Abstract
Hemiascomycetes are species of yeasts within the order Saccharomycetales. The order encompasses disparate genera with a variety of life styles, including opportunistic human pathogens (e.g., Candida albicans), plant pathogens (e.g., Eremothecium gossypii), and cosmopolitan yeasts associated with water and decaying vegetation. To analyze the phylogeny of medically important species of yeasts, we selected 38 human pathogenic and related strains in the order Saccharomycetales. The DNA sequences of six nuclear genes were analyzed by maximum likelihood and Bayesian phylogenetic methods. The maximum likelihood analysis of the combined data for all six genes resolved three major lineages with significant support according to Bayesian posterior probability. One clade was mostly comprised of pathogenic species of Candida. Another major group contained members of the family Metschnikowiaceae as a monophyletic group, three species of Debaryomyces, and strains of Candida guilliermondii. The third clade consisted exclusively of species of the family Saccharomycetaceae. Analysis of the evolution of key characters indicated that both codon reassignment and coenzyme Q9 likely had single origins with multiple losses. Tests of correlated character evolution revealed that these two traits evolved independently.
The Ascomycota include three major groups, the monophyletic Euascomycetes (molds), the Hemiascomycetes (yeasts), and the Archiascomycetes, which contains taxa that likely arose before the other lineages (35). According to the taxonomic study of the yeasts by Kurtzman and Fell (23), the Hemiascomycetes comprise one large order, the Saccharomycetales, which was established in 1960 by Kudrjavzev (17). All members of this order are characterized by (i) no or only rudimentary hyphae, (ii) vegetative cells that proliferate by budding or fission, (iii) cell walls that lack chitin, and (iv) asci that are formed singly or in chains (17). Phylogenetic analyses of ribosomal DNA (rDNA) and RNA polymerase II (RPB2) gene sequences support a single evolutionary origin of the Saccharomycetales (1, 3, 29). However, to date, only a few representatives of the order have been included in molecular phylogenetic studies of the higher-level relationships among the Ascomycota.
The order Saccharomycetales contains many species of practical importance and scientific interest, including several species that are pathogenic for humans, such as Candida parapsilosis, Candida tropicalis, and Candida albicans, which is the most common human pathogenic fungus (4). Other species of the Saccharomycetales are exploited by industry to produce secondary metabolites and fermentative by-products (23). During the translation of mRNA to polypeptides, several species of Candida exhibit alternative codon usage. The reassignment of the codon CUG from leucine to serine was first described by Kawaguchi et al. (16) for Candida cylindraceae. Sugita and Nakase (44) applied phylogenetic methods to investigate relationships among Candida species with alternative use of the CUG codon. They showed that only 11 of 78 species of Candida used CUG as a codon for leucine. The remaining 67 species translated CUG as serine, but they did not form a monophyletic group. The authors suggested a correlation between codon reassignment and coenzyme Q9 (Co-Q9) as the predominant ubiquinone in these species based on chemotaxonomy. Co-Q is a mitochondrial electron carrier with various numbers of isoprene units. The length of the isoprene chain is usually consistent within a monophyletic group (49) and has therefore been used in yeast taxonomy.
The order Saccharomycetales is purported to comprise 11 families and 55 genera (23). However, the family assignments of several genera of budding yeasts, e.g., Pichia and Debaryomyces, remain questionable (23). Because of a lack of distinctive morphological characters, molecular methods are invaluable for clarifying the phylogenetic relationships among ascomycetous yeast species. Previous phylogenies were based on single genes (7, 21, 22), sampled only one family (25), or were restricted to the most common pathogenic species of Candida (14, 30, 31, 50). The single-gene studies have focused on the actin gene (ACT1) (7) or the large (26S rDNA) or small (18S rDNA) ribosomal subunit (21) and generally lacked good statistical support, especially for relationships along the backbone of the trees. Relationships among different genera or families of the Saccharomycetales were rarely resolved. Recently, Kurtzman and Robnett (25) presented a multigene phylogeny based on three nuclear rDNA genes (18S rDNA, 26S rDNA, and ITS [internal transcribed spacer]), three protein-coding genes (EF-1α, ACT1, and RPB2), and two mitochondrial genes (small-subunit rDNA and COX2). This impressive study included 75 species but focused on members of the “Saccharomyces complex” and included only one pathogenic species of Candida (25).
The major goal of the present study was to generate a multigenic phylogeny of the families of Saccharomycetales that included the clinical and related species. We used maximum likelihood inference and a Bayesian metropolis-coupled Markov chain Monte Carlo analysis to evaluate the statistical support for the observed evolutionary relationships. We used the phylogeny to examine the evolution of alternative codon usage and isoprene chain length in Co-Q. Although others have correlated these two traits (44), our extensive data set of DNA sequences allowed the application of different statistical methods of character mapping, reconstruction of the ancestral character state, and evaluation of correlated character evolution. We used six nuclear genes (four encoding proteins, 18S rDNA, and 26S rDNA) and included 38 species from 5 of the 11 families recognized by Kurtzman and Fell (23). Several criteria guided the selection of strains: (i) a broad taxonomic sampling that included environmental and clinical isolates, (ii) taxa with different Co-Q systems, and (iii) Candida species with reassigned codon usage. We selected strains that permitted each of these features to be analyzed independently and allowed evaluation of putative associations among them.
MATERIALS AND METHODS
Taxa and extraction of DNA.
We sampled 38 strains, representing 11 genera and 5 families of the order Saccharomycetales (23). The identity, taxonomic classification, and source of each strain are listed in Table 1, which includes the GenBank accession numbers for the sequenced genes of each taxon. All fungi were cultured on yeast extract-peptone-dextrose (Difco) agar at room temperature. Each strain was suspended in 400 μl of 2× cetyltrimethyl ammonium bromide buffer, and genomic DNA was extracted as described by Gardes and Bruns (10) and stored at −20°C. Table 1 also lists the GenBank accession numbers of rRNA sequences of Schizosaccharomyces pombe and Neurospora crassa, representative taxa of Euascomycetes and Archiascomycetes, which were compared with species of Saccharomycetales to assess their monophyly. We also included two basidiomycetes to root the Ascomycota, Althelia bombacina and Coprinus cinereus.
TABLE 1.
Taxa and DNA sequences used in this investigationa
| Order or group and family | Species | Strain(s)b | GenBank accession no.c
|
|||||
|---|---|---|---|---|---|---|---|---|
| ACT1 | EF2 | RPB1 | RPB2 | 18S rDNA | 26S rDNA | |||
| Saccharomycetales | ||||||||
| Candidaceae | Candida albicans | CBS 562* | AJ389057 | AY497649† | AY497718† | AY497614† | AY497754† | AY497682† |
| Candida albicans | CBS 1905 | AY497580† | AY497639† | AY497708† | AY497604† | AY497744† | AY497673† | |
| Candida castellii | CBS 4332* | AY497583† | AY497647† | AY497717† | AY497612† | AY497752† | AY497680† | |
| Candida dubliniensis | CBS 7987 | AJ389058 | AY497659† | AY497730† | AY497625† | AY497766† | AY497694† | |
| Candida glabrata | CBS 138* | AJ389073 | AY497636† | AY497705† | AY497601† | AY497741† | AY497670† | |
| Candida guilliermondii | MMRL 1635 | AY497592† | AY497661† | AY497732† | AY497627† | AY497768† | AY497696† | |
| Candida guilliermondii | MMRL 1636 | AY497593† | AY497662† | AY497733† | AY497628† | AY497769† | AY497697† | |
| Candida guilliermondii | MMRL 1759 | AY497594† | AY497663† | AY497734† | AY497629† | AY497770† | AY497698† | |
| Candida intermedia | CBS 572* | AY497584† | AY497650† | AY497719† | AY497615† | AY497755† | AY497683† | |
| Candida maltosa | CBS 5611 | AY653540† | AY653542† | AY653536† | AY653538† | AY653544† | AY653546† | |
| Candida norvegica | CBS 4239* | AY497582† | AY497646† | AY497715† | AY497611† | AY497751† | AY497679† | |
| Candida parapsilosis | ATCC 96138 | AY497589† | AY497631† | AY497700† | AY497596† | AY497736† | AY497665† | |
| Candida parapsilosis | ATCC 96140 | AY497590† | AY497632† | AY497701† | AY497597† | AY497737† | AY497666† | |
| Candida parapsilosis | ATCC 96144 | AY497591† | AY497633† | AY497702† | AY497598† | AY497738† | AY497667† | |
| Candida parapsilosis | CBS 604* | AY497585† | AY497652† | AY497722† | AY497617† | AY497758† | AY497686† | |
| Candida tropicalis | CBS 94* | AY497587† | AY497660† | AY497731† | AY497626† | AY497767† | AY497695† | |
| Candida viswanathii | CBS 4024* | AJ389061 | AY497645† | AY497714† | AY497610† | AY497750† | AY497678† | |
| Candida zeylanoides | CBS 619* | AJ389069 | NDd | AY497724† | AY497619† | AY497760† | AY497688† | |
| Eremotheciaceae | Eremothecium gossypii | ATCC 10895 | AY497588 | AY497630 | AY497699 | AY497595 | AY497735 | AY497664 |
| Metschnikowiaceae | Clavispora lusitaniae | CBS 6936* | AJ389065 | AY497655† | AY497726† | AY497621† | AY497762† | AY497690† |
| Clavispora opuntiae | CBS 7068* | AJ389067 | AY497656† | AY497727† | AY497622† | AY497763† | AY497691† | |
| Metschnikowia pulcherrima | CBS 610 | AJ389071 | AY497653† | AY497723† | AY497618† | AY497759† | AY497687† | |
| Saccharomycetaceae | Debaryomyces carsonii | CBS 2285* | AY497687† | AY497643† | AY497712† | AY497608† | AY497748† | AY497676† |
| Debaryomyces etchellsii | CBS 2011* | AJ389068 | AY497641† | AY497710† | AY497606† | AY497746† | U45809 | |
| Debaryomyces hansenii | CBS 767 | AY497586† | AY497658† | AY497729† | AY497624† | AY497765† | AY497693† | |
| Issatchenkia orientalis | CBS 573 | AJ389087 | AY497651† | AY497720† | AY497616† | AY497756† | AY497684† | |
| Kluyveromyces lactis | CBS 683* | AJ389080 | AY497654† | AY497725† | AY497620† | AY497761† | AY497689† | |
| Kluyveromyces marxianus | CBS 712* | AJ389078 | AY497657† | AY497728† | AY497623† | AY497764† | AY497692† | |
| Lodderomyces elongisporus | CBS 2605 | AY653541† | AY653543† | AY653537† | AY653539† | AY653545† | AY653547† | |
| Pichia fermentans | CBS 1876 | AJ389083 | AY497638† | AY497707† | AY497603† | AY497743† | AY497672† | |
| Pichia guilliermondii | CBS 2030* | AJ389063 | AY497642† | AY497711† | AY497607† | AY497747† | AY497675† | |
| Pichia jadinii | CBS 1600* | AJ389090 | AY497637† | AY497706† | AY497602† | AY497742† | AY497671† | |
| Pichia membranifaciens | CBS 107* | AJ389088 | AY497634† | AY497703† | AY497599† | AY497739† | AY497668† | |
| Pichia norvegensis | CBS 1922 | AJ389085 | AY497640† | AY497709† | AY497605† | AY497745† | AY497674† | |
| Saccharomyces cerevisiae | CBS 1177* | AJ389075 | AY497635† | AY497704† | AY497600† | AY497740† | AY497669† | |
| Saccharomyces kluyveri | CBS 3082* | AJ389077 | AY497644† | AY497713† | AY497609† | AY497749† | AY497677† | |
| Dipodascaceae | Stephanoascus ciferrii | CBS 4856* | ND | AY497648† | AY497718† | AY497613† | AY497753† | AY497681† |
| Yarrowia lipolytica | CBS 599 | ND | ND | AY497721† | ND | AY497757† | AY497685† | |
| Outgroup for phylogenetic analysis of the Hemiascomycetes | ||||||||
| Schizosaccharomycetaceae | Schizosaccharomyces pombe | Y00447 | D83976 | X56564 | D13337 | L20414 | U40085 | |
| Sordiariaceae | Neurospora crassa | U78026 | AF258620 | AABX01000048.1 | AF107789 | X04971 | AF266411 | |
| Euascomycota | ||||||||
| Arthrodermataceae | Arthroderma benhamiae | AY083233.1 | ND | |||||
| Arthroderma ciferrii | AB015769.2 | AB04068.1 | ||||||
| Cochliobolus | Curvularia brachyspora | L36995 | AF279380 | |||||
| Cudoniaceae | Cudonia circinans | AF1074343 | AF279379 | |||||
| Discinaceae | Gyromitra esculenta | U42648 | AY640952 | |||||
| Dothideaceae | Dothidea insculpta | U42474 | AY640949 | |||||
| Gyalectaceae | Gyalecta ulmi | AF465464 | AF465463 | |||||
| Herpotrichiellaceae | Capronia pilosella | U42473 | AF279377 | |||||
| Exophiala jeanselmei | X80705 | AF050271 | ||||||
| Morchellaceae | Morchella esculenta | U42642 | AF279398 | |||||
| Neolectaceae | Neolecta irregularis | Z47721 | AF279401 | |||||
| Onygenaceae | Ajellomyces capsulatus | AF320009 | AF071952 | |||||
| Coccidioides immitis | M55627 | AB040702 | ||||||
| Paracoccidioides brasiliensis | AF22751 | U81263.1 | ||||||
| Peltigeraceae | Peltigera canina | AF356681 | AF286822 | |||||
| Peltulaceae | Peltula obscurans | AF356692 | AF356693 | |||||
| Pezizaceae | Peziza quelepidotia | AY640999 | AY640959 | |||||
| Rhytismataceae | Rhytisma acerinum | AF356695 | AF356696 | |||||
| Rocellaceae | Dendrographa minor | AF279381 | AF279383 | |||||
| Stereocaulaceae | Stereocaulon paschale | AF279412 | AF279413 | |||||
| Taphrinaceae | Taphrina pruni | AB000956 | Z49792 | |||||
| Trichocomaceae | Aspergillus fumigatus | AF397059.1 | AJ438344 | |||||
| Emericella nidulans | AB008403 | AF109337 | ||||||
| Hamigera avellanea | D14406 | AB000620 | ||||||
| Penicillium chrysogenum | AF411201.1 | AF034857 | ||||||
| Penicillium freii | AY640998 | AY640958 | ||||||
| Penicillium viridicatum | AY083222.1 | AF003358.1 | ||||||
| Umbilicariaceae | Lasallia pensylvanica | AF356664 | AF356665 | |||||
| Valsaceae | Diaporthe phaseolorum | L36985 | U47830 | |||||
| Verrucariaceae | Dermatocarpon luridum | AY640989 | AY640948 | |||||
| Verrucaria pachyderma | AF356667 | AF356668 | ||||||
| Xylariaceae | Seynesia erumpens | AF279409 | AF279410 | |||||
| Outgroup for phylogenetic analysis of Ascomycota | ||||||||
| Corticiacaea | Athelia bombacina | M55638 | AF279377 | |||||
| Psathyrellaceae | Coprinus cinereus | M92991 | AF041494 | |||||
The nomenclature was taken from Kurtzman and Fell (23) (Hemiascomycetes) or the National Center for Biotechnology Information TaxBrowser (Euascomycota) (www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html).
ATCC, American Type Culture Collection; CBS, Centraalbureau vor Schimmelcultures; MMRL, Medical Mycological Research Laboratory, Wiley Schell, Duke University Medical Center.
*, type strain. †, sequences were generated as part of this study.
ND, no data available.
Amplification and sequencing of gene fragments.
Using the primers indicated in Table 2, we amplified and sequenced a portion of the following genes from each taxon: actin (ACT1), elongation factor 2 (EF2), RNA polymerase B subunit (RPB1), small-subunit rRNA (18S rDNA), and large-subunit rRNA (26S rDNA). The resulting amplicons ranged in size from approximately 600 (EF2 and ACT1) to 1,400 (26S rDNA) nucleotides. The PCR mixture consisted of 25 μl of 75 mM Tris-HCl (pH 8.0), 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% Tween 20, 7.5 pmol of each primer, 100 μM deoxynucleoside triphosphates, 1.0 U of Taq polymerase (Marsh), and 20 ng of genomic DNA. An initial denaturation at 95°C for 5 min was followed by 35 to 45 cycles in which the annealing occurred at 50°C (or 58°C for primers EFIIF1 and EFIIR2) for 30 s, elongation at 72°C for 45 s, denaturation at 95°C for 60 s, and a final extension for 10 min at 72°C. To amplify the 1.2-kb fragment of the RPB2 gene, we used the PCR protocol of Liu et al. (29). Amplicons were sequenced by using BigDye terminator chemistry with separation and detection carried out on an ABI 3700 (Applied Biosystems) automated sequencer.
TABLE 2.
Primers used for PCR and cycle sequencing
| Gene | Primer | Sequence (5′ → 3′) | Positiona | Reference or sourceb |
|---|---|---|---|---|
| EF2 | EFIIF1 | AAGTCTCCAAACAAAGCATAAC | 2983-3005 | This study |
| EFIIR2 | GGGAAAGCTTGACCACCAGTAGC | 3602-3625 | This study | |
| EFII3F | TGAAAATGGTGTCATTAACC | 3054-3074 | This study | |
| EFII4R | TTCGTTAACTGGCAAGTAAG | 3551-3571 | This study | |
| ACT1 | ACT1 | TACCCAATTGAACACGGTAT | 1996-2015 | This study |
| ACT2 | TCTGAATCTTTCGTTACCAAT | 2575-2595 | This study | |
| ACT1-2F | TACAACGAATTGAGAGTTGC | 728-745 | This study | |
| ACT1-2R | GAAGAAGTTTGCATTTCTTG | 1329-1349 | This study | |
| RPB1 | Af | GARTGYCCDGGDCAYTTYGC | 224-244 | B. Hall |
| Cr | CCNGCDATNTCRTTRTCCATRTA | 951-974 | B. Hall | |
| RPB2 | RPB2-5F | GAYGAYMGWGATCAYTTYGG | 1159-1179 | 29 |
| RPB2-1014R | CCRCAIGCYTGICCYTCDGG | 994-1014 | F. Lutzoni | |
| RPB2-7R | CCCATWGCYTGCTTMCCCAT | 2349-2369 | 29 | |
| RPB2-980F | TCYCCIGCIGARACICCHGARGG | 980-1003 | F. Lutzoni | |
| 18S rDNA | nSSU 131 | CAGTTATCGTTTATTTGATAGTACC | 125-150 | 15 |
| NS3 | GCAAGTCTGGTGCCAGCAGCC | 598-619 | 48a | |
| NS1 | GTAGTCATATGCTTGTCTC | 20-38 | 48a | |
| NS22 | AATTAAGCAGACAAATCACT | 1307-1327 | R. Vilgalys | |
| SR4 | AAACCAACAAAATAGAA | 820-838 | R. Vilgalys | |
| 26S rDNA | LROR | ACCCGCTGAACTTAAGC | 26-42 | R. Vilgalys |
| LR5 | TCCTGAGGGAAACTTCG | 948-964 | R. Vilgalys | |
| LR7 | TACTACCACCAAGATCT | 1448-1432 | R. Vilgalys | |
| LR2R | AAGAACTTTGAAAAGAG | 374-389 | R. Vilgalys | |
| LR3 | CCGTGTTTCAAGACGGG | 651-635 | R. Vilgalys |
Position of the primer relative to C. albicans or S. cerevisiae (in italics). ND, no data available.
URLs for sources are as follows: B. Hall, http://faculty.washington.edu/benhall/; F. Lutzoni, http://www.lutzonilab.net/pages/primer.shtml; R. Vilgalys, http://www.biology.duke.edu/fungi/mycolab/primers.html.
Alignment of sequences and phylogenetic analyses. (i) Assembly and alignment of DNA sequences.
Sequence chromatograms were analyzed, assembled, and edited with Sequencher 4.1 (Gene Codes Corporation). After conversion to a FASTA format, the sequences of the EF2, ACT1, and RPB1 genes were automatically aligned by using the internet alignment program Multalin (6). RPB2 and the rDNA genes were aligned by using ClustalW 1.81 (48). Each gene alignment was imported into MacClade 4.05 (33) and manually edited. In addition, the four protein-coding genes were aligned according to the superimposed amino acid sequences and the rDNA genes were aligned with reference to the secondary structure, as described by Kjer (19). Secondary structure models for 18S and 26S rRNA subunits for Saccharomyces cerevisiae were obtained from the comparative RNA website (5). These models were used as templates for coding and alignment of the entire data set. In all six data sets, ambiguously aligned characters were excluded from the phylogenetic analyses.
(ii) Tests for substitutions and saturation of codon positions.
All three codon positions in the protein-coding genes were tested independently for saturation. This was achieved by plotting genetic distances in a two-parameter model (F84; described by Kishino and Hasegawa [18]) and the uncorrected p-distance estimates (20). Deviation from a 1:1 ratio of the two distances was visually evaluated.
(iii) Data set partitioning, maximum likelihood, and Bayesian analyses.
To appropriately model nucleotide substitutions, the data set was partitioned in several ways. In addition to analyzing all the data together, we analyzed separately the data for each gene. Each of the four protein-coding genes was analyzed for differences in the first two codon positions and the third codon position. The sequence data were also partitioned into protein-coding and rDNA genes, and the resolving powers of these data sets were compared by using maximum likelihood.
We initially analyzed each gene of the Saccharomycetales data set by using MrBayes v3.0B4 (12). Each protein-coding gene was partitioned into two character sets (first and second codon positions versus the third codon position), and the best-fitting evolutionary model was determined for each partition by using Modeltest 3.06 (40) (Table 3). Each Bayesian analysis consisted of six runs of 2,000,000 generations, each using the default, uniform priors, and a sample frequency of 100. Likelihood scores of each sampled generation were plotted by using Excel (Microsoft Corp.) and visually analyzed. The trees collected before the stationary phase of the chain was reached were discarded. The trees remaining from each of the six runs were combined, and a 95% consensus tree for each gene was generated by using PAUP* 4.0b10 (45). Consensus trees for the six different genes were then compared for topological congruence, as described by Kauff and Lutzoni (15). Taxa were considered to be in conflict when they showed different relationships in two genes, supported by posterior probabilities of ≥95%.
TABLE 3.
Overview of evolutionary substitution models applied to combined- and single-gene analyses and to different codon positions in the protein-coding genes
| Fungal data set | Substitution models applied to single or combined genes
|
Substitution models applied to different codon positions of a gene
|
||||
|---|---|---|---|---|---|---|
| Gene(s) | Alignment length (bp) | Modela | Gene | Codon position(s) | Modela | |
| Ascomycota | 18S and 26S rDNA | 3,057 | TrN+I+G | RPB1 | 1 and 2 | TrN+G |
| Saccharomycetales | 6 genes | 5,064 | TrN+I+G | RPB1 | 3 | TVM+I+G |
| RPB1 | 825 | TrNef+I+G | RPB2 | 1 and 2 | TVM+I+G | |
| RPB2 | 1,238 | TrN+I+G | RPB2 | 3 | GTR+I+G | |
| ACT1 | 489 | GTR+I+G | ACT1 | 1 and 2 | JC+G | |
| EF2 | 603 | TrN+G | ACT1 | 3 | GTR+I+G | |
| 18S rDNA | 1,149 | TrN+I+G | EF2 | 1 and 2 | TVM+I+G | |
| 26S rDNA | 760 | TIM+I+G | EF2 | 3 | TIM+I+G | |
Maximum likelihood analyses were conducted on the combined data for Saccharomycetales and Ascomycota and the protein-coding and rDNA genes within the Saccharomycetales, using model parameter estimates obtained from Modeltest (Table 3). For each maximum likelihood analysis, a heuristic search was conducted using a maximum parsimony tree as the starting tree in PAUP* 4.0b10 (45). Starting trees were obtained by a parsimony analysis with 1,000 random taxon-addition replicates, saving 10 trees per replicate.
Maximum likelihood analyses were applied to the combined data for all the Saccharomycetales in a homogeneous analysis as described above. We also conducted two heterogeneous Bayesian analyses using the same search options and combined model settings that were used for the single-gene analyses. In the first heterogeneous analysis, each of the six genes was accommodated with a different model (Table 3) without consideration of different codon positions (“6-model analysis”). For the second heterogeneous analysis, separate models were applied to the combined first and second codon positions and to the third codon positions of the four protein-coding genes (“10-model analysis”); the 18S and 26S rDNA data sets were not further partitioned (Table 3). We also compared the protein-coding and rDNA genes in separate heterogeneous analyses. Different evolutionary models were applied to the codon positions in the combined protein-coding genes as described above (“8-model analysis”). The rDNA genes were analyzed with one model for the 18S rDNA and another for the 26S rDNA (“2-model analysis”).
A likelihood ratio test was used to test whether the heterogeneous model was a significant improvement over the homogeneous model and to determine which heterogeneous analysis best fit the Saccharomycetales data (8). The likelihood of the observed data and the degrees of freedom were calculated using PAUP* 4.0b10 (45) for the homogeneous analysis and p4 v. 0.80 (9) for the 6- and 10-model analyses. To assess whether the likelihood of the more complex model was a significant improvement, the likelihood ratio was compared to a χ2 distribution.
(iv) Reconstruction of the ancestral character states and testing for the correlation of character evolution.
To study the evolutionary history of alternative codon usage and number of isoprene chain units in the Co-Q system within the Saccharomycetales, we reconstructed the ancestral character states of these traits. Data for translation of the CUG codon were obtained from the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) and an earlier study by Sugita and Nakase (44) and scored as leucine (1) or serine (0). Information on the Co-Q system was gathered from Kurtzman and Fell (23), supplemented by the CBS database (http://www.cbs.knaw.nl). For the Co-Q system, we coded Co-Qs with nine isoprene units as 1 and Co-Qs with any other isoprene chain length as 0. Each trait was mapped under the likelihood criterion as implemented in Mesquite v. 1.02 (32) on the maximum likelihood tree of the Saccharomycetales, using inferred branch lengths. The likelihood of the observed character state distributions was calculated by using two models: a one-parameter Markov k-state model (27), which is a generalization of the Jukes-Cantor model (13), and an asymmetrical two-parameter Markov k-state model, which allows two different rates of change (forward and reverse) (36). A likelihood ratio test (8) was applied, and the model with the best fit was chosen for ancestral character state reconstruction. At each node the ratios of likelihoods for both character states (0 and 1) were calculated. A likelihood ratio of at least 7:1 for a given character state at a node was considered to be significant (42).
To test whether the two characters evolved independently, we used a continuous Markov model in a maximum likelihood framework, as described by Pagel (36, 38, 39) and implemented in the program Discrete (37). Discrete uses the likelihood ratio test to compare likelihoods for a model with independent transition rates for each character and a model where the transition rate for one character is dependent on the state of the other characters. The null hypothesis of independent evolution is rejected if the model of correlated evolution fits the data significantly better than the simpler model of independent evolution.
RESULTS
Phylogeny of the Hemiascomycetes.
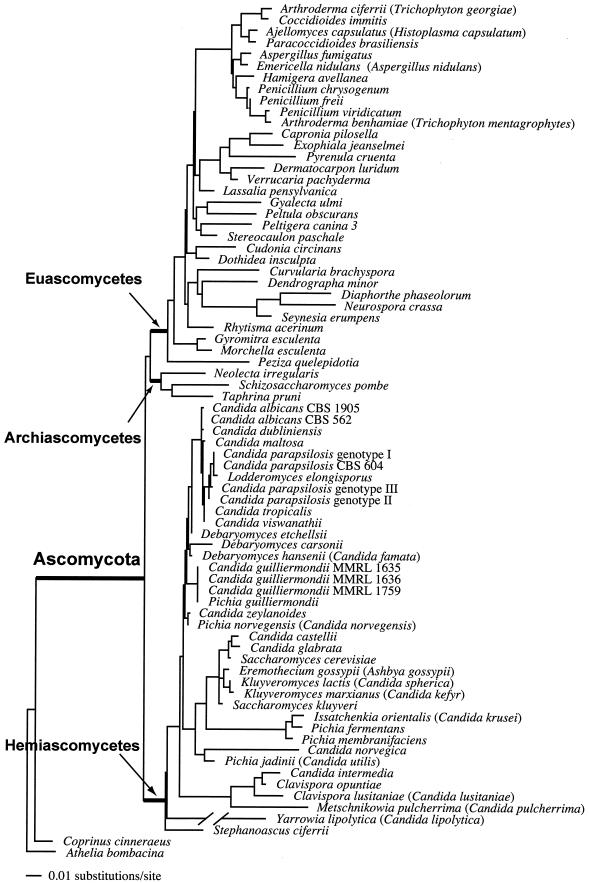
Analysis of 3,057 aligned rDNA characters of 73 ascomycetous taxa by using maximum likelihood revealed the monophyletic origin of the Hemiascomycetes (Saccharomycetales). With a likelihood value (−ln L) of 10,839, the phylogeny is supported by the data. The homogeneous Bayesian analysis supported this clade, with a significant posterior probability (100%) (Fig. 1). There was significant support for monophyly of the Euascomycetes (100%) and of the Archiascomycetes (97%). The Archiascomycetes and Euascomycetes form a monophyletic sibling (or “sister”) clade to the Hemiascomycetes, although support for this relationship is low. Within the Hemiascomycetes only a few terminal relationships were resolved with significant support (Fig. 1). No statistical support was observed for the backbone of the tree within the Hemiascomycetes. Although the topology differs from the tree obtained from the Saccharomycetales data (Fig. 2), none of these conflicts was supported in the analysis of the rDNA genes.
FIG. 1.
Single most likely tree based on a combined analysis of the nuclear 18S rDNA and 26S rDNA of 73 ascomycetous taxa, using two basidiomycetes as outgroups. The nodes marking the Ascomycota, Euascomycetes, Archiascomycetes, and Hemiascomycetes as well as many terminal branches are supported by homogeneous Bayesian posterior probabilities ≥95%. Arrows indicate the origins of the three classes of the phylum Ascomycota. The branch length for Y. lipolytica = 0.22906 U of expected substitutions per site.
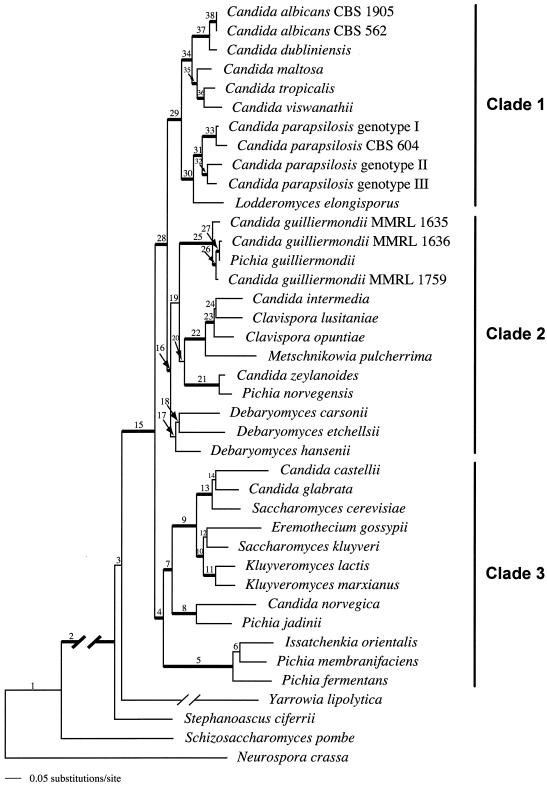
FIG. 2.
Combined maximum likelihood analysis of six genes (ACT1, EF2, RPB1, RPB2, 18S rDNA, and 26S rDNA) for 38 taxa of Hemiascomycetes and two outgroup species, an Archiascomycete (S. pombe) and a Euascomycete (N. crassa). Thickened lines denote heterogeneous Bayesian posterior probabilities ≥95% as calculated in the combined analysis. Node numbers are indicated above each branch. Refer to Table 4 for the statistical support of each node by each gene, as well as the combined multigenic support. Branch lengths leading to Y. lipolytica (0.19157) and the outgroup taxa (0.31967) were shortened to fit the figure. Black lines on the right indicate the three clades recognized in this study.
Analyses of single genes and topological congruence.
The phylogeny of the Hemiascomycetes inferred from each individual gene (data not shown) was compared with the phylogenetic tree from the heterogeneous Bayesian analysis of the combined genes (Fig. 2). For this comparison, we considered all clades that were statistically supported by posterior probabilities (≥95%) (28 groups; thick branches in Fig. 2). Although single-gene trees were compatible with the combined phylogeny in almost all cases, only 29 to 64% as many clades received significant support in the single-gene trees compared to the combined analysis. The individual genes and the number of groups with significant support are as follows: RPB1 (18 clades), RPB2 (15 clades), EF2 (10 clades), 18S rDNA (10 clades), 26S rDNA (10 clades), and ACT1 (8 clades). Table 4 lists each node recognized in the combined analysis (Fig. 2) and its statistical support as calculated in the single-gene tree. Twelve nodes were not supported by any of the genes. However, 5 of the 12 (nodes 4, 15, 16, 32, and 35) appeared in the combined analysis with significant support. The remaining seven were recognized but not significantly supported. Fifteen clades were supported when the rDNA genes were combined, but when the four protein-coding genes were analyzed, 35 groups were resolved with statistically significant support. In the combined analysis of protein-coding genes and rDNA genes, 7 of the 35 nodes lost their statistical support. These nodes can be found throughout the tree: node 3 is located on the backbone, nodes 19 and 20 define relationships of groups of 10 and 6 taxa, respectively, and nodes 6, 12, 14, and 24 show sibling relationships between two terminal taxa.
TABLE 4.
Single-gene posterior probabilities for nodes in the combined analysis of the Saccharomycetales shown in Fig. 2
| Group | Node | Posterior probabilitya
|
||||||
|---|---|---|---|---|---|---|---|---|
| Single-gene analyses
|
Combined analysis (28) | |||||||
| ACT1 (8) | EF2 (10) | RPB1 (18) | RPB2 (15) | 18S rDNA (10) | 26S rDNA (10) | |||
| 1 | <0.01 | <0.01 | <0.01 | <0.01 | 1.00 | <0.01 | <0.01 | |
| 2 | 1.0 | 1.00 | 1.00 | 0.31 | 0.77 | 1.00 | 1.00 | |
| 3 | <0.01 | <0.01 | <0.01 | 0.89 | 0.33 | <0.01 | 0.68 | |
| Clade 3 | 4 | <0.01 | <0.01 | 0.42 | 0.50 | 0.76 | <0.01 | 1.00 |
| 5 | <0.01 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| 6 | <0.01 | 0.88 | <0.01 | 0.54 | <0.01 | <0.01 | 0.86 | |
| 7 | <0.01 | <0.01 | 0.69 | 1.00 | <0.01 | <0.01 | 1.00 | |
| 8 | 0.99 | 1.00 | 1.00 | 1.00 | 0.95 | 0.33 | 1.00 | |
| 9 | <0.01 | 0.66 | 1.00 | 1.00 | 0.94 | 0.48 | 1.00 | |
| 10 | <0.01 | <0.01 | 1.00 | 0.90 | 0.99 | 0.68 | 1.00 | |
| 11 | 1.0 | 0.31 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| 12 | 0.29 | <0.01 | 0.77 | 0.16 | <0.01 | 0.39 | 0.63 | |
| 13 | <0.01 | <0.01 | 1.00 | 1.00 | <0.01 | 0.59 | 1.00 | |
| 14 | <0.01 | <0.01 | <0.01 | <0.01 | 0.25 | 0.64 | 0.31 | |
| 15 | <0.01 | <0.01 | 0.94 | <0.01 | 0.78 | <0.01 | 0.96 | |
| Clade 2 | 16 | <0.01 | <0.01 | 0.94 | 0.43 | <0.01 | <0.01 | 0.96 |
| 17 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.88 | |
| 18 | <0.01 | <0.01 | <0.01 | 0.83 | <0.01 | <0.01 | 0.55 | |
| 19 | <0.01 | 0.34 | <0.01 | <0.01 | <0.01 | <0.01 | 0.85 | |
| 20 | <0.01 | <0.01 | 0.98 | <0.01 | <0.01 | <0.01 | 0.75 | |
| 21 | <0.01 | <0.01 | 1.00 | 1.00 | <0.01 | 1.00 | 1.00 | |
| 22 | <0.01 | <0.01 | 0.48 | 0.99 | 1.00 | 0.64 | 1.00 | |
| 23 | <0.01 | <0.01 | <0.01 | <0.01 | 1.00 | 0.62 | 1.00 | |
| 24 | 0.21 | <0.01 | <0.01 | 0.28 | 0.97 | <0.01 | 0.73 | |
| 25 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| 26 | <0.01 | 0.78 | 0.74 | 1.00 | 0.20 | 0.20 | 1.00 | |
| 27 | 0.20 | 1.00 | 0.97 | 1.00 | 0.20 | 0.20 | 1.00 | |
| 28 | <0.01 | 1.00 | 0.99 | 0.32 | 0.47 | <0.01 | 0.96 | |
| Clade 1 | 29 | <0.01 | <0.01 | 0.85 | 0.51 | 1.00 | 0.98 | 1.00 |
| 30 | 1.00 | <0.01 | 1.00 | 0.51 | <0.01 | 0.99 | 1.00 | |
| 31 | 0.75 | 0.98 | 0.18 | 0.51 | <0.01 | 0.97 | 1.00 | |
| 32 | 0.24 | 0.33 | 0.62 | 0.93 | 0.20 | 0.40 | 1.00 | |
| 33 | 0.99 | 1.00 | 1.00 | 0.51 | 0.20 | 1.00 | 1.00 | |
| 34 | 0.40 | <0.01 | 1.00 | 1.00 | <0.01 | <0.01 | 1.00 | |
| 35 | 0.19 | 0.79 | <0.01 | 0.92 | <0.01 | <0.01 | 1.00 | |
| 36 | 0.36 | <0.01 | 1.00 | 1.00 | <0.01 | 0.34 | 1.00 | |
| 37 | 1.00 | 1.00 | 1.00 | 1.00 | <0.01 | 1.00 | 1.00 | |
| 38 | 1.00 | 0.98 | 1.00 | 1.00 | <0.01 | 0.84 | 1.00 | |
Significant nodes are shown in bold. The total numbers of significant nodes are shown in parentheses.
Among the protein-coding genes, the analysis revealed substitutional saturation in the third codon position (data not shown). For the first and second codon positions, we found no deviation from a 1:1 ratio in the distance plot, and therefore, these codon positions were unsaturated. Hence, evolutionary models were assigned to the first and second codon positions versus the third codon position in the analysis of single genes. In two cases, there was statistically significant conflict (posterior probability of 95%) among trees from the six genes. First, analysis of ACT1 grouped Issatchenkia orientalis with Pichia norvegensis, an association that was not observed in any other gene tree. In the analyses of the RPB1, RPB2, and the 26S rDNA gene sequences, P. norvegensis appeared to be closely related to Candida zeylanoides, with a posterior probability of 100%. BLAST searches of ACT1 sequences of I. orientalis, P. norvegensis, and C. zeylanoides in GenBank confirmed their identity. Second, in the EF2 gene phylogeny, P. norvegensis appeared most closely related to Candida intermedia. This result could be attributed to missing EF2 nucleotide data for C. zeylanoides, which was a sibling species to P. norvegensis in the other gene analyses.
Phylogenetic analysis of the combined data sets for Saccharomycetales. (i) Homogeneous and heterogeneous Bayesian analyses of the Saccharomycetales.
The maximum likelihood analysis of the combined data for the six genes (5,064 aligned nucleotides) resulted in the single tree shown in Fig. 2 (−ln L 48,580; 8 df). The heterogeneous analyses of the combined data set resulted in trees with −ln L 47,829 (6-model analysis) and −ln L 46,486 (10-model analysis) at 142 and 168 df, respectively. The likelihood ratio for the homogeneous analysis and the 6-model analysis revealed that the heterogeneous model significantly improved the likelihood of the data. The 6-model analysis was then compared with the 10-model analysis. Applying different evolutionary models to codon positions, as in the 10-model analysis, results in the highest likelihood and statistical support for the relationships within the Saccharomycetales.
(ii) Phylogenetic relationships among Saccharomycetales.
Three major clades were resolved within the Saccharomycetales with strong support. Clade 1 originates with node 29, clade 2 originates at node 16, and clade 3 originates at node 4 (Fig. 2). Yarrowia lipolytica was significantly supported as a sibling species to these three clades. Stephanoascus ciferrii formed the most basal taxon of the order, although the position of S. ciferrii was supported only by the analysis of the protein-coding genes. Clade 1 is comprised of six Candida species (C. albicans, C. dubliniensis, C. maltosa, C. tropicalis, C. viswanathii, and C. parapsilosis) and Lodderomyces elongisporus. They are most closely related to clade 2, which contains a monophyletic clade of the C. guilliermondii complex, the Metschnikowiaceae (i.e., species of Clavispora and Metschnikowia), and the sibling species C. zeylanoides and P. norvegensis. These taxa have a sibling relationship with a monophyletic clade of three Debaryomyces species. However, this association was significantly supported only by the analysis of the protein-coding genes, and there was no support for monophyly of the Debaryomyces clade. Clade 3 contains the Saccharomycetaceae and has a sibling relationship with clades 1 and 2 (Fig. 2). In clade 3, S. cerevisiae appears most closely related to Candida castellii and Candida glabrata. These three species are related to a clade composed of Eremothecium gossypii, Saccharomyces kluyveri, Kluyveromyces lactis, and K. marxianus, which together form the sibling group to Candida norvegica and Pichia jadinii. A clade of Issatchenkia orientalis, Pichia fermentans, and Pichia membranifaciens completes this sample of Saccharomycetaceae.
Ancestral character state evolution.
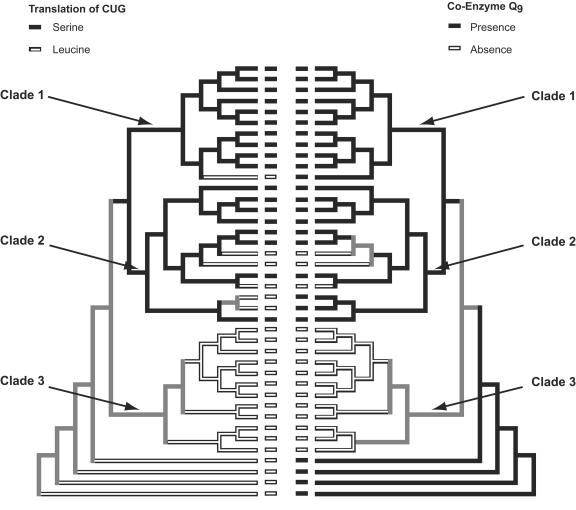
Ancestral character states for codon reassignment were reconstructed under the asymmetric two-parameter model. The logarithmic likelihood calculated under the model (−15.57) was significantly greater than that calculated under the one-parameter Markov k-state model (−20.57) at α = 0.05. The reconstruction indicated that codon reassignment occurred once in the evolutionary history of the Saccharomycetales in the most common ancestor of clades 1 and 2 (Fig. 3). The forward transition rate (0→1) was calculated to be 0.16, while the backward rate (1→0) was 4.26. Losses of the character are therefore more likely than gains. The left panel in Fig. 3 shows the reconstructed character states for each node. Gains or losses were assigned to nodes with statistical support higher than 87% support (black and white branches in Fig. 3). A character state could not be assigned unambiguously for nodes with less than 87% support (gray branches). The reconstruction indicates that codon reassignment occurred once, and there were at least five specific losses at the branches leading to Lodderomyces elongisporus, Clavispora opuntiae, Metschnikowia pulcherrima, Pichia norvegensis, and the Debaryomyces species (compare Fig. 2 and 3).
FIG. 3.
Diagrammatical representation of phylogram in Fig. 2 with character states for the translation of CUG (left side) and the presence or absence of Co-Q9 (right side) in the terminal taxa. The small rectangles in the center denote each species in Fig. 2. Different branch shading demarks reconstruction of ancestral character states. Black and white branches represent branches for which the presence or absence of the character could be reconstructed unambiguously with a statistical support value of ≥87%. Uncertainty in character reconstruction is indicated by gray branches (<87%).
For the Co-Q9 reconstruction, the likelihood ratio test showed no significant improvement for the asymmetry two-parameter model (−ln L = 16.68) over the one-parameter Markov k-state model (−ln L = 16.69). Hence, the latter was chosen to assess the evolution of Co-Q9. Transitions from 0 to 1 and vice versa were calculated to have occurred at a rate of 1.093. Character states of nodes in the tree were assigned as described above for codon recapture. The reconstruction of the evolution of nine isoprene subunits of Co-Q suggests that the most common ancestor of all three clades had nine subunits and that multiple losses occurred within clades 2 and 3 (Fig. 2 and 3).
When correlated evolution between codon recapture and Co-Q9 was tested, the null hypothesis of independent evolution could not be rejected (P = 0.1430). Likelihoods for two models of evolution (independent versus dependent) were calculated and compared in a likelihood ratio statistic. The likelihood of the model of dependent evolution was not significantly higher than the likelihood of the independent model. Since no significant correlation between the evolution of these characters could be detected by these reconstruction methods, we conclude that they evolved independently.
DISCUSSION
The phylogenetic results provide statistically significant support for the monophyletic origin of two families of Saccharomycetales, the Metschnikowiaceae and Saccharomycetaceae. Previous studies also concluded that these families are monophyletic, albeit with low statistical support (7, 21). The combination of multiple-gene and statistical approaches provided strong support for these two families. Kurtzman and Fell (23) were uncertain about the placement of the genera Issatchenkia and Debaryomyces within the Saccharomycetaceae. Our results suggest that Issatchenkia is a basal genus of the Saccharomycetaceae (clade 3). The three species of Debaryomyces appear to be closely related to the Metschnikowiaceae (clade 2).
Most, but not all, of the pathogenic species of Candida were placed within the well-supported clade 1. However, common pathogenic Candida species and emerging pathogenic yeasts (11) can be found in every major clade of the phylogram (Fig. 2); e.g., C. glabrata is in clade 3. C. viswanathii, a rare opportunistic pathogen, is closely related to C. tropicalis, as noted by Barns et al. (2). With the exception of C. glabrata (Torulopsis glabrata), the most prominent clinical species of Candida are clustered in clade 1. However, clade 1 also includes at least two nonpathogenic species (C. maltosa and L. elongisporus). This result, as well as the placement of other opportunistic pathogens in different clades (e.g., C. glabrata, C. lusitaniae, C. guilliermondii, I. orientalis [C. kruesei], and S. cerevisiae), suggests that pathogenicity evolved independently on multiple occasions. Indeed, the base of the phylogram includes two extremely rare pathogens, Y. lipolytica (anamorph, Candida lipolytica) and Stephanoascus ciferrii (anamorph, Candida ciferrii) (11).
Combining sequence data for six genes in a phylogenetic analysis enabled us to clarify several other relationships among the species and families of medical yeasts. (i) Previous studies analyzed single genes and failed to define the relationships among C. albicans, C. viswanathii, C. tropicalis, and C. parapsilosis with significant statistical support (7, 14, 21, 50). This investigation resolved the phylogeny of these species. (ii) S. kluyveri was thought to be most closely related to S. cerevisiae, despite low statistical support (7). However, our analysis determined that S. kluyveri belongs in the Saccharomycetales but is more closely related to the genus Kluyveromyces than Saccharomyces. (iii) The results also support transferring Eremothecium gossypii from the family Ermotheciaceae to the Saccharomycetaceae, where it is closely related to S. kluyveri and Kluyveromyces species (25). This finding warrants further analysis of the Ermotheciaceae family, which contains four other species of Eremothecium. (iv) We included strains of the three recognized genotypes of C. parapsilosis (28) and confirmed that genotype I (ATCC 96138) most resembles the type strain (CBS 604). This analysis provides statistical support for a sibling relationship between genotypes II (ATCC 96140) and III (ATCC 96144) within a clade that includes genotype I and the type strain. (v) Our data confirm the polyphyletic composition of both Pichia and the anamorphic genus, Candida (7, 21). As neither genus is monophyletic, we recommend a reexamination and revision of these multifarious genera.
A discrepancy of minor importance involves the placement of P. norvegensis, a member of the polyphyletic genus Pichia. Kurtzman and Robnett's comparison of 26S rDNA sequences of 500 species of ascomycetous yeasts put C. zeylanoides in the Debaryomyces clade, and P. norvegensis was grouped with Pichia and Issatchenkia (24). Similarly, an analysis of the actin gene phylogeny placed P. norvegensis with the other Pichia species, some distance from C. zeylanoides (7). In agreement with these studies, our actin gene tree also grouped P. norvegensis with other Pichia species. However, analyses of the RPB1, RPB2, and 26S rDNA genes (Table 4, node 21) provided excellent support for the arrangement in our combined tree (Fig. 2). The previous studies used the type strain of P. norvegensis (CBS 6564), whereas we used a different strain, P. norvegensis var. zeylanoides (CBS 1922).
We also investigated the evolutionary associations of two traits common among the Saccharomycetales: codon usage and the Co-Q9 system. Although an examination of these character states among the terminal taxa suggested a correlation between codon recapture and Co-Q9 (44), our comparative analyses did not find significant support for the coevolution of these characters. Of course, this analysis may have been affected by the taxa we sampled, and the inclusion of more taxa might lead to a different conclusion. However, there was significant support for the monophyletic origin of CUG usage.
Most human pathogenic species of Saccharomycetales are equipped with alternative CUG usage, Co-Q9, or both. These attributes may be advantageous for pathogenicity of these organisms in mammals, which is a hypothesis that remains to be tested. Recent reviews of the CUG reassignment from leucine to serine indicate that the requisite serine-tRNACAG evolved in an ancestor to both Saccharomyces and Candida, and this gene was subsequently lost from Saccharomyces (34, 43). There is some experimental evidence that the redefinition of the CUG codon destabilized the proteome, leading to the overexpression of stress proteins that may have imparted an evolutionary advantage to pathogenic yeasts (43).
Regarding the type of Co-Q, our data significantly support a phylogeny with Co-Q9 as the ancestral character state for the entire Saccharomycetales, with a loss in the Saccharomycetaceae lineage. Co-Q9 is present in the two most distal taxa, Y. lipolytica and Stephanoascus ciferrii, as well as in the Archiascomycetes, Euascomycetes, and many basidiomycetes. Additional taxa will need to be analyzed to determine more precisely where, and how often, species within the Saccharomycetaceae replaced Co-Q9 (with Co-Q6 or Co-Q7 among the taxa in clade 3). Obviously, neither the redefinition of CUG or the presence of Co-Q9 is essential for pathogenicity, as many pathogenic yeasts and molds translate CUG as leucine and use other forms of Co-Q.
For a long time, rDNA genes were the only accessible source of data to investigate phylogenetic relationships among fungi. That situation has changed over the last several years with the increasing accumulation of protein-coding DNA sequences in databases and the completion of genome sequencing projects. This investigation demonstrates the value of including protein-coding sequences, which contributed significantly to the statistical support and resolution of the phylogeny. Protein-coding DNA sequences are easier to align than data derived from rDNA genes, especially when the investigated taxa span several families, as presented here. Another limitation of rDNA sequence data is that these genes are highly conserved, variation is limited, and multiple substitutions may occur. This study also illustrates the importance of analyzing relevant genes to elucidate specific relationships; for example, clade 1 was defined by the rDNA genes, but relationships within the clade were resolved by analyzing the RPB1 and RPB2 genes. As illustrated in Table 4, reliance on a single gene or two may yield inaccurate results. The availability of a well-supported multigene phylogeny also provides a valuable framework for assessing the results of ongoing genome projects and comparative genomics.
This multilocus phylogeny has clarified the evolutionary relationships among the medically important and related species of Saccharomycetales, defined taxa that require further investigation, and analyzed the origins of pathogen-related characters.
Acknowledgments
For insightful comments on the manuscript and discussion we thank H. E. O'Brien, T. Y. James, M. L. Berbee, and F. S. Dietrich, who also generously provided DNA sequence data for E. gossypii ATCC 10895. We thank W. Schell, W. Meyer, and G. Bulmer for providing cultures. We are grateful to F. M. Lutzoni for assistance with the rDNA alignment. The EF2 and ACT1 primers were designed by G. Luo. We appreciate the expert technical assistance of L. Buckovnik and J. A. Jackson. The informative comments of two anonymous reviewers were most helpful.
This study was funded by Public Health Service grant AI 28836 from the National Institutes of Health and the German Academic Exchange Service in the form of a diploma stipend for S.D.
REFERENCES
- 1.Alexopoulos, C. J., C. W. Mims, and M. Blackwell. 1996. Introductory mycology, 4th ed. John Wiley & Sons, Inc., New York, N.Y.
- 2.Barns, S. M., D. J. Lane, M. L. Sogin, C. Bibeau, and W. G. Weisburg. 1991. Evolutionary relationships among pathogenic Candida species and relatives. J. Bacteriol. 173:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berbee, M. L., and J. W. Taylor. 1992. Detecting morphological convergence in true fungi, using 18S rRNA gene sequence data. Biosystems 28:117-125. [DOI] [PubMed] [Google Scholar]
- 4.Calderone, R. A. (ed.). 2001. Candida and candidiasis. ASM Press, Washington, D.C.
- 5.Cannone, J., S. Subramanian, M. Schnare, J. Collett, L. D'Souza, Y. Du, B. Feng, N. Lin, L. Madabusi, K. Muller, N. Pande, Z. Shang, N. Yu, and R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel, H. M., T. C. Sorrell, and W. Meyer. 2001. Partial sequence analysis of the actin gene and its potential for studying the phylogeny of Candida species and their teleomorphs. Int. J. Syst. Evol. Microbiol. 51:1593-1606. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1981. Evolutionary trees from DNA-sequences—a maximum-likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 9.Foster, P. 11. June 2004, accession date. p4—software for heterogeneous models of molecular sequences. [Online.] www.nhm.ac.uk/zoology/home/foster.htm.
- 10.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes—applications to the identification of mycorrhizae and rusts. Mol. Evol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 11.Hazen, K. 1995. New and emerging yeast pathogens. Clin. Microbiol. Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 13.Jukes, T. H., and C. Cantor. 1969. Evolution of protein molecules. Academic Press, New York, N.Y.
- 14.Kato, M., M. Ozeki, A. Kikuchi, and T. Kanbe. 2001. Phylogenetic relationship and mode of evolution of yeast DNA topoisomerase II gene in the pathogenic Candida species. Gene 272:275-281. [DOI] [PubMed] [Google Scholar]
- 15.Kauff, F., and F. Lutzoni. 2002. Phylogeny of the Gyalectales and Ostropales (Ascomycota, Fungi): among and within order relationships based on nuclear ribosomal RNA small and large subunits. Mol. Phylogenet. Evol. 25:138-156. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi, Y. H., J. Honda, and S. Taniguchi-Morimura. 1989. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature 341:164-166. [DOI] [PubMed] [Google Scholar]
- 17.Kirk, P. M., P. F. Cannon, J. C. David, and J. A. Staplers (ed.). 2001. Ainsworth & Bisby's dictionary of the fungi, 9th ed. CABI Publishing.
- 18.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum-likelihood estimate of the evolutionary tree topologies from DNA-sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 19.Kjer, K. M. 1995. Use of rRNA secondary structure in phylogenetic studies to identify homologous positions: an example of alignment and data presentation from the frogs. Mol. Phylogenet. Evol. 4:314-330. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 21.Kurtzman, C., and C. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtzman, C. P. 1994. Molecular taxonomy of the yeasts. Yeast 10:1727-1740. [DOI] [PubMed] [Google Scholar]
- 23.Kurtzman, C. P., and J. W. Fell (ed.). 1998. The yeasts: a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 24.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 25.Kurtzman, C. P., and C. J. Robnett. 2003. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 3:417-432. [DOI] [PubMed] [Google Scholar]
- 26.Lanave, C., G. Preparata, C. Saccone, and G. Serio. 1984. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20:86-93. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, P. O. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50:913-925. [DOI] [PubMed] [Google Scholar]
- 28.Lin, D., L. C. Wu, M. G. Rinaldi, and P. F. Lehmann. 1995. Three distinct genotypes within Candida parapsilosis from clinical sources. J. Clin. Microbiol. 33:1815-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, Y. J., S. Whelen, and B. D. Hall. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16:1799-1808. [DOI] [PubMed] [Google Scholar]
- 30.Lott, T. J., B. M. Burns, R. Zancope-Oliveira, C. M. Elie, and E. Reis. 1998. Sequence analysis of the internal transcribed spacer 2 (ITS2) from yeast species within the genus Candida. Curr. Microbiol. 36:63-69. [DOI] [PubMed] [Google Scholar]
- 31.Lott, T. J., R. J. Kuykendall, and E. Reiss. 1993. Nucleotide sequence analysis of the 5.8S rDNA and adjacent ITS2 region of Candida albicans and related species. Yeast 9:1199-1206. [DOI] [PubMed] [Google Scholar]
- 32.Maddison, D. W., and W. P. Maddison. 11. June 2004, accession date. Mesquite—a modular system for evolutionary analysis (version 1.02). [Online.] http://www.mesquiteproject.org.
- 33.Maddison, W. P., and D. R. Maddison. 1997. MacClade. Analysis of phylogeny and character evolution. Sinauer Associates, Inc., Sunderland, Mass.
- 34.Massey, S. E., G. Moura, P. Beltrao, R. Almeida, J. R. Garey, M. F. Tuite, and M. A. S. Santos. 2003. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 13:544-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishida, H., and J. Sugiyama. 1994. Archiascomycetes: detection of a major new lineage within the ascomycota. Mycoscience 35:361-366. [Google Scholar]
- 36.Pagel, M. 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. Biol. Sci. 255:37-45. [Google Scholar]
- 37.Pagel, M. 12. June 2004, accession date. Discrete: a method for the analysis of discrete or categorical traits on phylogenies. Ancestral states, correlated evolution. [Online.] http://sapc34.rdg.ac.uk/meade/Mark/.
- 38.Pagel, M. 1999a. Inferring the historical patterns of biological evolution. Nature 401:877-884. [DOI] [PubMed] [Google Scholar]
- 39.Pagel, M. 1999b. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 48:612-622. [Google Scholar]
- 40.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez, F., J. L. Oliver, A. Marin, and J. R. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485-501. [DOI] [PubMed] [Google Scholar]
- 42.Schluter, D., T. Price, A. O. Mooers, and D. Ludwig. 1997. Likelihood of ancestor states in adaptive radiation. Evolution 51:1699-1711. [DOI] [PubMed] [Google Scholar]
- 43.Silva, R. M., I. Miranda, G. Moura, and M. A. S. Santos. 2004. Yeast as a model organism for studying the evolution of non-standard genetic codes. Briefings Funct. Genomics Proteomics 3:35-46. [DOI] [PubMed] [Google Scholar]
- 44.Sugita, T., and T. Nakase. 1999. Non-universal usage of the leucine CUG codon and the molecular phylogeny of the genus Candida. Syst. Appl. Microbiol. 22:79-86. [DOI] [PubMed] [Google Scholar]
- 45.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Inc., Sunderland, Mass.
- 46.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial-DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 47.Tavare, S. 1986. Some probalistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 17:57-86. [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.White, T. J., T. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies, p. 315-322. In M. A. Innis, D. H. Gelford, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 49.Yamada, Y., M. Nojiri, M. Matsuyama, and K. Kondo. 1976. Coenzyme Q system in the classification of the ascosporogenous yeast genera Debaryomyces, Saccharomyces, Kluyveromyces, and Endomycopsis. J. Gen. Appl. Microbiol. 22:325-337. [Google Scholar]
- 50.Yokoyama, K., S. K. Biswas, M. Miyaji, and K. Nishimura. 2000. Identification and phylogenetic relationship of the most common pathogenic Candida species inferred from mitochondrial cytochrome b gene sequences. J. Clin. Microbiol. 38:4503-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]