Abstract
Knowledge of the clonal expansion of Mycobacterium tuberculosis and accurate identification of predominant evolutionary lineages in this species remain limited, especially with regard to low-IS6110-copy-number strains. In this study, 170 M. tuberculosis isolates with ≤6 IS6110 insertions identified in Cape Town, South Africa, were characterized by principal genetic grouping, restriction fragment length polymorphism analysis, spoligotyping, IS6110 insertion site mapping, and variable-number tandem repeat (VNTR) typing. These analyses indicated that all but one of the isolates analyzed were members of principal genetic group 2 and of the same low-IS6110-copy-number lineage. The remaining isolate was a member of principal genetic group 1 and a different low-IS6110-copy-number lineage. Phylogenetic reconstruction suggests clonal expansion through sequential acquisition of additional IS6110 copies, expansion and contraction of VNTR sequences, and the deletion of specific direct-variable-repeat sequences. Furthermore, comparison of the genotypic data of 91 representative low-IS6110-copy-number isolates from Cape Town, other southern African regions, Europe, and the United States suggests that certain low-IS6110-copy-number strain spoligotypes and IS6110 fingerprints were acquired in the distant past. These clones have subsequently become widely disseminated and now play an important role in the global tuberculosis epidemic.
Sequence analysis of Mycobacterium tuberculosis strains collected from different geographical settings has shown that the frequency of mutation in this species is extremely low (31). Single-nucleotide polymorphism (SNP) and variable-number tandem repeat (VNTR)-based analyses have consistently indicated that the global population of M. tuberculosis is highly clonal (1, 15, 34). Within this clonal structure, strains can be assigned to one of three principal genetic groups according to SNPs in the katG and gyrA genes (31). Using an expanded array of synonymous SNPs (sSNPs), these principal genetic groups have been further divided into eight main clusters to depict the evolution of M. tuberculosis (15). Simulations have suggested that the branch points in the SNP-derived trees are accurate (1). However, subsequent clonal expansion occurring at each of these branch points remains largely unknown due to the lack of resolution of SNP-based analyses (1).
Various combinations of more variable markers, primarily used for molecular epidemiological studies, have been utilized to identify genotype families which fall into the different clusters defined by SNPs (15). The most commonly used marker is IS6110, a transposable element used as a probe in restriction fragment length polymorphism (RFLP) analysis of clinical isolates (35). Genetic relationships between strains have been inferred according to an IS6110 RFLP Dice similarity index of >65% and the inheritance of other specific polymorphisms. These data have been applied to depict clonal expansion in high-copy-number strains with >6 IS6110 copies (5, 37, 40, 42) and for their epidemiological analysis on a global scale (4).
However, only a limited amount of evolutionary data exists for low-IS6110-copy-number strains with ≤6 IS6110 hybridizing bands due to their intrinsically limited IS6110 RFLP polymorphism. One report has presented evidence to show evolution of the IS6110 banding pattern in the progeny of a low-IS6110-copy-number strain (22). More recently, a high degree of congruence was shown to exist between IS6110 banding patterns and other markers in strains with few IS6110 copies collected in London, United Kingdom (7). Accordingly, these strains have been classified into three different groups (7). One of these groups (group II) includes the principal genetic group 2 clusters IV and V defined by sSNP analysis, representing strains with one to three and four to six IS6110 copies, respectively (15). A second distinct group includes strains from principal genetic group 1 cluster I (15), which have been associated with patients from East Africa and Asia (6, 30). However, the process of clonal expansion within these groups of low-IS6110-copy-number strains remains largely unresolved. Moreover, the genetic relationships among strains within these groups from different geographical regions are poorly understood.
In this study, we used RFLP (42), principal genetic grouping (31), IS6110 insertion site mapping (6), spoligotyping (19), and PCR analysis of VNTRs interspersed in multiple loci (14, 23, 24, 33) to determine the genetic relationships among low-IS6110-copy-number strains collected in Cape Town, South Africa. These data have been compared to available genotypic data from low-IS6110-copy-number strains isolated from other geographical areas in order to better define the evolution of low-IS6110-copy-number strains and the impact of such evolution on the interpretation of molecular epidemiological data.
MATERIALS AND METHODS
Study setting.
Between January 1992 and December 1998, M. tuberculosis isolates were obtained from patients residing in two suburbs of Cape Town, South Africa (3), as well as a subset of patients residing in the adjoining suburbs. The two suburbs have a population of 38,500 residents within an area of 3.4 km2 and have two health care clinics. In this setting, the average annual incidence of new bacteriologically confirmed tuberculosis cases (culture and/or smear positive) was 313/100,000 (38).
DNA fingerprinting.
Genomic DNA from each isolate of M. tuberculosis was digested with either PvuII or HinfI, electrophoretically fractionated, and Southern transferred to Hybond N+ (Amersham, Little Chalfont, United Kingdom). The blots containing the PvuII-digested DNA were sequentially hybridized with probes labeled by the enhanced chemiluminescence method that were complementary to the 3′ domain of the IS6110 element (IS-3′) (35), the 5′ domain of the IS6110 element (IS-5′) (42), the direct repeat (DR) (42), and Marker X (Roche, Basel, Switzerland). Each probe was stripped by denaturation before the next probe was applied. The HinfI Southern blots were hybridized with the 32P-labeled MTB484(1) probe complementary to the polymorphic G+C-rich repeat sequences (PGRS) (41). The autoradiographs were normalized, and the IS-3′, IS-5′, and DR bands were assigned using GelCompar II software. Cluster analysis was done using the unweighted pair group method with arithmetic mean and the Dice coefficient (17). Mutations in the IS6110-flanking domains were determined as previously described (43). The band corresponding to the IS6110 insertion in the DR region was identified by aligning the DR and IS-3′ autoradiographs (42). The blots probed by MTB484 (1) were visually analyzed by two independent persons (42).
PCR amplification was used to determine the presence or absence of an IS6110 insertion in the genes Rv0403c, Rv1758, and Rv3018c according to the previously described method (6). IS6110 insertion in the gene Rv2787c was determined using the primer set 5′-TTCAACCATCGCCGCCTCTAC-3′ and 5′-GGCCAAATCCAGCACGGTGAAC-3′.
Mutation analysis.
The M. tuberculosis isolates were classified into three principal genetic groups according to polymorphisms in the katG and gyrA genes (31), using the dot blot hybridization method (39).
Spoligotyping.
DNA polymorphism in the DR locus was detected in isolates with ≤6 IS6110 insertion elements by spoligotyping according to a standardized protocol (19).
MIRU-VNTR typing.
M. tuberculosis isolates were genotyped by PCR amplification of the 12 loci containing VNTRs of elements called mycobacterial interspersed repetitive units (MIRUs) (33) and 9 loci containing VNTRs of other interspersed sequences (14, 21, 24; P. Supply, S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht, unpublished data) using both manual (33) and automated (32) techniques. The primers against the MIRU-VNTR flanking regions were the same as those previously described (33), except that Hex labeling was replaced by Vic labeling. The primers against the other loci are described in Table 1. The samples were subjected to electrophoresis using a 96-well ABI 377 automated sequencer as previously described (32). Sizing of the PCR fragments and assignment of the various VNTR alleles were done using the GeneScan and Genotyper software packages (PE Applied Biosystems) as previously described (32) and based on the data in Table 1. The tables used for VNTR allele scoring are available at http://www.ibl.fr/mirus/mirus.html. Allele assignments in the manual and automated methods were identical.
TABLE 1.
Conditions for multiplex PCRs of nine VNTR loci
| Multiplex | Conventional designationa | VNTR length (bp) | MgCl2 (mM) | PCR primer pairs (5′ to 3′, with labeling indicated) |
|---|---|---|---|---|
| Mix E | VNTR 2347 | 57 | 1.5 | GCCAGCCGCCGTGCATAAACCT (FAM) |
| AGCCACCCGGTGTGCCTTGTATGAC | ||||
| VNTR 2461 | 57 | ATGGCCACCCGATACCGCTTCAGT (VIC) | ||
| CGACGGGCCATCTTGGATCAGCTAC | ||||
| VNTR 3171 | 54 | GGTGCGCACCTGCTCCAGATAA (NED) | ||
| GGCTCTCATTGCTGGAGGGTTGTAC | ||||
| Mix F | VNTR 0424 | 51 | 1.5 | CTTGGCCGGCATCAAGCGCATTATT |
| GGCAGCAGAGCCCGGGATTCTTC (FAM) | ||||
| VNTR 0577 | 58 | CGAGAGTGGCAGTGGCGGTTATCT (VIC) | ||
| AATGACTTGAACGCGCAAATTGTGA | ||||
| VNTR 1895 | 57 | GTGAGCAGGCCCAGCAGACT (NED) | ||
| CCACGAAATGTTCAAACACCTCAAT | ||||
| Mix G | VNTR 2401 | 58 | 3.0 | CTTGAAGCCCCGGTCTCATCTGT (FAM) |
| ACTTGAACCCCACGCCCATTAGTA | ||||
| VNTR 3690 | 58 | CGGTGGAGGCGATGAACGTCTTC (VIC) | ||
| TAGAGCGGCACGGGGGAAAGCTTAG | ||||
| VNTR 4156 | 59 | TGACCACGGATTGCTCTAGT | ||
| GCCGGCGTCCATGTT (NED) |
Global dissemination.
To determine the geographical spread of the low-IS6110-copy-number strains, M. tuberculosis isolates collected from southern (Western Cape) (n = 47), central (Free State) (n = 4), and northern (Gauteng and Mpumalanga) (n = 4) regions in South Africa and from Harare and Gweru (n = 11) in Zimbabwe were subjected to spoligotyping, IS6110 RFLP, and IS6110 insertion mapping (6). These genotypic data were compared to previously published genotypic data (IS6110 banding patterns [visual comparison], IS6110 insertion points, and spoligotype patterns) for low-IS6110-copy-number strains from Europe (United Kingdom [n = 14] [7] and Denmark [n = 2] [2]) and the United States (Michigan [n = 70] [6] and CDC1551 [12]).
Genetic-relationship analysis.
The evolutionary state(s) for the RFLP data was assigned according to the presence (indicated by 1) or the absence (indicated by 0) of a hybridizing band. Spoligotypes were assigned according to the presence or absence of spacer sequences, while the VNTR alleles were assigned according to the number of repeats present in the different loci. The complete set of evolutionary states for the different markers was subjected to phylogenetic analysis using the neighbor-joining algorithm Phylogenetic Analysis Using Parsimony (*Other Methods) version 4b10 (Sinauer Associates, Sunderland, Mass.). Bootstrapping was performed to establish a degree of statistical support for nodes within each phylogenetic reconstruction (10). A consensus tree was generated using the program contree [Phylogenetic Analysis Using Parsimony (*Other Methods) version 4b10] in combination with the majority rule formula. The resulting trees were rooted to the principal genetic group 1 isolate [SA CT(67)], a designation derived from the isolate’s origin city of Cape Town, South Africa. Only branches which occurred in >50% of the bootstrap trees were included in the final tree, and all branches with a zero branch length were collapsed.
RESULTS
Selection and molecular characterization of strains.
Between January 1992 and December 1998, M. tuberculosis isolates were obtained from 1,030 patients resident in the suburbs adjoining Cape Town, South Africa. IS6110 RFLP analysis established that 186 (18.1%) of these patients were infected with a strain containing ≤6 IS6110 hybridizing bands. No isolate lacking the IS6110 element was identified in this study setting. Isolates were available from 170 (91%) of these patients for further genotypic analysis.
Analysis of the katG and gyrA gene sequences classified 169 of these isolates in principal genetic group 2, while only one isolate was classified as principal genetic group 1 (31). All isolates were then subjected to further analysis using Southern hybridization in combination with probes complementary to IS-3′ (35) (Fig. 1) and IS-5′ (42), spoligotyping (19) (Table 2), and VNTR allele typing based on 21 independent loci (14, 21, 25, 32, 33) and PGRS RFLP typing (41) (Table 3). This set included MIRU-VNTR loci 2, 4 (ETR-D), 10, 16, 20, 23, 24, 26, 27, 31 (ETR-E), 39, and 40 and VNTR loci 424, 577 (ETR-C), 1895 (QUB-1895), 2347, 2401, 2461(ETR-B), 3171, 3690, and 4156 (QUB-4156) (alias designations are in parentheses). In addition, IS6110 insertion into the genes Rv0403c, Rv1758, Rv3018c, and Rv2787c was determined by PCR amplification (Fig. 1A). Table 3 summarizes the genotypic data of the different isolates as defined by the combined markers. For this data set, the numbers of principal genetic group 2 genotypes obtained with the different methods were ranked as follows: IS-3′ (14 genotypes) < spoligotypes (19 genotypes) = IS-5′ (19 genotypes) < IS-3′ and IS-5′ (22 genotypes) < VNTR loci (38 genotypes) < PGRS (45 genotypes). In combination, these different genotyping methods identified a total of 66 distinct principal genetic group 2 genotypes (Table 3).
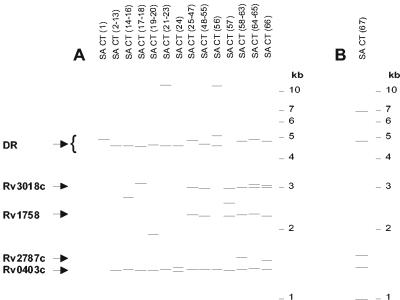
FIG. 1.
Southern blot analysis of Cape Town M. tuberculosis isolates with ≤6 IS6110 insertions. Genomic DNA was restricted with PvuII and electrophoretically fractionated on agarose gels, and Southern hybridization was done with an enhanced-chemiluminescence-labeled IS-3′ probe. (A) IS6110 banding pattern of distinct principal genetic group 2 isolates from Cape Town, South Africa. The lanes are labeled according to the isolate names in Table 3. The band representing the IS6110 insertion in the DR region and bands representing IS6110 insertions in genes Rv0403c, Rv1758, Rv2787c, and Rv3018c are indicated. (B) IS6110 banding pattern of the principal genetic group 1 isolate from Cape Town, South Africa. The lane is labeled according to the isolate name given in Table 3. Cohybridization between the IS6110 and DR probes could not be demonstrated due to the presence of a PvuII site situated between the 3′ domain of the IS6110 element and the flanking DR region.
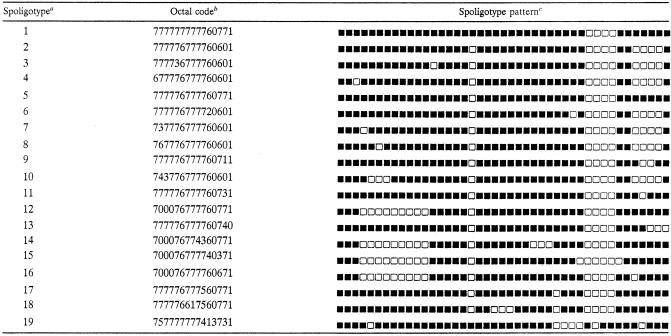
TABLE 2.
Spoligotype patterns of Cape Town M. tuberculosis isolates with ≤6 IS6110 elements
TABLE 3.
Genotypic classification of Cape Town M. tuberculosis isolates with ≤6 IS6110 elements
| Name | Principal genetic group | IS6110 copy no. | IS-3′ type | IS-5′ type | PGRS type | 12 MIRU-VNTR type (allele combination)a | 9 VNTR allele combinationb | Spoligotype (octal format)c |
|---|---|---|---|---|---|---|---|---|
| SA CT(1) | 2 | 1 | 1 | 1 | 1 | 1 (225125113322) | 144442353 | 1 (777777777760771) |
| SA CT(2) | 2 | 2 | 2 | 2 | 2 | 2 (223325153323) | 142442383 | 2 (777776777760601) |
| SA CT(3) | 2 | 2 | 2 | 2 | 3 | 3 (224325123422) | 242442343 | 3 (777736777760601) |
| SA CT(4) | 2 | 2 | 2 | 2 | 3 | 3 (224325123422) | 242442343 | 2 (777776777760601) |
| SA CT(5) | 2 | 2 | 2 | 2 | 4 | 4 (224325143223) | ND | 3 (777736777760601) |
| SA CT(6) | 2 | 2 | 2 | 2 | 4 | 4 (224325143223) | 442442333 | 2 (777776777760601) |
| SA CT(7) | 2 | 2 | 2 | 2 | 5 | 5 (224325143324) | 242442343 | 4 (677776777760601) |
| SA CT(8) | 2 | 2 | 2 | 2 | 5 | 5 (224325143324) | 242442343 | 3 (777736777760601) |
| SA CT(9) | 2 | 2 | 2 | 2 | 5 | 5 (224325143324) | 242442343 | 2 (777776777760601) |
| SA CT(10-11) | 2 | 2 | 2 | 2 | 6, 7 | 6 (224325153223) | 442442431 | 2 (777776777760601) |
| SA CT(12) | 2 | 2 | 2 | 2 | 2 | 7 (224325153323) | 142442383 | 2 (777776777760601) |
| SA CT(13) | 2 | 2 | 2 | 2 | 2 | 7 (224325153323) | 142442383 | 5 (777776777760771) |
| SA CT(14) | 2 | 3 | 3 | 3d | 8 | 8 (223325143323) | 2424422?3 | 6 (777776777720601) |
| SA CT(15) | 2 | 3 | 3 | 4 | 9 | 9 (224325133324) | 442442333 | 7 (737776777760601) |
| SA CT(16) | 2 | 3 | 3 | 4 | 9 | 10 (224325153323) | 442442333 | 7 (737776777760601) |
| SA CT(17) | 2 | 3 | 4 | 5 | 10 | 11 (223325163322) | 242442333 | 8 (767776777760601) |
| SA CT(18) | 2 | 3 | 4 | 5 | 10 | 12 (224325163322) | 242442333 | 8 (767776777760601) |
| SA CT(19) | 2 | 3 | 5 | 6 | 11 | 13 (224325153222) | 522442332 | 9 (777776777760711) |
| SA CT(20) | 2 | 3 | 5 | 7d | 12 | 14 (224325153324) | 252441343 | 10 (743776777760601) |
| SA CT(21) | 2 | 3 | 6d | 5 | 13 | 15 (223325153223) | 442442333 | 2 (777776777760601) |
| SA CT(22) | 2 | 3 | 6d | 5 | 14 | 10 (224325153323) | 442442333 | 2 (777776777760601) |
| SA CT(23) | 2 | 3 | 6d | 5 | 14 | 16 (224325153323) | 452442333 | 2 (777776777760601) |
| SA CT(24) | 2 | 3 | 7 | 8 | 15 | 17 (223325163433) | ND | 2 (777776777760601) |
| SA CT(25) | 2 | 4 | 8 | 9 | 16 | 18 (223325143322) | 432442333 | 5 (777776777760771) |
| SA CT(26-30) | 2 | 4 | 8 | 9 | 16, 17, 18, 19, 20 | 19 (223325143324) | 432442333 | 5 (777776777760771) |
| SA CT(31) | 2 | 4 | 8 | 9 | 19 | 20 (223325143325) | 432442333 | 5 (777776777760771) |
| SA CT(32-34) | 2 | 4 | 8 | 9 | 17, 21, 22 | 21 (223325153324) | 432442333 | 11 (777776777760731) |
| SA CT(35-36) | 2 | 4 | 8 | 10 | 23, 24 | 22 (223325153224) | 332442343 | 12 (700076777760771) |
| SA CT(37) | 2 | 4 | 8 | 10 | 23 | 23 (224325153322) | 432442343 | 12 (700076777760771) |
| SA CT(38-44) | 2 | 4 | 8 | 10 | 23, 25, 26, 27, 28, 29, 30 | 24 (224325153324) | 432442343 | 12 (700076777760771) |
| SA CT(45) | 2 | 4 | 8 | 10 | 31 | 24 (224325153324) | ND | 5 (777776777760771) |
| SA CT(46-47) | 2 | 4 | 8 | 11 | 32, 33 | 25 (224325153324) | 332442331 | 12 (700076777760771) |
| SA CT(48) | 2 | 4 | 9 | 12d | 34 | 26 (224325153324) | 432442333 | 13 (777776777760740) |
| SA CT(49-50) | 2 | 4 | 9 | 9 | 34, 35 | 21 (223325153324) | 432442333 | 13 (777776777760740) |
| SA CT(51-52) | 2 | 4 | 9 | 9 | 34, 35 | 27 (224325133324) | 432442333 | 13 (777776777760740) |
| SA CT(53) | 2 | 4 | 9 | 9 | 34 | 28 (226325153324) | 432442333 | 13 (777776777760740) |
| SA CT(54) | 2 | 4 | 9 | 10 | 36 | 24 (224325153324) | 432442343 | 14 (700076774360771) |
| SA CT(55) | 2 | 4 | 9 | 10 | 37 | 29 (224325153325) | 432442343 | 15 (700076777740371) |
| SA CT(56) | 2 | 4 | 10 | 13 | 38 | 30 (224325153434) | ND | 2 (777776777760601) |
| SA CT(57) | 2 | 4 | 11 | 14 | 39 | 31 (224225164434) | ND | 16 (700076777760671) |
| SA CT(58) | 2 | 5 | 12 | 15d | 40 | 32 (224325143324) | 432442334 | 5 (777776777760771) |
| SA CT(59) | 2 | 5 | 12 | 16 | 41 | 33 (224325153322) | 432442333 | 17 (777776777560771) |
| SA CT(60) | 2 | 5 | 12 | 16 | 42 | 33 (224325153322) | 432442333 | 15 (777776777760771) |
| SA CT(61) | 2 | 5 | 12 | 16 | 41 | 34 (234325133323) | 432442333 | 17 (777776777560771) |
| SA CT(62) | 2 | 5 | 12 | 16 | 41 | 35 (234325153323) | ND | 5 (777776777760771) |
| SA CT(63) | 2 | 5 | 12 | 17 | 41 | 33 (224325153322) | 432442333 | 18 (777776617560771) |
| SA CT(64) | 2 | 5 | 13 | 18 | 43 | 36 (224325153222) | 532442433 | 12 (700076777760771) |
| SA CT(65) | 2 | 5 | 13 | 18 | 44 | 37 (224325153222) | 532442333 | 12 (700076777760771) |
| SA CT(66) | 2 | 6 | 14 | 19 | 45 | 38 (224325164335) | ND | 12 (700076777760771) |
| SA CT(67) | 1 | 5 | 15 | 20 | 46 | 39 (254316?34613) | ND | 19 (757777777413731) |
MIRU-VNTR loci according to reference 33.
VNTR loci 424, 577, 1895, 2347, 2401, 2461, 3171, 3690, and 4156 (see Table 1 and references 14 and 24 and P. Supply et al., unpublished data). ND, not determined; ?, missing VNTR allele.
Spoligotypes are arbitrary designations to demonstrate genetic diversity, and octal formats are according to reference 8.
Mutations in the IS6110-flanking domains other than in the DR region (43).
Clonal expansion of strains.
Combined analysis of the above-mentioned different molecular markers strongly supported the close genetic relatedness of the principal genetic group 2 isolates and their clear distinctness from the single isolate from principal genetic group 1. Six out of 21 VNTR loci (MIRU-VNTR loci 2, 20, 23, and 24 and VNTR loci 2347 and 2401) were fully identical within principal genetic group 2 isolates analyzed (Table 3), while 7 other loci (MIRU-VNTR loci 4, 16, 27, and 39 and VNTR loci 1895, 2461, and 3171) displayed at most three variations compared to the predominant alleles among these isolates. PCR amplification of the Rv0403c region (Fig. 1A) showed that only the principal genetic group 2 variants with ≥2 and ≤6 IS6110 elements had an IS6110 in the same position, suggesting that these isolates were derived from a common ancestor. All principal genetic group 2 spoligotypes showed a deletion of direct-variable-repeat (DVR) sequences 33 to 36 (29), while isolates with ≥2 and ≤6 IS6110 insertions also all showed an additional DVR 18 deletion (Table 2). DVR 34 was deleted in the principal genetic group 1 isolate (Table 2).
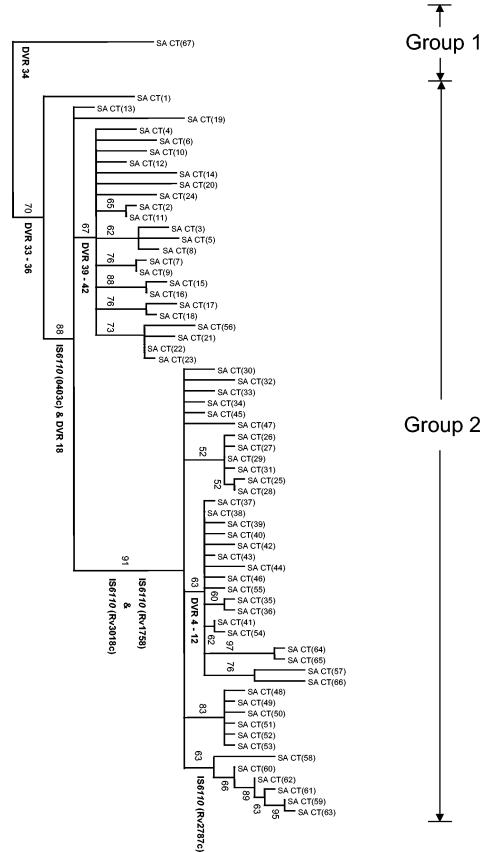
Phylogenetic analysis of the principal genetic group 2 isolates was done based on the whole set of markers, using the principal genetic group 1 member [SA CT(67)] as an outgroup (Fig. 2). The overall branching order of the tree suggests that the principal genetic group 2 isolates evolved from a common progenitor by sequential replicative transposition of IS6110, followed, in certain cases, by mutation in the regions flanking the IS6110 elements (Table 3). According to this tree, these clones first evolved by replicative transposition of the IS6110 element into Rv0403c (Fig. 1A) and by the deletion of DVR 18 to generate a strain with two IS6110 insertions (Fig. 2). These two genotypic characteristics, along with the six conserved VNTR loci, were subsequently inherited in all the progeny. In different branches of these progeny, subsequent IS6110 insertions were identified in Rv1758, Rv3018c, and Rv2787c to generate clonal variants with between three and six IS6110 insertions (Fig. 2). Along with these events occurred the deletion of DVRs 39 to 42 and 4 to 12 (Table 2 and Fig. 2). Broadly similar pictures of stepwise acquisition of IS6110 were obtained by phylogenetic analysis using either IS6110 RFLP, spoligotyping, and PGRS or VNTR genotypes alone (data not shown).
FIG.2.
Phylogenetic tree of Cape Town M. tuberculosis isolates with ≤6 IS6110 insertions. Genetic data from five different genotyping methods were subjected to phylogenetic analysis using the bootstrapping and neighbor-joining algorithms (see Materials and Methods). The tree was rooted to the principal genetic group 1 clone [isolate SA CT(67)]. Booststrap values are given at internal nodes. IS6110 insertions in defined genes, as well as DVR deletions, are indicated at the nodes where they occurred. All branches with zero length were collapsed. Isolate names are the same as in Table 3. Principal genetic groups 1 and 2 were assigned according to polymorphisms in the katG and gyrA genes (31). The scale indicates the number of steps per unit length.
Geographical distribution of strains.
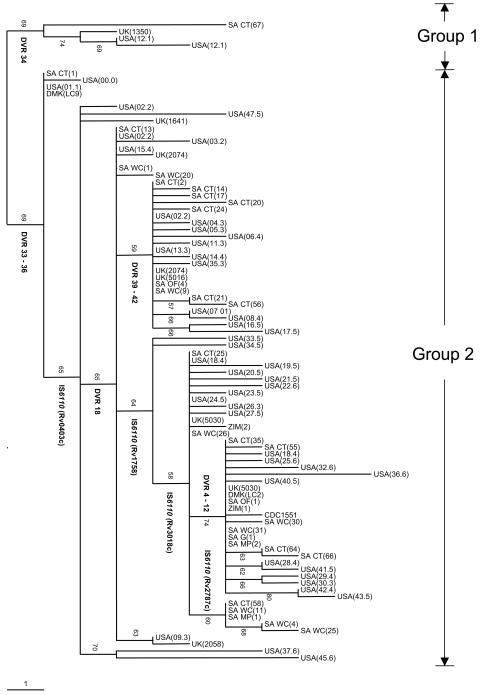
To determine the evolutionary relationships among the low-IS6110-copy-number strains in different geographical regions, the genotypic data from a representative set of the Cape Town isolates (n = 16) were compared with those of a representative set of low-IS6110-copy-number isolates from other regions of southern Africa (n = 17) (this study), Europe (London, United Kingdom [n = 8] [7] and Denmark [n = 2] [2]), and the United States (Michigan [n = 47] [6] and the CDC1551 reference strain [12]). Phylogenetic analysis using these data indicated a close evolutionary relationship among the principal genetic group 2 isolates from these different geographical regions (Fig. 3). Many of the isolates in the different settings shared identical IS-3′ banding patterns, IS6110 insertion points, and spoligotypes, and only in rare instances were IS6110 transposition and DVR deletion events found to be unique to a specific geographic region (Fig. 3; IS6110 insertion in Rv2787c of SA isolates). These observations suggest that these genotype properties were acquired in the distant past prior to global dissemination of the lineage. In contrast, comparison of MIRU-VNTR genotypes based on 12 loci, in common with those previously reported (6), failed to identify clones with IS-3′, spoligotype, and MIRU-VNTR genotypes identical between Cape Town and Michigan, indicating that the MIRU-VNTR loci are evolving more rapidly. A similar study could not be done for the PGRS genotypes, as the methodology has not been internationally standardized.
FIG. 3.
Phylogenetic tree of M. tuberculosis isolates with ≤6 IS6110 insertions obtained from different geographical regions. Representative isolates (n = 91) cultured in Cape Town, South Africa; Europe (2, 7); and the United States (6, 12) were included. Genetic data from IS6110 banding patterns, IS6110 insertion points, and spoligotyping were subjected to phylogenetic analysis using the bootstrapping and neighbor-joining algorithms. The tree was rooted to the principal genetic group 1 isolate [SA CT(67)]. Booststrap values are given at internal nodes. IS6110 insertions in defined genes, as well as DVR deletions, are indicated at the nodes where they occurred. All branches with zero length were collapsed. The Cape Town isolate names are the same as in Table 3, while the isolates from other regions of southern Africa are labeled as follows: SA WC (South Africa Western Cape), SA OF (South Africa Free Sate), SA G (South Africa Gauteng), SA MP (South Africa Mpumalanga), and ZIM (Zimbabwe). The isolates from Europe are labeled according to the system in references 7 and 2, while the isolates from the United States are labeled according to the system in references 6 and 12. Principal genetic groups 1 and 2 were assigned according to polymorphisms in the katG and gyrA genes (31). The scale indicates the number of steps per unit length.
DISCUSSION
This study provides evidence that nearly all M. tuberculosis isolates with ≤6 IS6110 elements collected in Cape Town, South Africa, are members of a lineage of the principal genetic group 2. Evidence for this is based on the inheritance of defined polymorphisms, which include (i) principal genetic group 2 classification according to mutations in the katG and gyrA genes (31) and concordant deletion of DVRs 33 to 36, known to be specific to principal genetic group 2 and 3 strains (29); (ii) the identification of a conserved IS6110 insertion in Rv0403c (13) and the deletion of DVR 18 from the DR region in principal genetic group 2 strains with ≥2 and ≤6 IS6110 insertions; and (iii) the presence of six fully conserved VNTR loci. Only one isolate from this setting was identified as being a member of a distinct low-IS6110-copy-number lineage of the principal genetic group 1, which corresponds to cluster I (15) or group I (7) and has been primarily associated with patients from East Africa and Asia (6, 30).
The principal genetic group 2 lineage studied here encompasses the groups referred to as groups II and III (7) or clade X (27) and clusters IV and V defined by sSNP analysis (15). Our phylogenetic analysis, based on fully independent markers in isolates from different geographical areas, supports the notion that strains in this principal genetic group 2 lineage evolved from a common progenitor containing a single IS6110 element by sequential acquisition of up to five additional IS6110 copies, as well as by expansion and contraction of VNTR sequences and the deletion of specific DVRs. Sequential acquisition of additional IS6110 copies is consistent with the direct evolutionary relationship between the sSNP clusters IV and V, which include strains with one to three and four to six IS6110 copies, respectively (15). Such congruence between phylogenies inferred from independent sets of markers (within our study or between our study and that of Gutacker et al. [15]) provides strong evidence for the robustness of the inferred phylogeny. Moreover, the deletion of DVRs suggested by our phylogenetic analysis (Fig. 2 and 3) is consistent with previous findings supporting the notion that evolution of the DR region is driven by loss of DVR sequences rather than by their duplication (9, 36, 44).
Interestingly, each IS6110 transposition event appeared to occur only once within the phylogenetic tree, suggesting divergent evolution. This is in sharp contrast to a previous suggestion that the IS6110 banding patterns of low-IS6110-copy-number strains could have evolved convergently due to the presence of preferential IS6110 integration sites (13). The limited number of IS6110 variants identified may suggest that IS6110 transposition is regulated in this lineage, raising the hypothesis of lineage-specific effects. Regulation of the number of transposable elements, referred to as taming, has been described in eukaryotic genomes and might be a specific mechanism against mutagenic effects induced by these elements (18).
The preservation of certain IS-3′ banding patterns and spoligotypes in isolates from Cape Town, other southern African regions, Europe, and the United States suggests that these markers have remained stable over a long period of time. Therefore, we hypothesize that these genotypes represent clones that evolved in the distant past and have become globally disseminated. Examination of the SpolD database (11) indicates that principal genetic group 2 isolates with the characteristic DVR 33-to-36 and DVR 18 deletions have been isolated in 27 different countries. By comparison, principal genetic group 1 clones with the characteristic DVR 34 deletion have been isolated in 26 countries, with a high prevalence in South Asia. Taken together, these findings suggest that in addition to other well-identified lineages, like W-Beijing (4), the principal genetic group 1 and 2 low-IS6110-copy-number lineages now play an important role in the global tuberculosis epidemic.
The inferred stability of the IS-3′ banding patterns and of certain spoligotypes for extended periods of time is likely too high to be informative for tracking ongoing transmission between patients in settings where this lineage is predominant. Conversely, comparison of our genotype data with those of Cowan et al. (6) failed to identify strains from Cape Town and Michigan in which the IS-3′ banding pattern, spoligotype, and MIRU-VNTR types were identical. Given the stability of MIRU-VNTR genotypes in epidemiologically linked isolates (16, 20, 23, 26), the absence of MIRU-VNTR matching between the two studies is in accordance with the above argument of distant relationships between shared IS-3′ types and spoligotypes. Moreover, it supports VNTR typing as a useful tool for epidemiological tracking across various epidemiological settings and bacterial populations. This is consistent with the contention that, as a multilocus-based method, VNTR typing is much less exposed to biases inherent in single loci or in copy numbers of a single genetic element, such as spoligotyping and IS6110-based typing, respectively.
Our study represents a step toward a better understanding of the evolutionary mechanisms modeling the genome in different M. tuberculosis lineages and of the different rates at which these events occur. This will provide new insights for the interpretation of molecular epidemiological data and enhance our understanding of how different strains contribute to the tuberculosis epidemic in specific regions and on a global scale.
Acknowledgments
This study was made possible by grants from the GlaxoSmithKline Action TB Initiative, IAEA (projects SAF6/003 and CRP 9925), the Harry Crossely Foundation, and the National Research Foundation (project 2054201) and from the Ministère de la Recherche and Ministère des Affaires Etrangères and the South African Medical Research Council (MRC). The work was also supported by Institut National de la Santé et de la Recherche Médicale (INSERM), Institut Pasteur de Lille, Région Nord-Pas-de-Calais. P.S. is a Chercheur du Centre National de Recherche Scientifique.
We thank E. Engelke, S. Carlini, M. De Kock, and Frederique De Matos for their technical assistance. We thank S. Charalambous for the provision of clinical isolates collected in the Free State, South Africa.
REFERENCES
- 1.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, J., A. B. Andersen, K. Kremer, and H. Miorner. 1999. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J. Clin. Microbiol. 37:2602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyers, N., R. P. Gie, H. L. Zietsman, M. Kunneke, J. Hauman, M. Tatley, and P. R. Donald. 1996. The use of a geographical information system (GIS) to evaluate the distribution of tuberculosis in a high-incidence community. S. Afr. Med. J. 86:40-44. [PubMed] [Google Scholar]
- 4.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 5.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. L. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreiswirth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-457. [PubMed] [Google Scholar]
- 6.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, J. W., H. Al Ghusein, S. Al Hashmi, P. Butcher, A. L. Dickens, F. Drobniewski, K. J. Forbes, S. H. Gillespie, D. Lamprecht, T. D. McHugh, R. Pitman, N. Rastogi, A. T. Smith, C. Sola, and H. Yesilkaya. 2003. Evolutionary relationships among strains of Mycobacterium tuberculosis with few copies of IS6110. J. Bacteriol. 185:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins, J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. Quitugua, N. Rastogi, R. A. Skuce, C. Sola, D. van Soolingen, and V. Vincent. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis. 5:216-219. [PubMed] [Google Scholar]
- 9.Fang, Z., N. Morrison, B. Watt, C. Doig, and K. J. Forbes. 1998. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J. Bacteriol. 180:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-793. [DOI] [PubMed] [Google Scholar]
- 11.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. A. Dang, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, M. L. Ho, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, J. W. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fomukong, N., M. Beggs, H. el Hajj, G. Templeton, K. Eisenach, and M. D. Cave. 1997. Differences in the prevalence of IS6110 insertion sites in Mycobacterium tuberculosis strains: low and high copy number of IS6110. Tuber. Lung Dis. 78:109-116. [DOI] [PubMed] [Google Scholar]
- 14.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 15.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms. Resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohamed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 41:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermans, P. W., F. Messadi, H. Guebrexabher, D. van Soolingen, P. E. de Haas, H. Heersma, H. de Neeling, A. Ayoub, F. Portaels, and D. Frommel. 1995. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J. Infect. Dis. 171:1504-1513. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, S., M. P. Gassama, and T. Heidmann. 1999. Taming of transposable elements by homology-dependent gene silencing. Nat. Genet. 21:209-212. [DOI] [PubMed] [Google Scholar]
- 19.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. Van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, H. S. Roahen, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Fleche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathema, B., P. J. Bifani, J. Driscoll, L. Steinlein, N. Kurepina, S. L. Moghazeh, E. Shashkina, S. A. Marras, S. Campbell, B. Mangura, K. Shilkret, J. T. Crawford, R. Frothingham, and B. N. Kreiswirth. 2002. Identification and evolution of an IS6110 low-copy-number Mycobacterium tuberculosis cluster. J. Infect. Dis. 185:641-649. [DOI] [PubMed] [Google Scholar]
- 23.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roring, S., D. Brittain, A. E. Bunschoten, M. S. Hughes, R. A. Skuce, J. D. van Embden, and S. D. Neill. 1998. Spacer oligotyping of Mycobacterium bovis isolates compared to typing by restriction fragment length polymorphism using PGRS, DR and IS6110 probes. Vet. Microbiol. 61:111-120. [DOI] [PubMed] [Google Scholar]
- 25.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebban, M., I. Mokrousov, N. Rastogi, and C. Sola. 2002. A data-mining approach to spacer oligonucleotide typing of Mycobacterium tuberculosis. Bioinformatics 18:235-243. [DOI] [PubMed] [Google Scholar]
- 28.Smittipat, N., and P. Palittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber. Lung Dis. 80:69-74. [DOI] [PubMed] [Google Scholar]
- 29.Soini, H., X. Pan, A. Amin, E. A. Graviss, A. Siddiqui, and J. M. Musser. 2000. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J. Clin. Microbiol. 38:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soini, H., X. Pan, L. Teeter, J. M. Musser, and E. A. Graviss. 2001. Transmission dynamics and molecular characterization of. Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J. Clin. Microbiol. 39:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 34.Supply, P., R. M. Warren, A. L. Banuls, S. Lesjean, G. D. van der Spuy, L. A. Lewis, M. Tibayrenc, P. D. van Helden, and C. Locht. 2003. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 47:529-538. [DOI] [PubMed] [Google Scholar]
- 35.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. Der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verver, S., R. M. Warren, Z. Munch, E. Vynnycky, P. D. van Helden, M. Richardson, G. D. van der Spuy, D. A. Enarson, M. W. Borgdorff, M. A. Behr, and N. Beyers. 2004. Transmission of tuberculosis in a high incidence urban community in South Africa. Int. J. Epidemiol. 33:351-357. [DOI] [PubMed] [Google Scholar]
- 39.Victor, T. C., A. M. Jordaan, A. van Rie, G. D. van der Spuy, M. Richardson, P. D. van Helden, and R. Warren. 1999. Detection of mutations in drug resistance genes of Mycobacterium tuberculosis by a dot-blot hybridization strategy. Tuber. Lung Dis. 79:343-348. [DOI] [PubMed] [Google Scholar]
- 40.Victor, T. C., A. van Rie, A. M. Jordaan, M. Richardson, G. D. Der Spuy, N. Beyers, P. D. van Helden, and R. Warren. 2001. Sequence polymorphism in the rrs gene of Mycobacterium tuberculosis is deeply rooted within an evolutionary clade and is not associated with streptomycin resistance. J. Clin. Microbiol. 39:4184-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren, R., M. Richardson, S. Sampson, J. H. Hauman, N. Beyers, P. R. Donald, and P. D. van Helden. 1996. Genotyping of Mycobacterium tuberculosis with additional markers enhances accuracy in epidemiological studies. J. Clin. Microbiol. 34:2219-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren, R. M., M. Richardson, S. L. Sampson, G. D. van der Spuy, W. Bourn, J. H. Hauman, H. Heersma, W. Hide, N. Beyers, and P. D. van Helden. 2001. Molecular evolution of Mycobacterium tuberculosis: phylogenetic reconstruction of clonal expansion. Tuberculosis 81:291-302. [DOI] [PubMed] [Google Scholar]
- 43.Warren, R. M., S. L. Sampson, M. Richardson, G. D. van der Spuy, C. J. Lombard, T. C. Victor, and P. D. van Helden. 2000. Mapping of IS6110 flanking regions in clinical isolates of M. tuberculosis demonstrates genome plasticity. Mol. Microbiol. 37:1405-1416. [DOI] [PubMed] [Google Scholar]
- 44.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]