Abstract
We investigated the acquisition and prevalence of Chlamydophila sp. infection in calves. Specimens were collected at weekly intervals from birth to week 12 postpartum from 40 female Holstein calf-dam pairs in a dairy herd. Real-time PCR detected, quantified, and differentiated Chlamydophila 23S rRNA gene DNA from vaginal cytobrush swabs and milk samples. Chemiluminescence enzyme-linked immunosorbent assay with lysed Chlamydophila abortus or Chlamydophila pecorum elementary body antigens quantified antibodies against Chlamydophila spp. in sera. Chlamydophila sp. DNA was found in 61% of calves and 20% of dams in at least one positive quantitative PCR. In calves, clinically inapparent C. pecorum infection with low organism loads was fivefold more prevalent than C. abortus infection and was most frequently detected by vaginal swabs compared to rectal or nasal swabs. In dams, C. abortus dominated in milk and C. pecorum dominated in the vagina. The group size of calves correlated positively (P < 0.01) with Chlamydophila infection in quadratic, but not linear, regression. Thus, a doubling of the group size was associated with a fourfold increase in frequency and intensity of Chlamydophila infection. For groups of 14 or 28 calves, respectively, logistic regression predicted a 9 or 52% probability of infection of an individual calf and a 52 or 99.99% probability of infection of the group. Anti-Chlamydophila immunoglobulin M antibodies in Chlamydophila PCR-positive calves and dams and in dams that gave birth to calves that later became positive were significantly higher than in PCR-negative animals (P ≤ 0.02). Collectively, crowding strongly enhances the frequency and intensity of highly prevalent Chlamydophila infections in cattle.
Intracellular bacteria of the order Chlamydiales were first associated with diseases of cattle (Bos taurus) when McNutt isolated such organisms from feedlot cattle with sporadic bovine encephalomyelitis (22). When chicken embryo and cell culture methods for Chlamydiales became widely used, around 1955, a number of studies worldwide documented chlamydial agents in many acute diseases of cattle. A prominent example is epizootic bovine abortion (35), which is similar to classic ovine chlamydial abortion. The same chlamydial agent also caused epididymitis and seminal vesiculitis and was excreted in bull semen (37). Chlamydial strains from ruminant abortion were identified as serotype 1, biotype 1, immunotype 1, or ompA type B577 of ruminant chlamydiae (19, 25, 30, 33). Recently, a reclassification as Chlamydophila abortus was proposed (13, 31). While the epithet is helpful because it separates this chlamydial species from avian Chlamydophila psittaci, the introduction of a new genus in the family Chlamydiaceae has created unnecessary confusion. C. abortus has also been associated with bovine mastitis (5, 29).
Another chlamydial agent has been associated worldwide with clinically severe bovine chlamydial disease manifestations other than abortion. The diseases include sporadic bovine encephalomyelitis, pneumonia, enteritis, polyarthritis, kerato-conjunctivitis, nephritis, or purulent endometritis (21, 22, 39, 41). This chlamydial strain was diagnosed as serotype 2, biotype 2, immunotype 2, or ompA type LW613 of ruminant chlamydiae (19, 25, 30, 33) and was classified as a separate chlamydial species (14). Recently, reclassification of this agent as Chlamydophila pecorum was proposed (13, 31).
Numerous studies confirmed the disease potential of C. abortus and C. pecorum by experimentally reproducing the acute and severe diseases listed above (2, 17, 36, 40) and demonstrating the effectiveness of antibiotics in preventing such diseases (28). Shewen (32) summarized in 1980 the status of our understanding of chlamydial infections in animals, including cattle: “Exceptionally, some animals may experience severe or even fatal disease as a result of chlamydial exposure. A well balanced host-parasite relationship represents the common nature of chlamydial infection. This long-lasting inapparent or ‘latent’ state has been documented in several species: birds, cattle, guinea pigs, sheep and humans. Under circumstances of stress, ‘carrier’ animals may shed the organisms in large numbers or may in fact lapse into clinical disease.”
Despite improvement in diagnostic techniques, most notably the introduction of the PCR, our understanding about the prevalence and pathogenetic significance of these infections has not substantially changed since Shewen's review in 1980. The major impediment has been the cumbersome nature and insensitivity of diagnostic procedures, particularly of the complement fixation test for determination of seroprevalence of chlamydial infection in cattle (18, 26). Several investigations reported a high prevalence of chlamydial infection or of antibodies against chlamydiae in cattle, and they linked these data with increased prevalence of diseases, such as endometritis, fertility disorders, and epizootic bovine abortion (4, 11, 38, 41).
Collectively, these data raise the question of whether chlamydial infections in cattle truly cycle between complete absence of the agents (latency) and clinical manifestation with high shedding, or if low-level clinically inapparent infections represent the norm and such infections occasionally aggregate into clinical manifestations. In the second case, our detection methods simply would not be sensitive and specific enough to detect such low levels of ongoing chlamydial infections of cattle. Recently, a highly sensitive real-time PCR method suitable for large-throughput routine detection, quantification, and differentiation of Chlamydophila DNA was established (7). By using vaginal cytobrush swabs of clinically normal virgin heifers, a 53% prevalence of C. abortus and C. pecorum infection was detected, supporting the notion of continuous low-level infection (6).
The high prevalence of genital chlamydial infection in heifers that had not had sexual intercourse prompted us to analyze the possible acquisition of such infections in cattle at an early age. Limited early studies using chicken embryo or cell culture detection of chlamydiae established juvenile onset of Chlamydophila infection in calves (12, 27). However, the prevalence of Chlamydophila infection in calves and the rates of acquisition and transmission have not been studied in detail. Newborn calves, which are highly susceptible to infectious agents because of the obstruction of uterine transfer of maternal antibodies by the syndesmochorial bovine placenta, represent an ideal population for the analysis of chlamydial infection. Thus, bovine neonates are immunologically naïve, so that most calves, particularly if they do not receive colostrum, are prone to contract diseases (1, 12). To analyze the acquisition and transmission of Chlamydophila infection, we sampled calves at weekly intervals for 12 weeks, beginning on the first week after delivery, and examined the specimens by Chlamydophila 23S rRNA quantitative PCR (qPCR) or Chlamydophila enzyme-linked immunosorbent assay (ELISA). We report here a high prevalence of clinically inapparent C. abortus and C. pecorum infection in calves and an increase in infection rates proportional to the square of the group size of newborn calves.
MATERIALS AND METHODS
Dairy herd and animals.
All animals used in this study were Holstein cattle. The mean ± standard deviation of the ages of mature animals was 4.15 ± 1.16 years at the beginning of the experiment. Animals were maintained at the E.V. Smith Research Center Dairy Unit, located in Shorter, Ala., in free-stall housing with mattresses, were fed a total mixed ration on corn silage base, and spent 6 h per day on a grass lot. Calves were separated from dams at birth and housed in individual calf hutches away from the dairy barn throughout the 12-week postpartum sampling period. Calves had direct physical contact only with their nearest neighbor calves housed in the left- and right-side adjacent hutches, but not with any other cattle on the farm. All calves were fed pooled colostrum from multiple dams for 3 days, starting immediately after birth, and were weaned at 12 weeks of age. All animal procedures were approved by Auburn University's Institutional Animal Care and Use Committee.
Specimens.
Samples from 40 dams and 41 calves were obtained for 46 study weeks once weekly from calving week to the 12th week postpartum. Dam samples included vaginal cytobrush swabs, blood, and milk; calf samples were nasal, vaginal, and rectal cytobrush swabs and blood. Each swab sample was collected by a 10-s rotation of the cytobrush (Puritan; Hardwood Products Company LP, Guilford, Maine), the cytobrush handle was clipped, and the swab was immediately transferred into 400 μl of RNA/DNA Stabilization Reagent for Blood/Bone Marrow (Roche Applied Science, Indianapolis, Ind.) in a 1.5-ml microcentrifuge tube with a screw cap. Swab samples were centrifuged at 250 × g for 1 min and stored at −80°C without the cytobrush. Blood was collected from the tail vein of the dam and from the jugular vein of the calf with a 7-ml blood collection tube (13 100-mm Vacutainer tubes with Hemogard closures; Becton Dickinson and Co., Franklin Lakes, N.J.). The serum was separated by centrifugation at 1,300 × g for 15 min and stored at −80°C in 2-ml microcentrifuge tubes with screw caps. For DNA extraction, 600 μl of milk was mixed with 600 μl of 6 M guanidine-HCl, 10 mM urea, 10 mM Tris-HCl, and 20% Triton X-100 (vol/vol), pH 4.4, in a 2.0-ml microcentrifuge tube with a screw cap.
DNA extraction.
Isolation of milk and swab sample nucleic acid for PCR was performed with a High Pure PCR Template Preparation kit (Roche Applied Science) according to the manufacturer's instructions. Forty or 120 μl of proteinase K (20 mg/ml in double distilled H2O) was added to swab sample or milk samples, respectively, and samples were incubated for 30 min at 72°C with shaking at 600 rpm. Three hundred microliters of isopropanol and 300 μl of chloroform were added to milk samples, and 100 μl of isopropanol was added to swab samples. After brief agitation, the sample solution was transferred to the DNA-binding glass fiber filter device, except for the lipophilic chloroform bottom phase of the milk samples. Samples were filtered by centrifugation at 3,000 × g for 3 min, followed by the addition of 500 μl of inhibitor removal buffer and centrifugation at 3,000 × g for 3 min. Samples were washed twice with 500-μl wash buffer and were centrifuged at 3,000 × g for 3 min. Traces of wash buffer were removed by centrifugation at 13,000 × g for 10 s, and 20 μl of elution buffer (10 mM Tris-HCl [pH 8.4], and 0.01 mM EDTA) prewarmed to 72°C was added to each sample filter inserted into the collection tube. The glass fiber filter devices were incubated for 2 min at 72°C with shaking at 600 rpm, and elution buffer was recovered by centrifugation at 13,000 × g for 1 min. After a second elution step with 20 μl of buffer, the eluted DNA stock (typically 35 μl per specimen) was stored at −80°C.
Chlamydophila ELISA antigen.
Prototype C. abortus strain B577 (VR-656; American Type Culture Collection, Manassas, Va.), isolated from the kidney of an aborted sheep fetus (34), and prototype C. pecorum strain E58 (VR-628; American Type Culture Collection), isolated from the brain of a calf with encephalomyelitis (22), were cultured in buffalo green monkey kidney cells (BioWhittaker, Walkersville, Md.) as described previously (15). Chlamydial elementary bodies (EB) harvested in cell culture medium were purified by step gradient centrifugation and suspended in sucrose-phosphate-glutamate buffer (8). Two hundred fifty microliters of EB stock was added to 750 μl of protein denaturation buffer (0.5 M Tris-HCl [pH 7.0], 20% sodium dodecyl sulfate (vol/vol), 20% glycerol (vol/vol), and 1 M dithiothreitol) and boiled for 10 min. Chlamydial lysates were concentrated by centrifugal ultrafiltration in an ultrafiltration device (Microcon YM-3; Fisher Scientific Co., Newark, Del.) with a molecular weight cutoff of 3,000, reconstituted in PBS-25 mM DTT, and stored at −80°C.
Chlamydophila enzyme-linked immunosorbent assay.
Sample sera, negative control sera from gnotobiotic calves challenged with bovine diarrhea virus, or positive control sera were analyzed in duplicate in C. abortus B577 and C. pecorum E58 EB lysate ELISAs (8). The protein content of EB lysate antigens was quantified by NanoOrange Protein fluorescence assay (Molecular Probes, Eugene, Oreg.). EB lysate antigen equivalent to 0.7 μg of EB protein per well, diluted to 100 μl in coating buffer (15 mM Na2CO3 and 35 mM NaHCO3 [pH 9.6]), was added per well to white C-bottom 96-well microtiter plates (White MaxiSorp; Fisher Scientific Co.). Plates were incubated overnight at 4°C, the coating solution was aspirated, and wells were washed five times with wash buffer (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl, and 0.1% Tween 20). Wells were blocked by adding 200 μl of assay diluent (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl, 0.1% Tween 20, and 10% normal rabbit serum) for 1 h at room temperature. The assay diluent was removed, and 100 μl of serum sample diluted 1:100 with the assay diluent was added for 1.5 h at room temperature. After five washes, 100 μl of alkaline phosphatase (AP)-conjugated antibodies against bovine immunoglobulin A (IgA), IgG, or IgM (Bethyl Laboratories, Inc., Montgomery, Tex.) per well was added, diluted in assay diluent (sheep anti-bovine IgM, 1:300; sheep anti-bovine IgG, 1:600; and sheep anti-bovine IgA, 1:500), and incubated for 1 h at room temperature. Plates were washed five times, 150 μl of freshly prepared AP substrate buffer solution (BM Chemiluminescence ELISA Substrate AP; Roche Applied Science) per well was added, and the luminescence emitted was read with a microplate reader (Spectrafluor Plus; Tecan, Inc., Durham, N.C.) after 10 min of shaking. Luminescence data were normalized between microtiter plates by subtracting the blank plus 2 standard deviations of values from samples for each plate, followed by multiplication with a normalization factor derived for each microtiter plate by division of the positive control sera with the mean positive control value of all microplates.
Real-time PCR.
Fluorescence resonance energy transfer (FRET) real-time qPCR for the Chlamydophila 23S rRNA gene, followed by melting curve analysis, detected, quantified, and differentiated Chlamydophila spp. on the LightCycler platform (Roche Applied Science) as described before (7). Each PCR in a glass capillary tube received 5 μl of extracted specimen DNA and 15 μl of master reaction mixture. Quantitative standards used were 104, 103, 102, and 10 copies of Chlamydophila abortus B577 and 10 copies of Chlamydophila pecorum LW613 DNA extracted from purified elementary body preparations by the High Pure method and quantified by a PicoGreen DNA fluorescence assay (Molecular Probes). Chlamydophila species were differentiated by melting curve analysis of the amplification products (6). C. abortus DNA in selected specimens was confirmed by C. psittaci B577 omp1 FRET-qPCR (7).
Statistical analysis.
All statistical analyses were performed with the Statistica 6.1 software package (StatSoft, Inc., Tulsa, Okla.). PCR genome copy data were log10 transformed, and the relative light unit values for anti-Chlamydophila antibodies were log2 transformed. The numbers of calves and infected calves investigated per study week were illustrated by line plots. The average number of Chlamydophila genomes per infected calf for each postpartum week was evaluated by mean plots ± 95% confidence intervals, and mean weeks of positive PCRs for four consecutive 9-week study periods were compared by using the Mann-Whitney U test. The influence of group size on rates and levels of Chlamydophila infection in calves was modeled by polynomial regression (10). Scatter plot and regression analyses confirmed the independence of the rate and level of Chlamydophila infection from the postpartum week. Anti-Chlamydophila antibody data were analyzed by repeated measures analysis of variance (ANOVA) (24). Comparisons of means within and among categories under the assumption of no a priori hypothesis were performed post hoc by the Tukey honest significant difference test. The normal distribution of continuous data was confirmed by the Shapiro-Wilk W test, and the homogeneity of variances was confirmed by the Levene test (10). The binomial outcome Chlamydophila infection (PCR positivity-negativity) versus group size or log2 anti-Chlamydophila IgM was modeled with logistic regression (20). Log (odds of infection) was modeled as log (odds) = b0 + b1x, where b0 is the intercept, b1 is the regression coefficient, and x is the independent variable group size or log2 IgM. Probability of infection was calculated as probability = odds/(1 + odds).
RESULTS
High prevalence of Chlamydophila spp. infection in postpartum calves.
In the present study, we investigated the prevalence and the characteristics of acquisition of Chlamydophila infection by calves early in their lives. All female calves, a total of 41 born to 40 dams, were entered into the study. Tables 1 and 2 show the detection rates and numbers of C. abortus and C. pecorum in calves and dams, respectively. Sixty-one percent (61%) of all calves were infected, as indicated by at least one positive qPCR in one of the specimens at one time point. However, most positive animals were positive with multiple specimens and at multiple time points. The prevalence of C. pecorum in calves was approximately five times as high as that of C. abortus, with the highest detection rate being with vaginal swabs, compared to rectal or nasal swabs. The numbers of Chlamydophila genomes detected per PCR were low, typically 2 to 10 C. pecorum genomes or 1 to 5 C. abortus genomes. These numbers represent approximately one-seventh of the number of Chlamydophila genomes in the specimen. The highest numbers of Chlamydophila genomes were detected with vaginal swabs.
TABLE 1.
Prevalence of C. abortus and C. pecorum DNA in calves (n = 41)a
| Chlamydophila species, % positive (n)b | Specimen type | No. of positive specimens (%)c | No. of Chlamydophila genomes per positive qPCRd |
|---|---|---|---|
| C. abortus, 12.2 (5) | Nasal | 0 (0) | 0 |
| Vaginal | 9.8 (4) | 2.69 ± 1.67 | |
| Rectal | 4.9 (2) | 2.03 ± 1.02 | |
| C. pecorum, 58.5 (24) | Nasal | 29.3 (12) | 6.16 ± 5.70 |
| Vaginal | 51.2 (21) | 10.41 ± 6.05 | |
| Rectal | 39.0 (16) | 3.39 ± 2.15 |
Sixty-one percent of calves (n = 25) tested positive in one or more qPCRs.
The number of Chamydophila-positive calves is not identical to the sum of calves testing positive for the Chlamydophila species, because four calves had a dual infection.
The number of Chamydophila-positive specimens is not identical to the sum of calves testing positive, because calves had multiple positive specimens. Values in parentheses are n values.
Values are means ± standard deviations.
TABLE 2.
Prevalence of C. abortus and C. pecorum DNA in dams (n = 40)a
| Chlamydophila species, % positive (n)b | Specimen type | No. of positive specimens (%)c | No. of Chlamydophila genomes per positive qPCRd |
|---|---|---|---|
| C. abortus, 15 (6) | Milk | 15 (6) | 1.71 ± 1.27 |
| Vaginal | 2.5 (1) | 2 | |
| C. pecorum, 7.5 (3) | Milk | 0 (0) | 0 |
| Vaginal | 7.5 (3) | 4.07 ± 2.07 |
Twenty percent of dams (n = 8) tested positive in one or more qPCRs. The total numbers of calves and dams are inconsistent because one dam gave birth to twins.
The number of Chlamydophila-positive dams is not identical to the sum of dams testing positive for the Chlamydophila species, because one cow showed a dual infection.
The number of Chamydophila-positive specimens is not identical to the sum of dams testing positive, because two specimens from one dam were positive. Values in parentheses are n values.
Values are means ± standard deviations.
Chlamydophila detection rates in dams were lower, with 20% of animals being infected. Interestingly, C. abortus was the dominant Chlamydophila species in dams and was the only species detected in milk samples, the main source of positive specimens. C. pecorum was more often detected in vaginal cytobrush specimens from dams.
Neither calves nor dams showed specific signs of severe Chlamydophila-induced disease throughout the study period. However, we did see signs of minor vaginal irritation, such as erythema and small subepithelial vaginal granulomas, of calves. Chlamydophila-infected dams showed signs of mastitis and prolonged postpartum vaginal secretion. Collectively, the high prevalence of Chlamydophila infections in calves, compared to the lower prevalence in dams, indicates that calves in the first weeks of their lives are more susceptible to Chlamydophila infection than adult cattle, most likely due to less effective immunity.
The number of calves per weekly group correlates with the frequency and intensity of Chlamydophila infection.
Figure 1 shows the numbers of calves for each study week. After low enrollment during the first study weeks, the number of calves born rose rapidly, to a peak of 29 calves enrolled in study week 20, and then decreased steadily. The number of calves positive for Chlamydophila DNA in each weekly group paralleled the total number of calves sampled (Fig. 1A). Figure 1B indicates that the number of positive calves does not represent a constant percentage rate but that the percentage of positive calves strongly increases with increasing size of the weekly group.
FIG. 1.
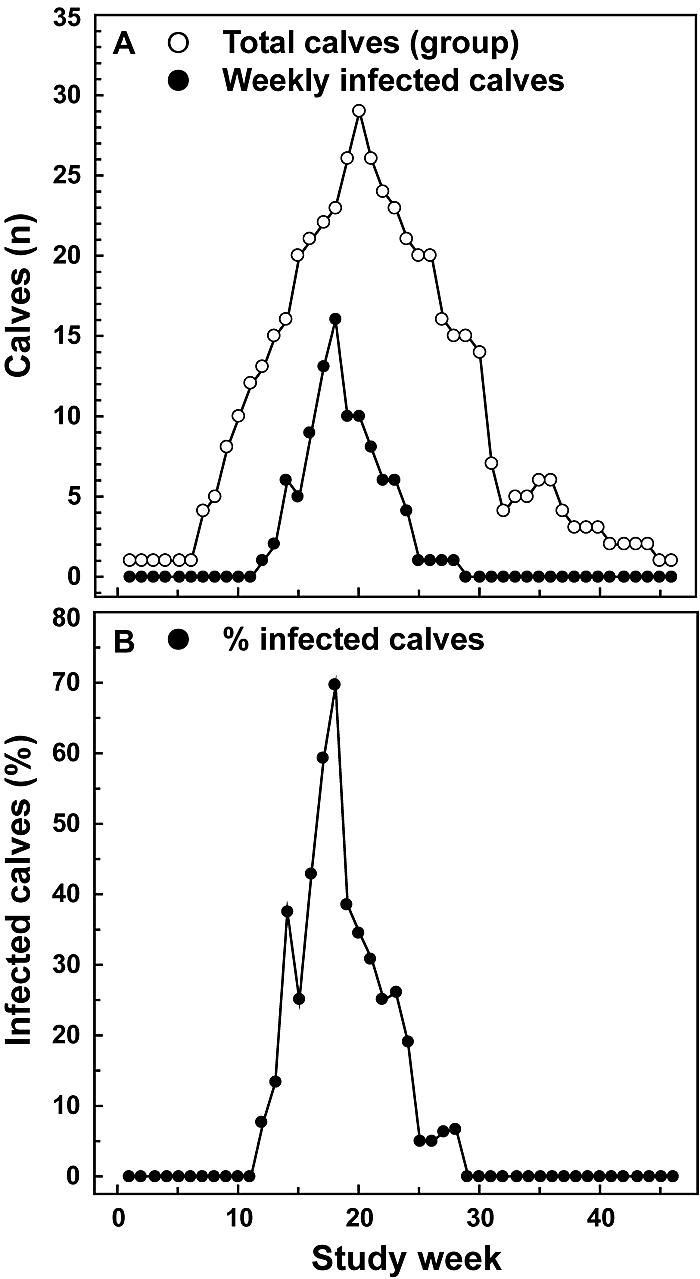
Number of calves examined and calves found to be infected with Chlamydophila spp. during each week over the course of the investigation. Each calf born during a 36-week time period was entered at birth into the study and sampled weekly for 12 consecutive weeks. (A) Total number of calves sampled and calves diagnosed as qPCR-positive for Chlamydophila DNA (infected) in any of the specimens (nasal, vaginal, and rectal swabs) during each study week. (B) Infected calves expressed as a percentage of all calves examined. The percentage of infected calves increases with group size (total number of calves examined in a week).
Figure 2 shows the time course, infection intensity expressed as the number of Chlamydophila genomes detected, and detection site of Chlamydophila infection of calves and displays these parameters for calves born during each of four consecutive 9-week enrollment periods (birth cohorts) (Fig. 2). The peak infection frequencies shift highly significantly (P ≤ 0.01) over the course of the study, from a mean of 9.6 weeks for Chlamydophila-positive specimens in the first 9-week birth cohort to mean detection at 4.8 weeks postpartum for the third birth cohort. The Chlamydophila infection was not detected in calves enrolled in the fourth 9-week period; however, the size of the birth cohort was small. Maximum infection intensity, expressed as the log10 of Chlamydophila genomes per positive PCR, coincides approximately with peak detection frequency.
FIG. 2.
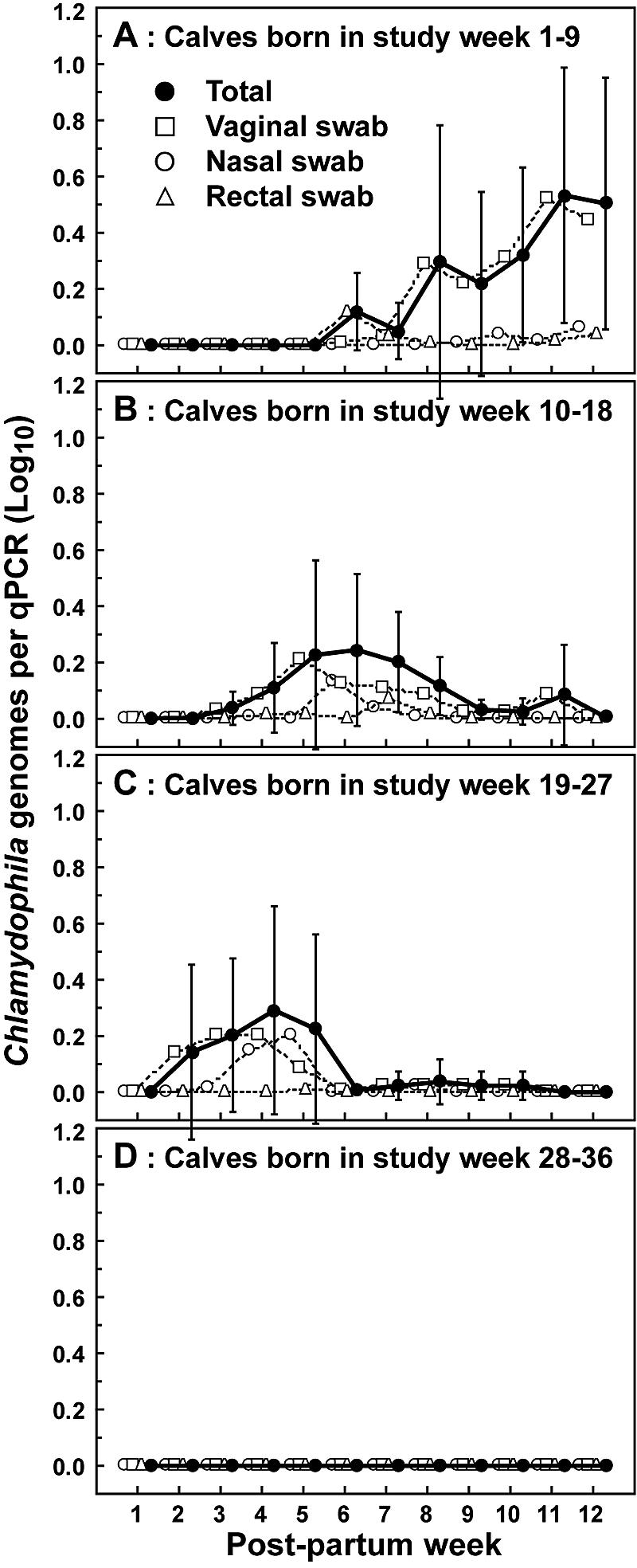
Time course of detection, and quantity of Chlamydophila genomes detected per qPCR of infected calves. Chlamydophila genomes were detected and quantified in samples collected from calves over the first 12 weeks postpartum. Calves were categorized into four birth cohorts according to birth week relative to the 36-week study period. (A) Calves born during study weeks 1 to 9 (n = 8); (B) calves born during study weeks 10 to 18 (n = 16); (C) calves born during study weeks 19 to 27 (n = 14); (D) calves born during study weeks 28 to 36 (n = 3). Five-microliter aliquots out of a total of 35 μl of nucleic acids extracted from nasal, vaginal, or rectal cytobrush specimens were subjected to Chlamydiaceae 23S rRNA FRET-qPCR. The log10 of the mean of Chlamydophila genomes (± 95% confidence interval) indicates genomes detected only per positive qPCR in each sampling week. A log10 of 0 indicates negative birth cohorts with zero Chlamydophila genomes detected by qPCR. The average postpartum sampling week for positive qPCRs is week 9.63 for the birth cohort shown in panel A, week 6.57 for the birth cohort shown in panel B, and week 4.79 for the birth cohort shown in panel C. These means are all significantly different (P < 0.01, Mann-Whitney U test).
Cytobrush specimens from the vagina, the dominant site of Chlamydophila infection in female calves, with approximately 80% of all detected Chlamydophila genomes, allowed tracking of time course and intensity of these infections. The trends of average Chlamydophila detection frequencies and quantities, evident in Fig. 2A to C, also were observed in individual calves. In postpartum week 1, for all calves at all sampling sites, only two PCRs were positive for less than one Chlamydophila genome, indicating that calves were essentially born noninfected. Thus, the kinetics of the subsequent infection likely characterize the course of primary infection in animals that are immunologically naïve, except for colostrum-acquired anti-Chlamydophila antibodies. Vaginal Chlamydophila infection typically was first detected 2 to 6 weeks postpartum, with low levels of one to five Chlamydophila genomes per PCR. These numbers peaked over the next 1 to 3 weeks of the infection to approximately 50 to 500 genomes per PCR and then decreased to 0 over the next 2 weeks. These results indicate a 3- to 5-week course of naturally acquired, primary, subclinical Chlamydophila infection in calves with generally low numbers of the agent. They further indicate a high susceptibility of calves to Chlamydophila infection and slow elimination of the organism by the emerging adaptive immunity in response to the infection.
Regression models correlate the square of group size with frequency and intensity of Chlamydophila infection in calves.
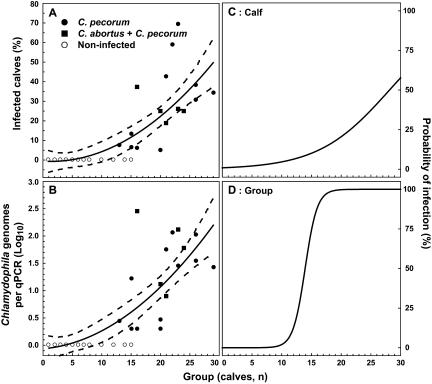
We used polynomial regression analysis to quantify the relationship between group size and percentage of calves found infected each study week or Chlamydophila genomes per positive PCR. Only the quadratic exponent, but not linear regression or any other exponent for group size, described these relationships significantly (P values were 0.02 and 0.04 for the percentage of infected calves and the number of Chlamydophila genomes per qPCR, respectively, relative to group size) in highly significant equations (P < 10−4) (Fig. 3A and B). Thus, the square of the number of animals in a group of newborn calves best explains the percentage of Chlamydophila-infected calves and the intensity of the infection expressed as the log10 of Chlamydophila genomes per positive PCR. The equations indicate that in a group of 14 calves, 1 to 2 calves (10%) will be infected with 3.6 Chlamydophila genomes per PCR; however, in a group of 28 calves, approximately 13 calves (45%) will be infected with 209 Chlamydophila genomes per PCR.
FIG. 3.
Influence of group size on rate and intensity of weekly Chlamydophila infection in calves. The percentage of infected calves (A) and the number of Chlamydophila genomes detected per qPCR of infected calves (B) were plotted for each week and group size observed in the study. Melting curve analysis of the qPCR identified Chlamydophila spp., C. pecorum infection in calves, both C. pecorum and C. abortus infection in calves, and qPCR-negative calves in a study week. The log10 of the mean of Chlamydophila genomes (± 95% confidence interval) indicates genomes detected only per positive qPCR in each sampling week. A log10 of 0 indicates negative groups with zero Chlamydophila genomes detected by qPCR. Polynomial regression equations composed of intercept and the square of group size, but not group size, highly significantly describe (A) the relationship between group size (x) and the percentage of infected calves (percent infected calves = −1.215 + 0.059 x2; P < 0.01, r = 0.80, r2 = 0.64); and (B) and group size (x) and the level of Chlamydophila infection per infected calf (log10 Chlamydophila genomes per positive PCR = −0.032 + 0.003 x2; P < 0.01, r = 0.83, r2 = 0.69). These results indicate that frequency and intensity of Chlamydophila infection in calves increases fourfold with each doubling of group size. Logistic regression analysis indicates that group size is an excellent predictor of infection of both individual calves (C) and the group as whole (D). The graphs represent the logistic regression equation: the percent probability of infection for individual calves = 100 [e−4.69 + 0.17 x/(1 + e−4.69 + 0.17 x)], and the percent probability of infection for the group as a whole = 100 [e −13.76 + 0.99 x/(1 + e −13.76 + 0.99 x)], where x equals the group size. (C) The odds ratio for infection (Wald 95% confidence interval) of individual calves is 1.18 (1.13 to 1.24; P < 0.01) per one-calf increase in group size. The group size with a 50% probability of infection of any given calf in this group is approximately 28. (D) The odds ratio for infection of the group (infection of a calf of the group) is 2.68 (1.95 to 3.69; P < 0.01) per one-calf increase. The group size with a 50% probability of group infection is approximately 14.
Using logistic regression modeling, we analyzed whether group size predicted the probability of Chlamydophila infection of individual calves or of the group (Fig. 3C and D). An increase of one calf per group highly significantly (P < 10−4) associates with a 1.18-fold (range, 1.13 to 1.24; 95% confidence interval) increased odds ratio for infection of individual calves and a 2.68-fold (1.95 to 3.69) increased odds ratio for infection of the group (any calf in the group infected). These logistic regression models predict for a group of 14 calves a 9% probability of infection of an individual calf and a 52% probability of infection of the group. Conversely, for a group of 28 calves they predict a 52% probability of infection of an individual calf and a 99.99% probability of infection of the group.
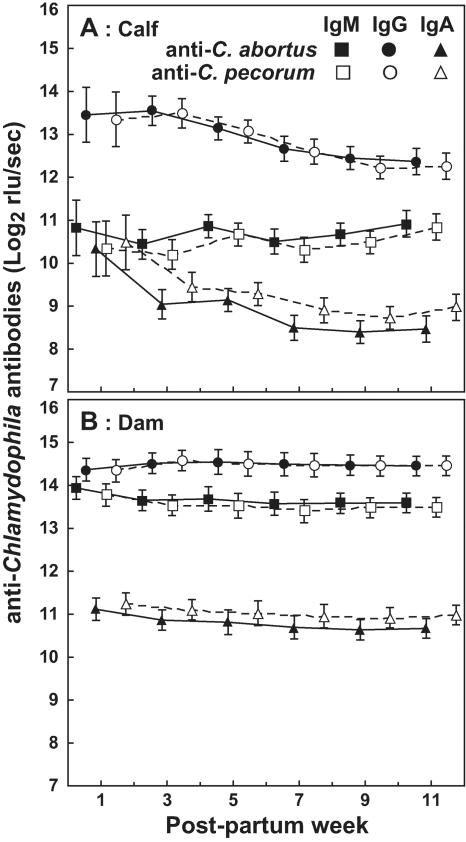
Calf anti-Chlamydophila IgG and IgA, but not IgM, decline over the 12-week postpartum sampling period.
Anti-C. abortus and C. pecorum serum IgM and IgG antibodies in calves or in dams were essentially identical (Fig. 4). IgA antibodies against C. pecorum in calves and dams were consistently higher than anti-C. abortus IgA antibodies, but these differences failed to reach significance (P = 0.06 and 0.14, respectively). Because of these virtually identical concentrations, we averaged C. abortus and C. pecorum serum antibodies as anti-Chlamydophila antibody concentrations in all subsequent analyses. Concentration differences between anti-Chlamydophila antibody isotypes were significant in calves or dams throughout the 12-week postpartum study period (P < 10−4). The means of all dam anti-Chlamydophila antibody concentrations, but particularly of IgM, were significantly higher than those of the calves (P < 10−4). When compared by the Tukey honest significant differences test, anti-Chlamydophila IgG and IgA in calves were significantly lower in week 11 than in week 1 postpartum (P < 10−4). IgM antibodies in calves (Fig. 4A) and all antibody isotypes in dams (Fig. 4B) remained unchanged throughout the postpartum sampling period. This lack of decline in IgM antibodies in calves and of all antibodies in dams suggests a steady-state equilibrium of antibody elimination and regeneration. Presumably, the stimulus by continuous subclinical Chlamydophila infection maintains serum anti-Chlamydophila antibody concentrations. Conversely, calf serum IgG and IgA initially decline from their colostrum-derived peaks because isotype switch in newly producing plasma cells has not yet occurred to replenish calf anti-Chlamydophila IgG and IgA serum antibody concentrations.
FIG. 4.
Anti-Chlamydophila antibodies detected for calves and dams throughout the 12-week postpartum sampling period. Antibodies of calf (A) or dam (B) sera bound to C. abortus or C. pecorum elementary body lysate antigen were detected by chemiluminescence ELISA. Data are shown as means of log2-transformed chemiluminescent signal (relative light units [rlu]) per second ± 95% confidence interval. Repeated measures ANOVA indicates that the concentrations of the three anti-C. abortus or anti-C. pecorum antibody isotypes differ highly significantly throughout the 12 postpartum weeks in both calf and dam sera (for calves, F(5, 233) = 203.95 and P < 10−4; for dams, F(5, 216) = 345.40 and P < 10−4), and the concentration of each isotype is always higher in dams than in calves (P < 10−4). IgM or IgG concentrations are not different for anti-C. abortus and anti-C. pecorum calf or dam antibodies. Anti-C. pecorum IgA is consistently higher than anti-C. abortus IgA in both calves and dams, but these differences are not statistically significant (for calves, F(1, 78) = 3.62 and P = 0.06; for dams, F(1, 72) = 2.25 and P = 0.14). Post hoc comparison using the Tukey honest significant differences test reveals that IgG (P < 10−4) and IgA (P < 10−4) in calves decrease from postpartum week 1 to week 11 while IgM remains unchanged. All antibody isotypes in dams remain unchanged throughout the 12 postpartum weeks.
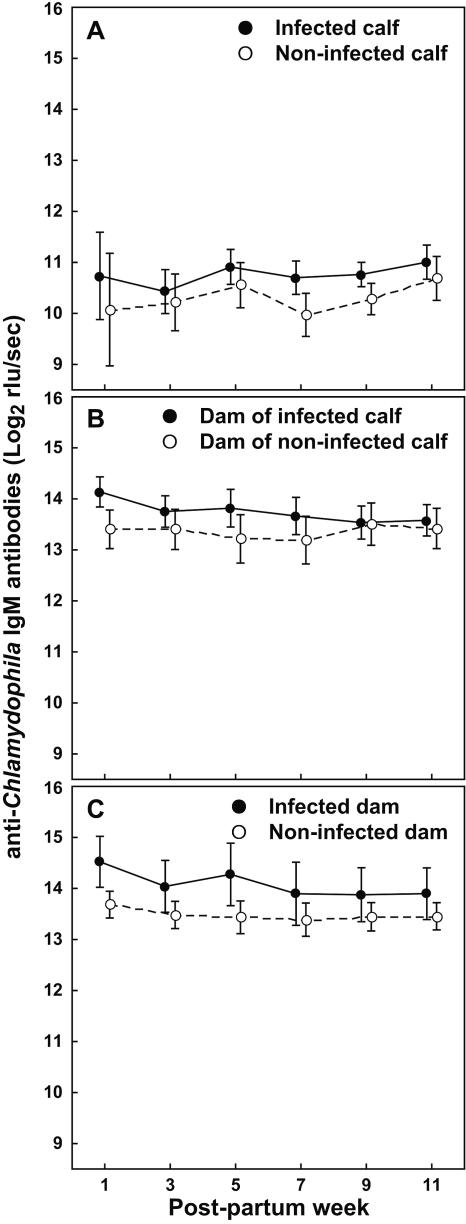
High anti-Chlamydophila IgM antibodies predict Chlamydophila infection in calf and dam.
We analyzed the anti-Chlamydophila serum antibody concentrations of infected (PCR-positive at any time point in any specimen) versus noninfected calves or dams (Fig. 5). In calves, increased IgM, but not IgG and IgA (data not shown), associated significantly with Chlamydophila infection (Fig. 5) [F(1, 38) = 5.49 and P = 0.02 by repeated measures ANOVA].
FIG. 5.
Anti-Chlamydophila serum IgM antibodies of calves and dams diagnosed as Chlamydophila-infected or noninfected during the 12 postpartum weeks. Concentrations of anti-C. abortus and C. pecorum IgM antibodies were averaged, and mean concentrations (± 95% confidence interval) for infected and noninfected calves or dams are shown for the postpartum sampling period. Calves or dams that were qPCR positive for Chlamydophila DNA in any of the specimens (nasal, vaginal, or rectal swab or milk) during any of the sampling weeks were considered infected. (A) Throughout the sampling period, anti-Chlamydophila IgM, but not IgG and IgA antibodies (data not shown), were significantly higher in infected than in noninfected calves [For IgM, F(1, 38) = 5.49 and P = 0.02 by repeated measures ANOVA]. (B) Anti-Chlamydophila IgM antibody concentrations, as well as IgG and IgA (data not shown), are significantly higher in dams of infected calves than in dams of noninfected calves (by repeated measures ANOVA, F(1, 35) = 4.29 and P = 0.04 for IgM; F(1, 35) = 9.08 and P = 0.01 for IgG; and F(1, 35) = 10.13 and P < 0.01 for IgA). (C) Anti-Chlamydophila IgM antibody concentrations, as well as IgG and IgA (data not shown), are significantly higher in infected than noninfected dams (by repeated measures ANOVA, F(1, 35) = 9.88 and P < 0.01 for IgM; F(1, 35) = 9.58 and P < 0.01 for IgG; and F(1, 35) = 9.05, P < 0.01 for IgA).
Next, we examined if antibody levels in the dam correlated with Chlamydophila-infection of the calf. Anti-Chlamydophila IgM antibodies, as well as IgG and IgA antibodies (data not shown), of dams of Chlamydophila-infected calves were significantly higher than those of dams of noninfected calves (Fig. 5B) (by repeated measures ANOVA, F(1, 35) = 4.29 and P = 0.04 for IgM, F(1, 35) = 9.08 and P = 0.01 for IgG, and F(1, 35) = 10.13 and P < 0.01 for IgA).
Finally, we tested for correlation between dam antibody concentrations and dam Chlamydophila infection. Again, all anti-Chlamydophila antibody isotypes of dams with Chlamydophila infection were significantly higher than those of noninfected dams (Fig. 5C) (P ≤ 0.01 by repeated measures ANOVA).
The previous analyses of anti-Chlamydophila antibody levels had indicated that IgM antibodies were consistently associated with Chlamydophila infection of both calf and dam. Using logistic regression analyses, we tested if IgM antibody levels predicted the probability of Chlamydophila infection. Calf anti-Chlamydophila IgM of week 7, and less significantly those of week 9, but not of earlier postpartum weeks, indicated increased probability of Chlamydophila infection in calves throughout the 12 postpartum weeks, with a 3.58 (1.18 to 10.86) odds ratio per doubling of anti-Chlamydophila IgM (P = 0.02). Anti-Chlamydophila IgM of the dam in postpartum week 1 estimated the probability of Chlamydophila infection of both calf and dam in the subsequent 12 postpartum weeks with odds ratios of 5.98 (1.42 to 25.20) and 6.94 (1.35 to 35.61) per doubling of dam anti-Chlamydophila IgM, respectively (P < 0.02). Collectively, these logistic regression models imply, under an assumption of 50% prevalence of Chlamydophila infection in a herd, that an animal with an anti-Chlamydophila serum IgM concentration of half of the population mean has a 13 to 22% probability of PCR-detectable Chlamydophila infection. An animal with an IgM concentration of twice the population mean is at 78 to 87% probability Chlamydophila-infected.
DISCUSSION
Classic methods for the detection of Chlamydophila agents and of antibodies against these agents have indicated that these bacteria are highly prevalent in cattle and have uncovered their contribution to numerous disease conditions (32). These methods demonstrated acute Chlamydophila-induced diseases, such as sporadic bovine encephalomyelitis and epizootic bovine abortion, as well as variable, but generally high, Chlamydophila seroprevalence worldwide. However, it was impossible to consistently detect low levels of these organisms, which were suspected in conjunction with the sporadic occurrence of serious acute disease. In the present investigation, we used highly sensitive real-time PCR and chemiluminescence ELISA with a wide dynamic range to re-examine in a prospective cohort study the prevalence characteristics of Chlamydophila sp. infection in newborn female calves and their dams. The results of this investigation have the potential to shift the focus from Chlamydophila infection as a rare, severe disease to Chlamydophila infection as a pervasive, low-level infection of cattle without apparent disease (silent epidemic) or with only a subtle expression of disease, one that impacts herd health and fertility but is difficult to recognize in individual animals.
Our virtual inability to demonstrate infection of calves in the first week after birth strongly suggests that the calves were born free of chlamydial infection and acquired subsequent infections from the environment as immunologically naïve animals. Typically, vaginal infections in calves started with very low initial chlamydial loads, followed by peak loads about 2 weeks into the infection and subsequent clearance 1 to 3 weeks later. This course of Chlamydophila infection indicates that the original inoculum was low and that the organism load slowly accumulated to peak Chlamydophila levels, which then represented a stimulus strong enough to elicit an immune response that cleared the infection. Presumably, the first inoculum into the continuously maintained cohort of calves originated from the herd of adult animals by direct contact after birth or by transfer during feeding. However, the profound influence of calf group size on Chlamydophila infection, a fourfold infection frequency and intensity with doubling of group size, strongly implies that subsequent infections were mostly exchanged between calves. These findings suggest a pivotal importance for repeated low-level inoculation through social interaction, such as mutual licking, as the main source of Chlamydophila transmission in cattle. Calves in the study herd had little opportunity to contract chlamydial infection from their herd mates, being reared in hutches separate from the herd, with physical contact only with the nearest neighbor calf. Thus, infections presumably spread mainly by linear contact from calf to calf, rather than by shared contact between all animals in the herd. Most likely, transmission frequencies in herds with more contact between animals will be higher, and smaller groups than in this study will have infection incidences like those observed for large groups here. The present investigation clearly indicates the importance of crowding on spread and intensity of Chlamydophila infection in cattle, suggesting that the increased probability of shedding animals and the increased contact frequency in large herds is a major epidemiological determinant of bovine Chlamydophila infection.
While we did not specifically investigate the association of disease with chlamydial infection, we did see in this study signs of minor vaginal inflammation in infected calves, such as subepithelial granulomas and diffuse reddening, and catarrhal mastitis and prolonged postpartum vaginal secretion in infected dams. Understanding the possible association of Chlamydophila infection in cattle with disease, subclinical or manifest, will require carefully designed investigations. It was recently shown that there is a high frequency of natural Chlamydophila vaginal infection in heifers and that C. abortus infection at breeding, via the cervical or cohort route, depresses fertility of heifers (8, 9). Our study here demonstrates unambiguously that female calves acquire such vaginal infections very early in their lives by nonsexual transmission.
High constant serum antibody levels throughout the 12 postpartum weeks indicate that all dams had experienced Chlamydophila infection. Detection of Chlamydophila organisms in dams, albeit at low prevalence, indicates that specific immunostimulation by Chlamydophila infection maintains the steady-state antibody concentrations. All antibody levels in calves, but particularly IgM levels, were significantly (P < 10−4) lower than in dams. The absence of immediate postpartum Chlamydophila infection in calves and the relatively low organism levels indicate that these antibodies were colostrum-derived rather than produced by the calves after specific stimulation of the immune response in utero. The significant decline of Chlamydophila-specific serum IgG and IgA, as indicators of mucosal IgA (23), over the 12 postpartum weeks supports this notion. Conversely, the anti-Chlamydophila IgM isotype, with the shortest half-life of the antibody isotypes (16), did not decline in calves. Presumably, the antigenic stimulus from Chlamydophila infection detected in PCR-positive calves, but likely also present at lower levels in PCR-negative calves, triggered the adaptive immune response of the calves. Chlamydophila-specific first-response IgM, replenishing the declining pool of colostrum-derived IgM, apparently maintained constant IgM serum concentration in calves.
The prediction of calf infection by dam anti-Chlamydophila IgM suggests that host genetics might explain the shared susceptibility to Chlamydophila infection of dam and calf. Calves receive all maternal antibodies through colostrum fed in the first 24 h after birth (1, 3). A direct immunological influence of the dam on the infection of the calf is unlikely, since calves were fed pooled colostrum and since preweek-7 calf anti-Chlamydophila IgM failed to predict calf infection. The evidence in this study argues against direct prepartum or immediate postpartum transmission of Chlamydophila infection from dam to calf, since first infections typically were detected later than 1 week after birth, while dam and calf had been separated immediately after birth. Thus, most likely a shared genetic background explains similar susceptibilities of dam and calf to Chlamydophila infection.
An intriguing aspect of this study was the dominance of C. pecorum in mucosal infections, particularly of the vagina, in contrast to the dominance of C. abortus in milk of dams. The propensities of C. pecorum for mucosal infection and of C. abortus for systemic infection are in agreement with the consistently, albeit not statistically significantly (P ≥ 0.06), higher anti-C. pecorum IgA than anti-C. abortus IgA, but similar IgG and IgM antibody levels. Carefully designed field studies are necessary to confirm the present finding in calves, and more interestingly, elucidate potential associations between low-level Chlamydophila infection in cattle and subtle disease manifestations. Given the extremely high prevalence of Chlamydophila infection in cattle, such manifestations have the potential to continuously incur large losses in the cattle industry. Of particular interest will be the influence of C. pecorum infections on health and growth rates of calves and on fertility in adult cattle and the influence of C. abortus infection on fertility and milk production in cows.
Acknowledgments
We thank Dongya Gao for excellent technical assistance and Stuart Price and James Wright for careful reading of the manuscript.
This research was supported by grants from the Food Animal Health and Disease Research Program of the College of Veterinary Medicine, Auburn University, and from Bayer AG Animal Health Research, Monheim, Germany.
REFERENCES
- 1.Barrington, G. M., and S. M. Parish. 2001. Bovine neonatal immunology. Vet. Clin. N. Am. Food Anim. Pract. 17:463-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen, R. A., P. Spears, J. Storz, and G. E. Seidel, Jr. 1978. Mechanisms of infertility in genital tract infections due to Chlamydia psittaci transmitted through contaminated semen. J. Infect. Dis. 138:95-98. [DOI] [PubMed] [Google Scholar]
- 3.Butler, J. E. 1998. Immunoglobulin diversity, B-cell and antibody repertoire development in large farm animals. Rev. Sci. Tech. 17:43-70. [DOI] [PubMed] [Google Scholar]
- 4.Cavirani, S., C. S. Cabassi, G. Donofrio, B. De Iaco, S. Taddei, and C. F. Flammini. 2001. Association between Chlamydia psittaci seropositivity and abortion in Italian dairy cows. Prev. Vet. Med. 50:145-151. [DOI] [PubMed] [Google Scholar]
- 5.Corner, A. H., G. L. Bannister, and D. P Hill. 1968. A histological study on the effects of enzootic abortion of ewes virus in the lactating bovine mammary gland. Can. J. Comp. Med. Vet. Sci. 32:259-268. [PMC free article] [PubMed] [Google Scholar]
- 6.DeGraves, F. J., D. Gao, H. R. Hehnen, T. Schlapp, and B. Kaltenboeck. 2003. Quantitative detection of Chlamydia psittaci and C. pecorum by high-sensitivity real-time PCR reveals high prevalence of vaginal infection in cattle. J. Clin. Microbiol. 41:1726-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeGraves, F. J., D. Gao, and B. Kaltenboeck. 2003. High-sensitivity quantitative PCR platform. BioTechniques 34:106-115. [DOI] [PubMed] [Google Scholar]
- 8.DeGraves, F. J., T. Kim, J. Jee, T. Schlapp, H. R. Hehnen, and B. Kaltenboeck. Reinfection with Chlamydophila abortus by uterine and indirect cohort routes reduces fertility in cattle preexposed to Chlamydophila. Infect. Immun. 72:2538-2545. [DOI] [PMC free article] [PubMed]
- 9.DeGraves, F. J., K. Stemke-Hale, J. Huang, S. A. Johnston, K. F. Sykes, T. Schlapp, H. R. Hehnen, and B. Kaltenboeck. 2002. Vaccine identified by in vivo genomic screening enhances fertility in cattle during environmental challenge with Chlamydia, p. 265-268. In J. Schachter (ed.), Chlamydial infections. Proceedings of the 10th International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, San Francisco, Calif.
- 10.Dohoo, I., W. Martin, and H. Stryhn. 2003. Veterinary epidemiologic research. AVC Inc., Charlottetown, Canada.
- 11.Domeika, M., A. Ganusauskas, M. Bassiri, G. Froman, and P. A. Mardh. 1994. Comparison of polymerase chain reaction, direct immunofluorescence, cell culture and enzyme immunoassay for the detection of Chlamydia psittaci in bull semen. Vet. Microbiol. 42:273-280. [DOI] [PubMed] [Google Scholar]
- 12.Eugster, A. K., and J. Storz. 1971. Effect of colostral antibodies on the pathogenesis of intestinal chlamydial infections in calves. Am. J. Vet. Res. 32:711-718. [PubMed] [Google Scholar]
- 13.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 14.Fukushi, H., and K. Hirai. 1992. Proposal of Chlamydia pecorum sp. nov. for Chlamydia strains derived from ruminants. Int. J. Syst. Bacteriol. 42:306-308. [DOI] [PubMed] [Google Scholar]
- 15.Huang, J., F. J. DeGraves, S. D. Lenz, D. Gao, P. Feng, D. Li, T. Schlapp, and B. Kaltenboeck. 2002. The quantity of nitric oxide released by macrophages regulates Chlamydia-induced disease. Proc. Natl. Acad. Sci. USA 99:3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Husband, A. J., M. R. Brandon, and A. K. Lascelles. 1972. Absorption and endogenous production of immunoglobulins in calves. Aust. J. Exp. Biol. Med. Sci. 50:491-498. [DOI] [PubMed] [Google Scholar]
- 17.Jones, G. E., A. Donn, J. Machell, B. Biolatti, and S. Appino. 1998. Experimental infections of the genital tract of cattle with Chlamydia psittaci and Chlamydia pecorum, p. 446-449. In R. S. Stephens (ed.), Chlamydial infections. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, San Francisco, Calif.
- 18.Kaltenboeck, B., D. Heard, F. J. DeGraves, and N. Schmeer. 1997. Use of synthetic antigens improves detection by enzyme-linked immunosorbent assay of antibodies against abortigenic Chlamydia psittaci in ruminants. J. Clin. Microbiol. 35:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaltenboeck, B., K. G. Kousoulas, and J. Storz. 1993. Structures of and allelic diversity and relationships among the major outer membrane protein (ompA) genes of the four chlamydial species. J. Bacteriol. 175:487-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinbaum, D. G., and M. Klein. 2002. Logistic regression: a self-learning text, 2nd ed. Springer, New York, N.Y.
- 21.Kölbl, O., and A. Psota. 1968. Miyagawanellen-Isolierungen bei Polyarthritis, Pneumonie, Encephalomyelitis and interstitieller Herdnephritis (Fleckniere) der Kälber. Wien. Tierärztl. Monat. 55:443-445. [PubMed] [Google Scholar]
- 22.McNutt, S. H., and E. F. Waller. 1940. Sporadic bovine encephalomyelitis. Cornell Vet. 30:437-448. [Google Scholar]
- 23.Näslund, K., M. Tråvén, B. Larsson, A. Silván, and N. Linde. 2002. Capture ELISA systems for the detection of bovine coronavirus-specific IgA and IgM antibodies in milk and serum. Vet. Microbiol. 72:183-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien, R. G., and M. K. Kaiser. 1985. MANOVA method for analyzing repeated measures designs: an extensive primer. Psychol. Bull. 97:316-333. [PubMed] [Google Scholar]
- 25.Perez-Martinez, J. A., and J. Storz. 1985. Antigenic diversity of Chlamydia psittaci of mammalian origin determined by microimmunofluorescence. Infect. Immun. 50:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Martinez, J. A., N. Schmeer, and J. Storz. 1986. Bovine chlamydial abortion: serodiagnosis by modified complement-fixation and indirect inclusion fluorescence tests and enzyme-linked immunosorbent assay. Am. J. Vet. Res. 47:1501-1506. [PubMed] [Google Scholar]
- 27.Reggiardo, C., T. J. Fuhrmann, G. L. Meerdink, and E. J. Bicknell. 1989. Diagnostic features of chlamydia infection in dairy calves. J. Vet. Diagn. Investig. 1:305-330. [DOI] [PubMed] [Google Scholar]
- 28.Rodolakis, A., A. Souriau, J. P. Raynaud, and G. Brunault. 1980. Efficacy of a long-acting oxytetracycline against chlamydial ovine abortion. Ann. Rech. Vet. 11:437-444. [PubMed] [Google Scholar]
- 29.Rønsholt, L. U., and A. Basse. 1981. Bovine mastitis induced by a common intestinal Chlamydia psittaci strain. A pathogenetic and serological investigation. Acta Vet. Scand. 22:9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schachter, J., J. Banks, N. Sugg, M. Sung, J. Storz, and K. F. Meyer. 1975. Serotyping of Chlamydia: isolates of bovine origin. Infect. Immun. 11:904-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schachter, J., R. S. Stephens, P. Timms, C. Kuo, P. M. Bavoil, S. Birkelund, J. Boman, H. Caldwell, L. A. Campbell, M. Chernesky, G. Christiansen, I. N. Clarke, C. Gaydos, J. T. Grayston, T. Hackstadt, R. Hsia, B. Kaltenboeck, M. Leinonnen, D. Ocjius, G. McClarty, J. Orfila, R. Peeling, M. Puolakkainen, T. C. Quinn, R. G. Rank, J. Raulston, G. L. Ridgeway, P. Saikku, W. E. Stamm, D. T. Taylor-Robinson, S. P. Wang, and P. B. Wyrick. 2001. Radical changes to chlamydial taxonomy are not necessary just yet. Int. J. Syst. Evol. Microbiol. 51:249. [DOI] [PubMed] [Google Scholar]
- 32.Shewen, P. E. 1980. Chlamydial infection in animals: a review. Can. Vet. J. 21:2-11. [PMC free article] [PubMed] [Google Scholar]
- 33.Spears, P., and J. Storz. 1979. Biotyping of Chlamydia psittaci based on inclusion morphology and response to diethylaminoethyl-dextran and cycloheximide. Infect. Immun. 24:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storz, J. 1966. Psittacosis-lymphogranuloma infection of sheep. Antigenic structures and interrelations of PL agents associated with polyarthritis, enzootic abortion, intrauterine and latent intestinal infections. J. Comp. Pathol. 76:351-362. [DOI] [PubMed] [Google Scholar]
- 35.Storz, J., D. G. McKercher, J. A. Howarth, and O. C. Straub. 1960. The isolation of a viral agent from epizootic bovine abortion. J. Am. Vet. Med. Assoc. 137:509-514. [Google Scholar]
- 36.Storz, J., E. J. Carroll, E. H. Stephenson, L. Ball, and A. K. Eugster. 1976. Urogenital infection and seminal excretion after inoculation of bulls and rams with chlamydiae. Am. J. Vet. Res. 37:517-520. [PubMed] [Google Scholar]
- 37.Storz, J., E. J. Carroll, L. Ball, and L. C. Faulkner. 1968. Isolation of a psittacosis agent (Chlamydia) from semen and epididymis of bulls with seminal vesiculitis syndrome. Am. J. Vet. Res. 29:549-555. [PubMed] [Google Scholar]
- 38.Wang, F.-I., H. Shieh, and Y.-K. Liao. 2001. Prevalence of Chlamydophila abortus infection in domesticated ruminants in Taiwan. J. Vet. Med. Sci. 63:1215-1220. [DOI] [PubMed] [Google Scholar]
- 39.Wilson, M. R., and R. G. Thomson. 1968. Chlamydia pneumonia of calves. Res. Vet. Sci. 9:467-473. [PubMed] [Google Scholar]
- 40.Wittenbrink, M. M., H. A. Schoon, D. Schoon, R. Mansfeld, and W. Bisping. 1993. Endometritis in cattle experimentally induced by Chlamydia psittaci. J. Vet. Med. B 40:437-450. [DOI] [PubMed] [Google Scholar]
- 41.Wittenbrink, M. M., H. Horchler, and W. Bisping. 1988. Investigations into the incidence of Chlamydia psittaci in the genital tract and feces of female cattle. J. Vet. Med. B 35:237-246. [PubMed] [Google Scholar]