Abstract
Streptomyces strains are known for their capability to produce a lot of different compounds with various bioactivities. Cultivation under different conditions often leads to the production of new compounds. Therefore, production cultures of the strains are extracted with ethyl acetate and the crude extracts are analyzed by HPLC. Furthermore, the extracts are tested for their bioactivity by different assays. For structure elucidation the compound of interest is purified by a combination of different chromatography methods.
Genome sequencing coupled with genome mining allows the identification of a natural product biosynthetic gene cluster using different computer programs. To confirm that the correct gene cluster has been identified, gene inactivation experiments have to be performed. The resulting mutants are analyzed for the production of the particular natural product. Once the correct gene cluster has been inactivated, the strain should fail to produce the compound.
The workflow is shown for the antibacterial compound polyketomycin produced by Streptomyces diastatochromogenes Tü6028. Around ten years ago, when genome sequencing was still very expensive, the cloning and identification of a gene cluster was a very time-consuming process. Fast genome sequencing combined with genome mining accelerates the trial of cluster identification and opens up new ways to explore biosynthesis and to generate novel natural products by genetic methods. The protocol described in this paper can be assigned to any other compound derived from a Streptomyces strain or another microorganism.
Keywords: Genetics, Issue 119, natural products, polyketomycin, Streptomyces diastatochromogenes Tü6028, extraction, purification, HPLC, biosynthetic gene cluster, genome sequencing, genome mining, single crossover.
Introduction
Natural products from plants and microorganisms have always been an important source for clinical drug development and research. The first antibiotic Penicillin was discovered in 1928 from a fungus by Alexander Fleming1. Nowadays, many more natural products are used in clinical treatment.
One genus known for its capability of producing various kinds of secondary metabolites with different bioactivities is Streptomyces. Streptomyces are Gram-positive bacteria and belong to the class of Actinobacteria and the order Actinomycetales. Almost two-thirds of the clinically used antibiotics are derived from Actinomycetales, mainly from Streptomyces, like amphotericin2, daptomycin3 or tetracycline4. Two Nobel Prizes have been awarded in the field of Streptomyces antibiotic research. The first one went to Selman Waksman for the discovery of Streptomycin, the first antibiotic effective against tuberculosis.5 In 2015, as part of the Nobel Prize in Physiology and Medicine, the discovery of avermectin from S. avermitilis was awarded as well. Avermectin is used for the treatment of parasitic diseases6,7.
The traditional approach for the discovery of natural products in microorganisms such as Streptomyces generally involves cultivation of the strain under different growth conditions, as well as extraction and analysis of secondary metabolites. Bioactivity assays (e.g. assays for antibacterial and anticancer activity) are performed to detect the activity of the compound. Finally, the compound of interest is isolated and the chemical structure is elucidated.
The structures of natural products are often composed of single moieties which are forming complex molecules. There are a few, but limited, major biosynthetic pathways leading to building blocks, which are used for the biosynthesis of natural products. The major biosynthetic pathways are the polyketide pathways, pathways leading to terpenoids and alkaloids, pathways using amino acids, and pathways leading to sugar moieties. Each pathway is characterized by a set of specific enzymes8. Based on the structure of the compound, these biosynthetic enzymes can be predicted.
Nowadays, the detailed structural analysis of a compound in combination with next generation sequencing and bioinformatic analysis can help to identify the responsible biosynthetic gene cluster. The cluster information opens up new ways for further natural product research. This includes heterologous expression to increase the yield of the natural product, targeted compound modification by gene deletion or alteration and combinatorial biosynthesis with genes from other pathways.
Polyketomycin was isolated independently from the culture broth of two strains, Streptomyces sp. MK277-AF19 and Streptomyces diastatochromogenes Tü602810. The structure was elucidated by NMR and X-ray analysis. Polyketomycin is composed of a tetracyclic decaketid and a dimethyl salicylic acid, linked by the two deoxysugar moieties β-D-amicetose and α-L-axenose. It displays cytotoxic and antibiotic activity, even against Gram-positive multidrug-resistant strains such as MRSA11.
A genomic cosmid library of S. diastatochromogenes Tü6028 was generated and screened many years ago. Using specific gene probes the polyketomycin gene cluster with a size of 52.2 kb, containing 41 genes, was identified after several months of intense work12. Recently, a draft genome sequence of S. diastatochromogenes was obtained leading to the fast identification of the polyketomycin biosynthetic gene cluster. In this overview, a method helping to identify a natural product and elucidate its biosynthetic gene cluster will be described, using polyketomycin as an example.
Here we explain the single steps which lead from a natural product to its biosynthetic gene cluster shown for polyketomycin produced by Streptomyces diastatochromogenes Tü6028. The protocol comprises the identification and purification of a natural product with antibiotic properties. Further structural analysis and comparison with results from genome mining lead to the identification of the biosynthetic gene cluster. This procedure can be applied to any other compound derived from a Streptomyces strain or any other microorganism.
Protocol
1. Identification of a Natural Product with Antibiotic Property
Cultivate a microorganism under different conditions (e.g. time, temperature, pH, media) following the "OSMAC (one strain-many compounds) approach"13. Select one medium in which the production of a compound is observed.
- Cultivate Streptomyces diastatochromogenes Tü6028 under the following conditions
- Grow Streptomyces diastatochromogenes Tü6026 strain on MS plates (soy flour 20 g, D-mannitol 20 g, MgCl2 10 mM, agar 1.5%, tap water 1 L). Inoculate a small loop of the spores of this strain into 20 mL liquid TSB medium (tryptic soy broth 30 g, tap water 1 L, pH 7.2; preculture medium) in an Erlenmeyer flask with a spiral. Shake the flask on a rotary shaker (28 °C, 180 rpm, 2 days).
- Inoculate main culture in 100 mL HA medium (glucose 4 g, yeast extract 4 g, malt extract 10 g, tap water 1 L, pH 7.2) with 2 mL of the preculture. Culture the strain at 28 °C for 6 days on a rotary shaker at 180 rpm.
- Preparation of the crude extract
- Harvest cells by centrifugation (10 min, 3,000 x g).
- For next steps handle organic solvents under a fume hood.
- For compound extraction from the mycelium, resuspend cells in a twofold volume of acetone and shake in a tube for 30 min, 180 rpm. Filter the liquid through commercial filter paper and evaporate acetone by rotary evaporator at 40 °C and 550 bar. Dissolve the extract in 20 mL water:ethyl acetate (1:1) and shake it in a separating funnel for 30 min at 180 rpm.
- For compound extraction from the culture medium, adjust the culture broth to pH 4.0 by the addition of 1 M HCl. Add 100 mL ethyl acetate and shake it in a separating funnel for 30 min, 180 rpm.
- Collect ethyl acetate phase and evaporate by rotary evaporation at 40 °C and 240 bar.
- Analysis of the crude extract by HPLC
- Dissolve the extracts in 1 mL MeOH, filter them through a 0.45 µm pore size filter and run high-performance liquid chromatography (HPLC)14.
- In the case of polyketomycin, equip the HPLC system with a C18 precolumn (4.6 x 20 mm2) and a C18 column (4.6 x 100 mm2). Use a linear gradient of acetonitrile + 0.5% acetic acid ranging from 20% to 95% in H2O + 0.5% acetic acid (flow rate: 0.5 mL min-1). NOTE: Polyketomycin has a retention time of 25.9 min (see Figure 1A). The UV/vis spectrum has maxima at 242 nm, 282 nm, 446 nm and minima at 262 nm and 359 nm (see Figure 1B). In the negative modus a mass of 863.2 [M-H]- is detectable (see Figure 1C).
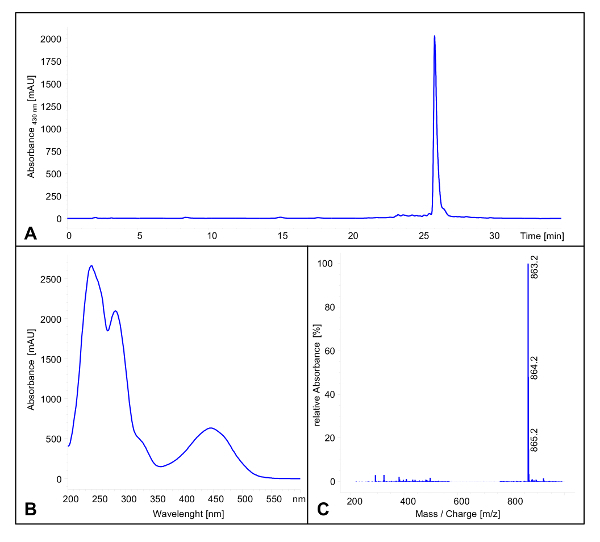 Figure 1: LC/MS Analysis of Polyketomycin. (A) HPLC chromatogram (λ = 430 nm) of the crude extract after cultivation of Streptomyces diastatochromogenes Tü6028 for 6 days. Polyketomycin has a retention time of 25.9 min (B) UV/vis spectra of Polyketomycin. (C) Mass spectra of Polyketomycin in the negative modus. Main peak with m/z 863.2 [M-H]-.Please click here to view a larger version of this figure.
Figure 1: LC/MS Analysis of Polyketomycin. (A) HPLC chromatogram (λ = 430 nm) of the crude extract after cultivation of Streptomyces diastatochromogenes Tü6028 for 6 days. Polyketomycin has a retention time of 25.9 min (B) UV/vis spectra of Polyketomycin. (C) Mass spectra of Polyketomycin in the negative modus. Main peak with m/z 863.2 [M-H]-.Please click here to view a larger version of this figure.
- Identification of the antibiotic activity using disc diffusion assay
- Inoculate test strains like Gram-positive bacteria Bacillus subtilis (in LB medium; LB 20 g, tap water 1 L, pH 7.4) and other Streptomyces strains (in TSB medium; tryptic soy broth 30 g, tap water 1 L, pH 7.2), Gram-negative bacteria Escherichia coli (in LB medium) and fungal strains (in media such as YPD medium; yeast extract 10 g, peptone 20 g, glucose 20 g, tap water 1 L, pH 7.4). Take 100 µL of test strain preculture and spread them onto respective agar plates.
- Dissolve the crude extract or purified compound in 500 µL of methanol (alternatively water or DMSO) and pipette 20-50 µL onto sterile paper discs. Dry paper discs for 30 min under the work bench and put them onto the plates with test cultures. Prepare a negative control (solvent) and a positive control (antibiotic, e.g. apramycin [1 mg]).
- Incubate the plates under adequate conditions until the test strains are visibly grown and determine inhibition zone, if apparent. Incubate E. coli and Bacillus sp. at 37 °C for 16 h, Streptomyces sp. at 28 °C for 2-4 days, fungal strain (mainly dependent on exact test strain) at 30 °C for 2 days).
2. Large Scale Extraction, Purification and Structure Elucidation of the Compound
Cultivate S. diastatochromogenes in 5 L HA medium (glucose 4 g, yeast extract 4 g, malt extract 10 g, tap water 1 L, pH 7.2). Inoculate 2 mL of the preculture in 30 x 500 mL Erlenmeyer flasks containing 150 mL HA medium. Incubate the strain at 28 °C for 6 days on a rotary shaker at 180 rpm.
Harvest cells by centrifugation (10 min, 15,000 x g, RT) and extract compounds using ethyl acetate (see section 1.3).
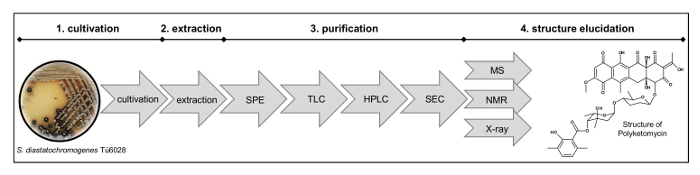 Figure 2: Workflow for Structure Elucidation. The process comprises (1) cultivation of the strain, (2) extraction, (3) purification by solid phase extraction (SPE), thin layer chromatography (TLC), preparative high-performance liquid chromatography (HPLC), size exclusion chromatography (SEC) and (4) structure elucidation by mass analysis (MS), nuclear magnetic resonance (NMR) and X-ray measurements. Please click here to view a larger version of this figure.
Figure 2: Workflow for Structure Elucidation. The process comprises (1) cultivation of the strain, (2) extraction, (3) purification by solid phase extraction (SPE), thin layer chromatography (TLC), preparative high-performance liquid chromatography (HPLC), size exclusion chromatography (SEC) and (4) structure elucidation by mass analysis (MS), nuclear magnetic resonance (NMR) and X-ray measurements. Please click here to view a larger version of this figure.
- Purification of Polyketomycin NOTE: The process of purification and structure elucidation is shown in Figure 2.
- Fractionate crude extract by a C18 solid phase extraction (SPE) column using a 10%-stepwise MeOH gradient ranging from 30% to 100% MeOH in H2O, 100 mL for each condition.
- Purify the compound-containing fraction further by preparative thin-layer chromatography (TLC)15 using CH2Cl2/MeOH (7:1) as elution system.
- Purify the compound by preparative HPLC16. Equip the HPLC system with a C18 precolumn (5 µm; 9.4 x 20 mm) and a main column (5 µm; 9.4 x 150 mm). Use a gradient of acetonitrile + 0.5% acetic acid ranging from 20% to 95% in H2O + 0.5% acetic acid (flow rate: 2.0 mL/min).
- To remove solvents and other small impurities, perform a last purification step by size exclusion chromatography using a column in MeOH17. Collect the final pure compound and evaporate MeOH by rotary evaporation at 40 °C and 240 bar.
- Structure elucidation
- Dissolve the pure compound (more than 2 mg) in 600-700 µL (dependent on the machine) of DMSO-d6, record 1D NMR (1H, 13C) and 2D NMR (HSQC, HMBC,1H-1H COSY) spectra on a NMR spectrometer18. Express chemical shifts in δ values (ppm) by using DMSO-d6.
- Record a high-resolution mass spectrum (HRESI-MS)19 of polyketomycin using a high-resolution mass spectrometer.
- Elucidate the structure by interpretation of the results of NMR and MS data analysis10.
3. Propose Biosynthetic Model of the New Isolated Compound
Analyze the structure of the isolated compound and predict enzymes, which might be involved in its biosynthesis. Assign them to polyketide (type I, II or III), non-ribosomal peptide synthesis, lantipeptide, terpenoid, or sugar metabolism pathway8.
- Example polyketomycin
- Subdivide the structure into obvious single moieties: tetracyclic moiety, two monosaccharides and the dimethyl salicylic acid.
- Find out, where the moieties might be derived from: The tetracyclic moiety might be derived from a polyketide synthase type II and the dimethyl salicylic acid moiety might be derived by an iterative polyketide synthase type I. The two sugar moieties, which are 6-deoxysugars, might be synthesized from glucose involving a TDP-glucose-4,6-dehydratase during biosynthesis and attached probably by two glycosyltransferases (see Figure 3)8.
- Predict putative genes in the cluster. Think of enzymes which are involved in the synthesis of the single moieties, in connecting the units, as well as in modifying and tailoring the molecule. The biosynthetic gene cluster of Polyketomycin most probably contains genes encoding a polyketide synthase type II (therefore minimum an ACP, KSα und KSβ for connecting extender units), an iterative polyketide synthase type I (at least ACP, AT, KS), a TDP-glucose-4,6-dehydratase (necessary for step: glucose → deoxyglucose) and two glycosyltransferases (attaching two sugar monomers to the aglycone)8.
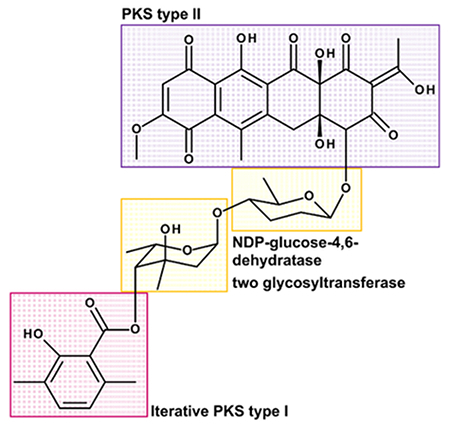 Figure 3: Structure of Polyketomycin Divided into Single Building Blocks. Polyketomycin is composed of a tetracyclic decaketid (PKS type II) and a dimethyl salicylic acid (iterative PKS type I), linked by the two deoxysugar moieties β-D-amicetose and α-L-axenose (NDP-glucose-4,6-dehydratase and two glycosyltransferase required). Please click here to view a larger version of this figure.
Figure 3: Structure of Polyketomycin Divided into Single Building Blocks. Polyketomycin is composed of a tetracyclic decaketid (PKS type II) and a dimethyl salicylic acid (iterative PKS type I), linked by the two deoxysugar moieties β-D-amicetose and α-L-axenose (NDP-glucose-4,6-dehydratase and two glycosyltransferase required). Please click here to view a larger version of this figure.
4. Genome Sequencing/Mining
- Next generation sequencing
- Sequence the genomic DNA by next generation sequencing technologies like Illumina, 454 pyrosequencing or SOLiD20. Align single reads to a reference sequence or assemble de novo. NOTE: The genome of S. diastatochromogenes Tü6028 was sequenced at the Center for Biotechnology (CeBiTec) at the University of Bielefeld. All reads were assembled to a draft genome of 7.9 Mb.
- Genome mining
- Search for open reading frames (ORFs) by the usage of for example NCBI Prokaryotic Genome Annotation Pipeline21,22, RAST (rapid annotation using subsystem technology)23,24,25, Prokka (rapid prokaryotic genome annotation)26 or GenDB27. These programs also propose their functions. The analysis of the S. diastatochromogenes Tü6028 draft genome sequence led to the identification of more than 7,000 ORFs.
- Run specific BLAST (Basic Local Alignment Search Tool) analysis to get more information like alignments with other similar genes and catalytic domains28,29,30.
- For the identification of the secondary metabolite gene cluster run programs like antiSMASH31,32,33, NaPDoS34 and NRPSpredictor35,36. In the draft genome of Streptomyces diastatochromogenes antiSMASH31,32,33 identified 22 gene clusters.
- Analyze the putative gene clusters for their enzymatic pathway(s) (polyketide synthase (type I, II or III), non-ribosomal peptide synthetase, lanthipeptide, terpenoid, sugar metabolism …). Search for cluster(s) containing genes which might be involved in the synthesis of the compound (see section 3). In S. diastatochromogenes annotated cluster 2 contains polyketide synthase type II genes, three polyketide synthase type I genes, a TDP-glucose-4,6-dehydratase gene and two glycosyltransferase genes (see Figure 4).
- Focus on single genes within the cluster. For PKS type I and NRPS the specificity of acyltransferases and adenylation domains, and thus the incorporation of single extender units, may be predicted. Also compare the order of the incorporated extender unit with the molecule. antiSMASH31,32,33 also shows similar clusters of already known compounds, with a link to MIBiG database37.
- Compare the structure of the compound with these other compounds and check for similarities.
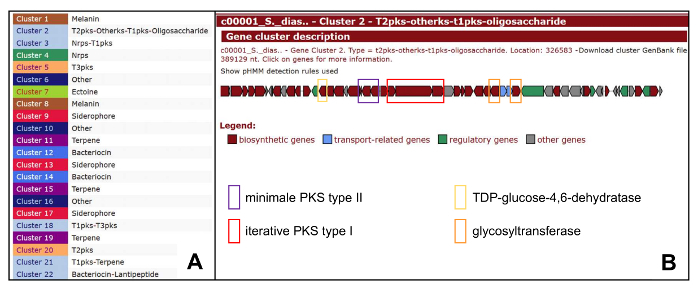 Figure 4: antiSMASH Output of Polyketomycin Biosynthetic Gene Cluster and Overview of Other Clusters in S. diastatochromogenes Tü6028. (A) Overview of predicted biosynthetic gene clusters in the genome of S. diastatochromogenes Tü6028; (B) Cluster 2 Polyketomycin biosynthetic gene cluster with targeted genes. Please click here to view a larger version of this figure.
Figure 4: antiSMASH Output of Polyketomycin Biosynthetic Gene Cluster and Overview of Other Clusters in S. diastatochromogenes Tü6028. (A) Overview of predicted biosynthetic gene clusters in the genome of S. diastatochromogenes Tü6028; (B) Cluster 2 Polyketomycin biosynthetic gene cluster with targeted genes. Please click here to view a larger version of this figure.
5. Verification of the Biosynthetic Gene Cluster
Search for a gene with obvious and important (essential) functions for biosynthesis of the compound. For verification of the polyketomycin gene cluster the gene pokPI, coding for the ketosynthase α from the PKS type II, was selected and inactivated by an out of frame-deletion (see Figure 5B). Alternatively, interrupt the gene by cloning an internal fragment (see Figure 5C).
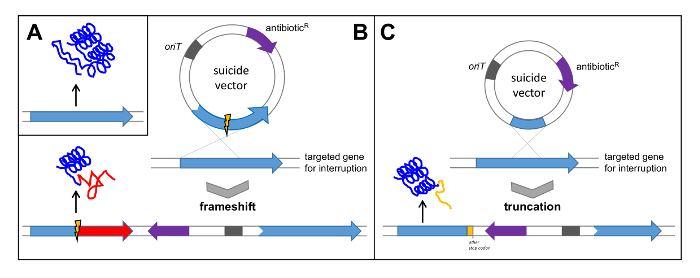 Figure 5: Verification of a Gene Cluster by Single Crossover. (A) Native gene leads to the translation of a functional protein; (B) Cloning of the gene with internal deletion into a suicide vector leads to a single crossover resulting in a frame shift in the targeted gene and subsequent translation of non-functional protein; (C) Cloning of an internal fragment of the gene into a suicide vector leads to a truncation of the gene and subsequent translation of a non-functional protein. oriT: origin of transfer; antibioticR: antibiotic resistance. Please click here to view a larger version of this figure.
Figure 5: Verification of a Gene Cluster by Single Crossover. (A) Native gene leads to the translation of a functional protein; (B) Cloning of the gene with internal deletion into a suicide vector leads to a single crossover resulting in a frame shift in the targeted gene and subsequent translation of non-functional protein; (C) Cloning of an internal fragment of the gene into a suicide vector leads to a truncation of the gene and subsequent translation of a non-functional protein. oriT: origin of transfer; antibioticR: antibiotic resistance. Please click here to view a larger version of this figure.
- Cloning of the single-crossover construct
- Amplify a fragment containing pokPI by PCR38 with primers pokPI_for and pokPI_rev.
- Clone PCR product into suicide vector pKC113239 (apramycinR [50 mg/mL]). The suicide vector is not able to replicate in the Streptomyces strain and thus has to integrate into the targeted gene by homologous recombination to provide apramycin-resistance. Transfer the vector into E. coli cloning host by heat shock transformation40.
- Isolate the vector by alkaline lysis41.
- Digest the vector DNA with a single cutter restriction enzyme that cuts within the fragment. The pokPI gene was digested withenzyme KpnI.
- Treat digested vector DNA with large DNA polymerase I (klenow) fragment, which has 5'→3' polymerase activity and 3'→5' exonuclease-activity. Blunting of the sticky ends and subsequent religation leads to loss of four bases. Check for loss of these bases by the steps transformation into E. coli XL1 Blue, picking single colonies, isolating their plasmid DNA,41 and analyzing further by restriction digestion and sequencing.
- Conjugation of the single-crossover construct into Streptomyces
- Transfer the suicide vector containing the pokPI gene (pKC1132_pokPIdel) with deletion into E. coli ET12567 pUZ800242 (kanamycinR) by heat shock transformation40. Incubate the cells in 100 mL LB media (kanamycin 50 µg mL-1, apramycin 50 µg mL-1) overnight at 37 °C. Harvest cells by centrifugation (3,000 x g, 10 min, 4 °C) and wash cells twice by resuspending in 50 mL LB media. Finally, resuspend cells in 2 x 500 µL of LB media.
- Mix 500 µL E. coli pUZ8002 pKC1132_pokPIdel with 500 µL Streptomyces diastatochromogenes culture (alternatively use spores). Spread the mixture on MS plates (soy flour 20 g, D-mannitol 20 g, MgCl2 10 mM, agar 1.5%, tap water 1 L). Incubate the plates for 20 h at 28 °C.
- Overlay each plate with apramycin (1.25 mg) and fosfomycin (5 mg) dissolved in 1 mL of water and let them dry. Incubate the plates for several days at 28 °C until exconjugants are visible.
- Check single-crossover mutants
- Inoculate single exconjugants in 20 mL TSB medium (apramycin 50 mg mL-1) and incubate them at 28 °C for three days and 180 rpm.
- Isolate genomic DNA43 and check interruption of the pokPI gene by PCR.
- Inoculate single clones with verified gene interruption and Streptomyces wildtype strain in 100 mL HA medium and incubate them for 6 days at 28 °C and 180 rpm. Extract the crude extract (see section 1.3). Check compound production by HPLC-MS analysis (see section 1.4). The corresponding peak in the HPLC chromatogram and the mass of the compound should not be detectable any more (see Figure 6).
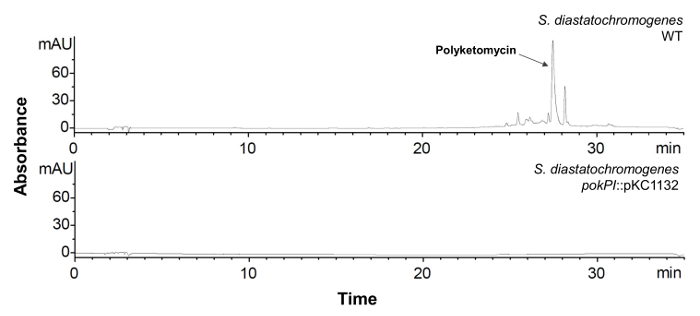 Figure 6: HPLC Analysis of S. diastatochromogenes with Inactivated pokPI Gene. HPLC chromatogram (λ = 430 nm) of crude extract of S. diastatochromogenes WT (top) and mutant strain with interrupted pokPI-Gen (below). The mutant strain does not produce polyketomycin anymore. Please click here to view a larger version of this figure.
Figure 6: HPLC Analysis of S. diastatochromogenes with Inactivated pokPI Gene. HPLC chromatogram (λ = 430 nm) of crude extract of S. diastatochromogenes WT (top) and mutant strain with interrupted pokPI-Gen (below). The mutant strain does not produce polyketomycin anymore. Please click here to view a larger version of this figure.
Representative Results
In this overview we describe the single steps from identification of an antibiotic leading to its biosynthetic gene cluster. Many years ago we cloned a cosmid library, packaged them into phages, transduced E. coli host cells, and had to screen thousands of colonies to identify the clones having overlapping regions of polyketomycin cluster. Sequencing of the cosmids was also a difficult and expensive process12.
In order to conduct further studies on the strain we sequenced the whole genome of Streptomyces diastatochromogenes Tü6028. With the draft genome sequence we easily identified the biosynthetic gene cluster of polyketomycin and other clusters encoding promising compounds. Figure 7 compares the "old" method of identifying the biosynthetic cluster by cloning of a cosmid library and elaborative screening, and the "new" method by whole genome sequencing with subsequent genome mining on a rough time scale. The new sequencing technologies and new genome mining programs speed up the whole process.
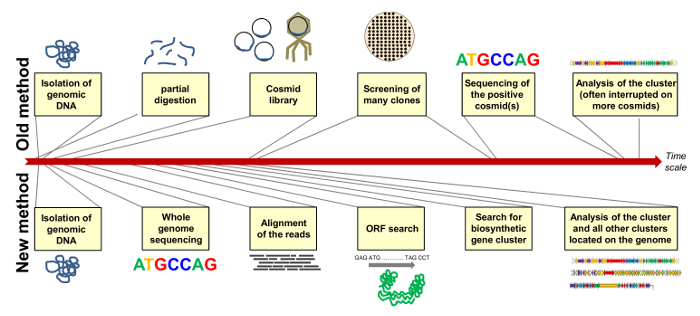 Figure 7: Comparison of the "Old" and "New" Method of Assigning a Biosynthetic Gene Cluster. The "old" method comprises cloning of a cosmid library with selection of positive clones and sequencing of the respective cosmid(s) (above); The "new" method includes whole genome sequencing and -mining to identify all secondary metabolite gene clusters located on the genome of the strain (below). Duration of single steps are shown on a rough time scale. Please click here to view a larger version of this figure.
Figure 7: Comparison of the "Old" and "New" Method of Assigning a Biosynthetic Gene Cluster. The "old" method comprises cloning of a cosmid library with selection of positive clones and sequencing of the respective cosmid(s) (above); The "new" method includes whole genome sequencing and -mining to identify all secondary metabolite gene clusters located on the genome of the strain (below). Duration of single steps are shown on a rough time scale. Please click here to view a larger version of this figure.
Discussion
In this lab a genomic cosmid library of Streptomyces diastatochromogenes was generated and screened many years ago, resulting in the identification of the polyketomycin gene cluster through an extremely time-consuming process. Characterization of single genes were possible using targeted gene deletions and analysis of the resulting mutants12. Recently, a draft genome sequence of S. diastatochromogenes was obtained allowing the fast identification of the polyketomycin biosynthetic gene cluster. We could easily detect the biosynthetic genes, although the draft genome sequence contains still many contigs. The described process can be achieved within months. However, the procedure comprises many steps. Single steps may fail several times, preventing progression to subsequent steps:
The genus Streptomyces is known for its capacity to produce bioactive compounds. While they carry often more than 20 biosynthetic gene clusters, usually only one or two compounds are produced under laboratory conditions. The application of the OSMAC approach (cultivation of one strain under different conditions) to wake up silent gene clusters may sometimes not be enough. Genetic manipulation of regulatory genes, such as the pleiotropic regulator genes adpA44 and bldA45,46, is also an effective method to activate the production of other secondary metabolites.
For the elucidation of the compound's structure, e.g. by NMR analysis, usually more than 2 mg of purified compound is necessary. Therefore, fermentation of more than 10 L culture is often required. Without a fermenter that is able to maintain oxygen, pH and temperature conditions, it might be challenging in a small lab to handle this amount of culture and subsequent extraction. During purification, the compound may be changed due to oxidation, radiation or temperature. Also, the more purification steps are used, the higher the chance of degradation.
When analyzing the structure of the natural product, and the clusters in the genome, sometimes it is not that easy to identify the appropriate cluster. Firstly, if there is only a draft genome sequence some part of the cluster may be missing. Secondly, not all genes which are required for the biosynthesis are in the cluster. Thirdly, sometimes a cluster is split into two parts separated from each other by many kilobases. Fourthly, it may be difficult to decide which one is the appropriate gene cluster. In case of large PKS type I or NRPS systems, where it is possible to calculate the number of extender units based on the number of modules, or even identify the single extender units by analysis of the selecting domains, it turns up easily. However, in the case of iteratively working enzymes the prediction of the synthesized compounds is often not possible, especially if the strain has more than 40 gene clusters. Fifthly, nature is highly complex and full of yet unknown compounds. Often the biosynthesis is a mixture of different pathways. If the new compound is not identified yet, or not related to another compound, it may be difficult to identify the cluster, to propose a biosynthesis model and to prove it.
Once the cluster is identified, the single crossover technique is a good and quick method to verify the hypothesis. PCR, cloning into a suicide vector, conjugation, selection of positive clones, and production assay are the only steps required. One disadvantage of this technique is that the integration of the vector into the chromosome is not stable due to further recombination events. Therefore, in order to further analyze single genes, precise gene deletions are required. Also it may be tricky to manipulate Streptomyces strains on the genetic level.
The described procedure can be assigned to any other compound produced by a Streptomyces strain or another microorganism. The knowledge about a biosynthetic gene cluster and its synthesized compound gives us further opportunities to modify already existing molecules with the aim of improving them for the fight against multidrug-resistant pathogens.
Disclosures
The authors have nothing to disclose.
Acknowledgments
S. Zhang is funded by China Scholarship Council. The authors are very grateful to former people working on polyketomycin project in this lab and Prof. Dr. Hans-Peter Fiedler, University of Tübingen, for providing the polyketomycin producer. The research was supported by the DFG (RTG 1976).
References
- Fleming A. On the antibacterial action of cultures of a penicillin, with special reference to their use in isolation of B. influenzae. Br J Exp Pathol. 1929;10(3):226–236. [Google Scholar]
- Lemke A, Kiderlen AF, Kayser O. Amphotericin B. Appl. Microbiol. Biotechnol. 2005;68(2):151–162. doi: 10.1007/s00253-005-1955-9. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick P, Raja A, LaBonte J, Lebbos J. Daptomycin. Nat. Rev. Drug Discov. 2003;2(12):943–944. doi: 10.1038/nrd1258. [DOI] [PubMed] [Google Scholar]
- Zakeri B, Wright GD. Chemical biology of tetracycline antibiotics. Biochem. cell Biol. 2008;86(2):124–136. doi: 10.1139/O08-002. [DOI] [PubMed] [Google Scholar]
- Schatz A, Bugle E, Waksman SA. Streptomycin, a Substance Exhibiting Antibiotic Activity Against Gram-Positive and Gram-Negative Bacteria. Exp. Biol. Med. 1944;55(1):66–69. doi: 10.1097/01.blo.0000175887.98112.fe. [DOI] [PubMed] [Google Scholar]
- Burg RW, et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 1979;15(3):361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerton JR, Ostlind DA, et al. Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob. Agents Chemother. 1979;15(3):372–378. doi: 10.1128/aac.15.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J. Am. Chem. Soc. 2010;132(8):2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose I, et al. Polyketomycin, a new antibiotic from Streptomyces sp. MK277-AF1. II. Structure determination. J. Antibiot. (Tokyo) 1998;51(1):26–32. doi: 10.7164/antibiotics.51.26. [DOI] [PubMed] [Google Scholar]
- Paululat T, Zeeck A, Gutterer JM, Fiedler HP. Biosynthesis of polyketomycin produced by Streptomyces diastatochromogenes.Tü6028. J. Antibiot. (Tokyo) 1999;52(2):96–101. doi: 10.7164/antibiotics.52.96. [DOI] [PubMed] [Google Scholar]
- Momose I, et al. Polyketomycin, a new antibiotic from Streptomyces. sp. MK277-AF1. I. Taxonomy, production, isolation, physico-chemical properties and biological activities. J. Antibiot. (Tokyo) 1998;51(1):21–25. doi: 10.7164/antibiotics.51.21. [DOI] [PubMed] [Google Scholar]
- Daum M, et al. Organisation of the biosynthetic gene cluster and tailoring enzymes in the biosynthesis of the tetracyclic quinone glycoside antibiotic polyketomycin. Chembiochem. 2009;10(6):1073–1083. doi: 10.1002/cbic.200800823. [DOI] [PubMed] [Google Scholar]
- Bode HB, Bethe B, Höfs R, Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 2002;3(7):619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Wolfender J-L. HPLC in Natural Product Analysis: The Detection Issue. Planta Med. 2009;75(07):719–734. doi: 10.1055/s-0028-1088393. [DOI] [PubMed] [Google Scholar]
- Stahl E. Thin-layer chromatography. A laboratory handbook. 1967. 2nd edition.
- Snyder LR, Kirkland JJ, Glajch JL. Preparative HPLC Separation. Pract. HPLC Method Dev. 2012. pp. 616–642.
- Granath KA, Kvist BE. Molecular weight distribution analysis by gel chromatography on sephadex. J. Chromatogr. A. 1967;28:69–81. doi: 10.1016/s0021-9673(01)85930-6. [DOI] [PubMed] [Google Scholar]
- Jackman LM, Sternhell S. Application of Nuclear Magnetic Resonance Spectroscopy in Organic Chemistry. Int. Ser. Org. Chem. 1969.
- Xian F, Hendrickson CL, Marshall AG. High resolution mass spectrometry. Anal. Chem. 2012;84(2):708–719. doi: 10.1021/ac203191t. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Tatusova T, et al. Prokaryotic Genome Annotation Pipeline. NCBI Handb. 2nd Ed. 2013.
- Angiuoli SV, et al. Toward an online repository of Standard Operating Procedures (SOPs) for (meta)genomic annotation. OMICS. 2008;12(2):137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin T, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Meyer F. GenDB--an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31(8):2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Madden T. The BLAST Sequence Analysis Tool. NCBI Handb. 2nd Ed. 2003. Chapter 16.
- Marchler-Bauer A, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2014;43:222–226. doi: 10.1093/nar/gku1221. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:339–346. doi: 10.1093/nar/gkr466. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, et al. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:204–212. doi: 10.1093/nar/gkt449. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, et al. antiSMASH 3.0 - a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:237–243. doi: 10.1093/nar/gkv437. (W1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, et al. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012;7(3):34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33(18):5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röttig M, et al. NRPSSpredictor2--a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39:362–367. doi: 10.1093/nar/gkr323. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015;11(9):625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis K, et al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. Pt 1. [DOI] [PubMed] [Google Scholar]
- Bierman M, et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116(1):43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Froger A, Hall JE. Transformation of plasmid DNA into E. coli.using the heat shock method. J. Vis. Exp. 2007. p. e253. [DOI] [PMC free article] [PubMed]
- Bimboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil DJ, et al. Analysis of Streptomyces avermitilis. genes required for avermectin biosynthesis utilizing a novel integration vector. Gene. 1992;111(1):61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 1995;11(6):217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- Makitrynskyy R, et al. Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol. 2013;3(10):130121. doi: 10.1098/rsob.130121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalan L, et al. A cryptic polyene biosynthetic gene cluster in Streptomyces calvus is expressed upon complementation with a functional bldA gene. Chem. Biol. 2013;20(10):1214–1224. doi: 10.1016/j.chembiol.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Gessner A, et al. Changing Biosynthetic Profiles by Expressing bldA in Streptomyces Strains. ChemBioChem. 2015;16(15):2244–2252. doi: 10.1002/cbic.201500297. [DOI] [PubMed] [Google Scholar]


