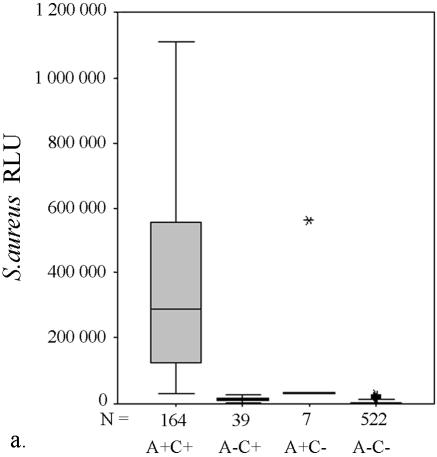
To improve the clinical usability of the S. aureus AccuProbe test, ROC analysis was used to find a cutoff value that would increase the sensitivity of the test without excessively compromising the specificity of the test. When the cutoff value was set to ∼16,000 RLU, an equal number of false-positive and false-negative cases were found. Setting the cutoff value higher than 16,000 RLU therefore increased the likelihood of a positive test result being a true-positive case. When the cutoff value was elevated to 30,000 RLU (Fig. 1a), 17 more culture-positive cases were detected by the AccuProbe at the expense of six more false-positive results. The median RLU value for these false-positive S. aureus samples was 31,472 RLU (Table 1), i.e., very close to the cutoff value.
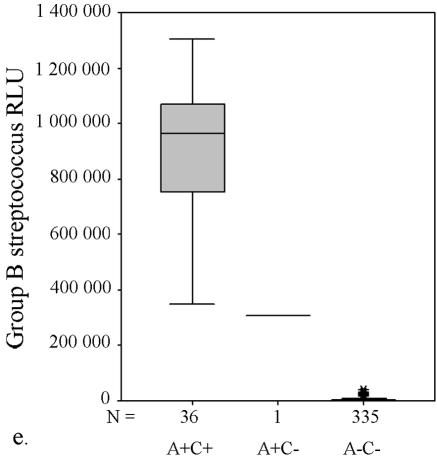
FIG. 1.
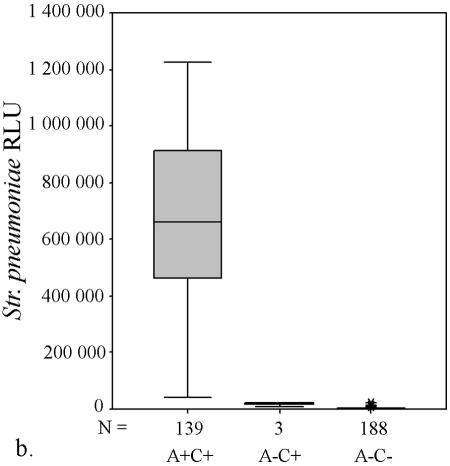
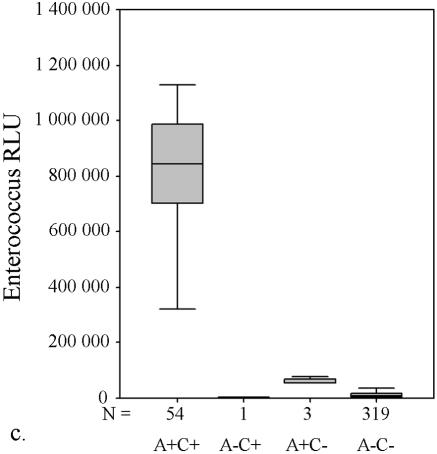
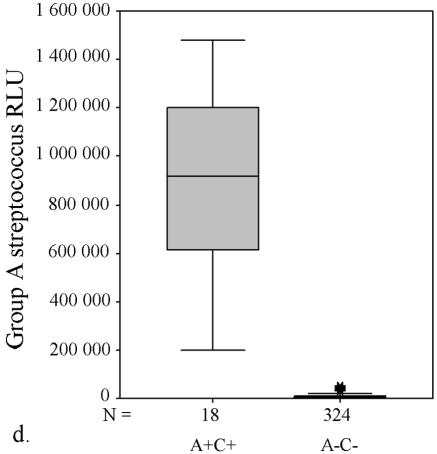
AccuProbe test results with adjusted cutoff values (30,000 RLU with S. aureus and S. pneumoniae probes and 55,000 RLU with enterococcus, group A and group B streptococcus probes) versus culture. (a to e) Box and whisker plot representations show the distribution of true-positive and -negative and false-positive and -negative test results. The central box represents the distance between the first and third quartiles with the median RLU value between them marked with a horizontal line. The minimum RLU value is the origin of the lower whisker, and the maximum is the limit of the upper whisker. A+/−, AccuProbe positive/negative; C+/−, culture positive/negative.
TABLE 1.
Contradictory results between AccuProbe tests with adjusted cutoff values versus culture
| AccuProbe test (organism and result) | No. of samples | RLUa
|
||
|---|---|---|---|---|
| Min | Max | Med | ||
| S. aureus | ||||
| False positive | 7 | 30,093 | 562,252 | 31,472 |
| False negative | 39 | 2,271 | 28,350 | 12,055 |
| S. pneumoniae | ||||
| False positive | 0 | |||
| False negative | 3 | 10,140 | 24,253 | 23,141 |
| Enterococcus | ||||
| False positive | 3 | 55,386 | 79,709 | 56,407 |
| False negative | 1 | 4,729 | 4,729 | 4,729 |
| Group A streptococcus | ||||
| False positive | 0 | |||
| False negative | 0 | |||
| Group B streptococcus | ||||
| False positive | 1 | 309,773 | 309,773 | 309,773 |
| False negative | 0 | |||
Abbreviations: Min, minimum RLU value for giving a false-positive or false-negative test result; Max, maximum RLU value for giving a false-positive or false-negative test result; Med, median RLU value for giving a false-positive or false-negative test result.
Similarly, with S. pneumoniae there were four false-negative AccuProbe results with 50,000 RLU as the cutoff value, and three false-negative results with 30,000 as the cutoff value (Fig. 1b and Table 1). For enterococcus, five false-positive AccuProbe results were found with a cutoff value of 50,000 RLU and two fewer when 55,000 was used as the cutoff value. In addition, one false-negative result was also found (Fig. 1c and Table 1). With group A streptococcus, there was one false-positive finding (50,750 RLU) with the cutoff value of 50,000 RLU but no false-negative or false-positive samples with a cutoff value of 55,000 RLU (Fig. 1d and Table 1). With group B (Fig. 1e) streptococcus there was only one false-positive sample, which turned out to be Streptococcus equi. The manufacturer reports cross-reaction with some isolates of S. equi in the package insert of the Group B Streptococcus Culture Identification Test.
The diagnostic characteristics of all AccuProbe tests with the selected ROC analysis based cutoff values are given in Table 2.
TABLE 2.
Performance characteristics of AccuProbe tests with adjusted cutoff values
| Cutoff RLU and organism | % (95% CI)
|
|||
|---|---|---|---|---|
| Sensitivity | Specificity | Positive predictive value | Negative predictive value | |
| 30,000 | ||||
| S. aureus | 80.8 (74.69-85.97) | 98.7 (97.29-99.47) | 95.9 (91.75-98.34) | 93.1 (90.62-95.01) |
| S. pneumoniae | 97.9 (93.95-99.56) | 100 (98.06-100) | 100 (97.38-100) | 98.4 (95.48-99.67) |
| 55,000 | ||||
| Enterococcus | 98.2 (90.28-99.95) | 99.1 (97.3-99.81) | 94.7 (85.38-98.9) | 99.7 (98.27-99.99) |
| Group A streptococcus | 100 (81.47-100) | 100 (98.87-100) | 100 (81.47-100) | 100 (98.87-100) |
| Group B streptococcus | 100 (90.26-100) | 99.7 (98.35-99.99) | 97.3 (85.84-99.93) | 100 (98.9-100) |
DISCUSSION
Our study showed that for streptococcal and enterococcal probes, the sensitivity, specificity, and positive and negative predictive values of the AccuProbe tests were already excellent when the cutoff values recommended by the manufacturer are used. However, with the S. aureus probe, although the specificity was very high (99.8%), the sensitivity of the test (72.4%) was low. It was clear that the sensitivity of the test was unacceptable.
The blood culture procedure in the present study included incubation in a continuous monitoring system, Gram staining, and use of the AccuProbe test when the stain demonstrated gram-positive cocci. According to this procedure, the sensitivity of the S. pneumoniae AccuProbe test for the direct identification was excellent. Pneumococci, however, are known to be subject in liquid cultures to autolysis, which first renders the cells gram negative and later brings about lysis of the culture. Consequently, it is possible that when autolysis has occurred, the Gram stain may not demonstrate gram-positive cocci, which may diminish the sensitivity of the blood culture procedure as a whole. However, we have never found a sample giving a positive signal in a continuous monitoring system that would have resulted in a negative Gram stain and then later positive subcultures on agar plates or positive acridine orange stains.
Defining cutoff levels for diagnostic tests is a difficult task that combines ethical and practical considerations with statistical evidence. The main difficulty is not statistical but rather the need to decide how good the test should be in order for it to be clinically relevant. Therefore, the “best clinical” cutoff value is not necessarily the same as the “best statistical” cutoff value. Considering the potentially life-threatening nature of the sepsis, we were willing to accept a moderate number of false-positive results if the sensitivity of the test could be improved. To study the effects of different cutoff values on the diagnostic performance of the AccuProbe tests, ROC curve analysis was used (1, 23). On the basis of our analysis, the cutoff values for individual AccuProbe tests were adjusted as indicated in Table 2. With these adjustments the performance of the AccuProbe tests, especially that for S. aureus, was significantly improved.
In our hospital, and in Finland generally, the most common blood culture pathogens are Escherichia coli, coagulase-negative staphylococci, S. aureus, S. pneumoniae, and Enterococcus spp. Along with enterococci, S. aureus and coagulase-negative staphylococci are among the causes of blood culture-positive nosocomial infections, the rate of which has increased during the past decade (4, 9). Our ROC analysis-based cutoff value adjustment improved the clinical usability of the S. aureus AccuProbe test, but the test still does not perform as well as the other AccuProbe tests. It may be that the sensitivity of the S. aureus AccuProbe test could be further improved with optimization of sample preparation protocol. A probe capable of identifying E. coli, as well as other gram-negative rods, at the species level would also be highly desirable. The development of such a probe has been hampered by the fact that the target sites are often detected not only in E. coli but also in Shigella spp. and in other enteric bacteria. However, an evaluation of a DNA probe matrix that contains a probe for E. coli has recently been described (11). That particular study examined the use of a Gen-Probe research prototype assay for direct identification of microorganisms from blood. Commercial application of this assay is not yet available.
Overall, as noted in the introduction, relatively few methods for direct identification of bacteria from blood culture broth have been described, and their performance in large-scale routine use has not been reported. Very recently, M. Sogaard et al. (Abstr. 14th Eur. Cong. Clin. Microbiol. Infect. Dis., abstr. O57, 2004) reported the use in a routine setting of a PNA FISH assay, which is now available as a kit. In the future, a study comparing the performances of the AccuProbe and the PNA FISH kit in routine clinical laboratory use would be of great interest.
Munson et al. recently reported that, as far as antimicrobial management is concerned, the most important information provided by the clinical microbiology laboratory is the first telephone call reporting positive blood culture and Gram stain results (13). Their data also demonstrated that release of antimicrobial susceptibility test results had little impact on antimicrobial usage. Since we have optimized the cutoff values for direct identification, on the basis of the high predictive values, we can reliably report these results, i.e., the species of the clinically important gram-positive cocci, at almost the same time (within 1 h) as the Gram stain results. In addition, a statistical antibiotic resistance pattern based on local data can be added to the report for guidance. Such a preliminary resistance pattern is indeed relevant, since the antibiotic resistance of the clinically most important bacteria, such as S. aureus, is still relatively stable in Finland (5, 9). However, the final decision about antimicrobial therapy is always made in conjunction with the use of other laboratory and clinical data.
For a method to be successfully implemented in a routine microbiology laboratory, it should be fast, reliable, cost-effective, and simple to perform. The modified microcentrifuge method takes only ∼30 min to perform and was easy to adopt in our normal daily routines with only a few positive blood cultures per day. The sample preparation needs no special expertise, and the results are objective since they are measured with a luminometer, which automatically calculates and prints out the test result.
The AccuProbe culture identification test is clearly more expensive than identification by conventional methods. The additional cost per sample consists of reagent costs (ca. 6$ per probe) and labor costs of ca. 30 min per sample (one or more probes). However, considering the clinical significance of blood culture and the timely identification provided by the direct AccuProbe method, the additional cost is justified.
In conclusion, on the basis of the present study we can now offer timely and accurate information to clinicians to assist them in the application of antimicrobial therapy in the early stages of infection.
REFERENCES
- 1.Altman, D. G., and J. M. Bland. 1994. Statistics notes: diagnostic tests. 3. Deceiver operating characteristic plots. BMJ 309:188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis, T. E., and D. D. Fuller. 1991. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J. Clin. Microbiol. 29:2193-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, T. E., D. D. Fuller, and E. C. Aeschleman. 1992. Rapid, direct identification of Staphylococcus aureus and Streptococcus pneumoniae from blood cultures using commercial immunologic kits and modified conventional tests. Diagn. Microbiol. Infect. Dis. 15:295-300. [DOI] [PubMed] [Google Scholar]
- 4.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 5.European Antibiotic Resistance Surveillance System. 2003. EARSS annual report 2002. [Online.] http://www.earss.rivm.nl.
- 6.Fredricks, D. N., and D. A. Relman. 1998. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J. Clin. Microbiol. 36:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassan-King, M., I. Baldeh, O. Secka, A. Falade, and B. Greenwood. 1994. Detection of Streptococcus pneumoniae DNA in blood cultures by PCR. J. Clin. Microbiol. 32:1721-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kempf, V. A. J., K. Trebesius, and I. B. Autenrieth. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyytikäinen, O., J. Lumio, H. Sarkkinen, E. Kolho, A. Kostiala, P. Ruutu, et al. 2002. Nosocomial bloodstream infections in Finnish hospitals during 1999-2000. Clin. Infect. Dis. 35:14-19. [DOI] [PubMed] [Google Scholar]
- 10.Marlowe, E. M., L. Gibson, J. Hogan, S. Kaplan, and D. A. Bruckner. 2003. Conventional and molecular methods for verification of results obtained with BacT/Alert nonvent blood culture bottles. J. Clin. Microbiol. 41:1266-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marlowe, E. M., J. J. Hogan, J. F. Hindler, I. Andruszkiewicz, P. Gordon, and D. A. Bruckner. 2003. Application of an rRNA probe matrix for rapid identification of bacteria and fungi from routine blood cultures. J. Clin. Microbiol. 41:5127-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald, C. L., and K. Chapin. 1995. Rapid identification of Staphylococcus aureus from blood culture bottles by a classic 2-hour tube coagulase test. J. Clin. Microbiol. 33:50-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munson, E. L., D. J. Diekema, S. E. Beekmann, K. C. Chapin, and G. V. Doern. 2003. Detection and treatment of bloodstream infection: laboratory reporting and antimicrobial management. J. Clin. Microbiol. 41:495-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikkari, S., I. J. McLaughlin, W. Bi, D. E. Dodge, and D. A. Relman. 2001. Does blood of healthy subjects contain bacterial ribosomal DNA? J. Clin. Microbiol. 39:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira, K., S. M. Brecher, A. Durbin, D. S. Shapiro, D. R. Schwartz, P. C. De Girolami, J. Dakos, G. W. Procop, D. Wilson, C. S. Hanna, G. Haase, H. Peltroche-Llacsahuanga, K. C. Chapin, M. C. Musgnug, M. H. Levi, C. Shoemaker, and H. Stender. 2003. Direct identification of Staphylococcus aureus from positive blood culture bottles. J. Clin. Microbiol. 41:889-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliveira, K., G. W. Procop, D. Wilson, J. Coull, and H. Stender. 2002. Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 40:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudolph, K. M., A. J. Parkinson, C. M. Black, and L. W. Mayer. 1993. Evaluation of polymerase chain reaction for diagnosis of pneumococcal pneumonia. J. Clin. Microbiol. 31:2661-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shrestha, N. K., M. J. Tuohy, G. S. Hall, C. M. Isada, and G. W. Procop. 2002. Rapid identification of Staphylococcus aureus and the mecA gene from BacT/ALERT blood culture bottles by using the LightCycler system. J. Clin. Microbiol. 40:2659-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder, J. W., K. S. Benzing, G. K. Munier, G. D. Bostic, P. S. Bozigar, R. Hanna, and M. J. Zervos. 2000. Clinical comparison of nonvented aerobic BacT/Alert blood culture bottle and standard aerobic bottle for detection of microorganism in blood. J. Clin. Microbiol. 38:3864-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speers, D. J., T. R. Olma, and G. L. Gilbert. 1998. Evaluation of four methods for rapid identification of Staphylococcus aureus from blood cultures. J. Clin. Microbiol. 36:1032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinstein, M. P., M. L. Towns, S. M. Quartey, S. Mirrett, L. G. Reimer, G. Parmigiani, and L. B. Reller. 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin. Infect. Dis. 24:584-602. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Y., D. J. Isaacman, R. M. Wadowsky, J. Rydquist-White, J. C. Post, and G. D. Ehrlich. 1995. Detection of Streptococcus pneumoniae in whole blood by PCR. J. Clin. Microbiol. 33:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zweig, M. H., and G. Campbell. 1993. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin. Chem. 39:561-577. [PubMed] [Google Scholar]