Abstract
Organisms producing CTX-M-β-lactamases are emerging around the world as a source of resistance to oxyiminocephalosporins such as cefotaxime (CTX). However, the laboratory detection of these strains is not well defined. In this study, a molecular detection assay for the identification of CTX-M-β-lactamase genes was developed and used to investigate the prevalence of these enzymes among clinical isolates of Escherichia coli and Klebsiella species in the Calgary Health Region during 2000 to 2002. In addition, National Committee for Clinical Laboratory Standards (NCCLS) recommendations were evaluated for the ability to detect isolates with CTX-M extended-spectrum β-lactamases (ESBLs). The PCR assay consisted of four primer sets and demonstrated 100% specificity and sensitivity for detecting different groups of CTX-M-β-lactamases in control strains producing well-characterized ESBLs. Using these primer sets, 175 clinical strains producing ESBLs were examined for the presence of CTX-M enzymes; 24 (14%) were positive for blaCTX-M-1-like genes, 95 (54%) were positive for blaCTX-M-14-like genes, and the remaining 56 (32%) were negative for blaCTX-M genes. Following the NCCLS recommendations for ESBL testing, all of the control and clinical strains were detected when screened with cefpodoxime and when both cefotaxime and ceftazidime with clavulanate were used as confirmation tests.
Resistance to the expanded-spectrum cephalosporins can occur in Escherichia coli and Klebsiella species via the production of extended-spectrum β-lactamases (ESBLs) that are capable of hydrolyzing the oxyiminocephalosporins and monobactams (11). Recently, a family of ESBLs which preferentially hydrolyze cefotaxime (CTX), the CTX-M-β-lactamases, have been recognized and reported in the literature with increasing frequency (3). This resistance mechanism is widespread throughout the world, with reports of clinical isolates producing these β-lactamases from Europe, Africa, Asia, South America, and most recently North America (3, 27).
CTX-M-β-lactamases are not closely related to TEM or SHV ESBLs (7) but share high amino acid identity with chromosomal β-lactamases from Kluyvera georgiana (34), Kluyvera cryocrescens (17) and Kluyvera ascorbata (24). In fact, the CTX-M-5 enzyme is identical to the chromosomal gene of K. ascorbata (3). According to a recent review and new data within GenBank, CTX-M-β-lactamases can be divided into five groups based on their amino acid sequence identities (3). Group I includes CTX-M-1, -3, -10 to -12, -15 (UOE-1), -22, -23, -28, -29, and -30. Group II includes CTX-M-2, -4 to -7, and -20 and Toho-1. Group III includes CTX-M-8. Group IV includes CTX-M-9, -13, -14, -16 to -19, -21, and -27 and Toho-2. Finally group V includes CTX-M-25 and -26. The members of these groups exhibit >94% amino acid identity within the group and ≤90% amino acid identity between groups (3).
The laboratory detection of organisms producing CTX-M-β-lactamases is not well defined. The guidelines published by the National Committee for Clinical Laboratory Standards (NCCLS) for the detection of ESBL-producing E. coli and Klebsiella spp. were first published in 1999 before organisms producing CTX-M-β-lactamases became recognized as an important cause of resistance to the newer cephalosporins (3, 29). It is unclear if these guidelines are capable of detecting organisms producing CTX-M-β-lactamases.
A study was designed to develop and evaluate a molecular detection assay for the identification of strains producing known CTX-M-β-lactamases and to investigate if genes encoding these enzymes were present among ESBL-producing strains of E. coli and Klebsiella species isolated from clinical specimens. Only E. coli and K. pneumoniae were examined because NCCLS guidelines for screening and confirmation of ESBL-production are established for these organisms. The ability of these guidelines to detect strains producing CTX-M-β-lactamases was also evaluated.
MATERIALS AND METHODS
Bacterial strains.
Strains with well-described β-lactamases were used as positive and negative controls (Table 1). Consecutive nonduplicate isolates of E. coli and Klebsiella spp. collected at Calgary Laboratory Services during January 2000 to December 2002 were included in this study. These organisms were screened for the presence of ESBLs and then investigated for the presence of CTX-M-β-lactamases. Strains were identified to the species level with Vitek (Vitek AMS; bioMérieux Vitek Systems Inc., Hazelwood, Mo.).
TABLE 1.
Control strains producing well-characterized β-lactamases
| Strain | Organism | β-Lactamase | Groupa | Source or reference |
|---|---|---|---|---|
| CF2 | Enterobacter cloacae | CTX-M-1 | I | 18 |
| Rio-4 | Proteus mirabilis | CTX-M-2 | II | 5 |
| VER-1 | E. cloacae | CTX-M-3 | I | 18 |
| Cfr2525/96 | Citrobacter freundii | CTX-M-3 | I | 21 |
| Eco3553/98 | E. coli | CTX-M-15 | I | 1 |
| 34 | Salmonella enterica serovar Typhimurium | CTX-M-5 | II | 9 |
| Rio-3 | Enterobacter aerogenes | CTX-M-8 | III | 5 |
| 785D | E. coli | CTX-M-9 | IV | 36 |
| EC97/38582 | E. coli | CTX-M-10 | I | 31 |
| EC984167 | E. coli | CTX-M-14 | IV | 26 |
| CF1 | E. coli | CTX-M-14 | IV | 18 |
| Rio-6 | E. coli | CTX-M-16 | IV | 4 |
| BM4493 | Klebsiella pneumoniae | CTX-M-17 | IV | 13 |
| ILT-2 | K. pneumoniae | CTX-M-18 | IV | 35 |
| ILT-3 | K. pneumoniae | CTX-M-19 | IV | 35 |
| CT1b | E. coli | Toho-1 | II | 25 |
| S1b | E. coli | SHV-2 | 20 | |
| S2b | E. coli | SHV-7 | 8 | |
| T1b | E. coli | TEM-3 | 20 | |
| T4b | E. coli | TEM-10 | 20 | |
| T5b | E. coli | TEM-50 | 37 |
Group I includes CTX-M-1, -3, -10 to -12, -15 (UOE-1), -22, -23, -28, -29, and -30. Group II includes CTX-M-2, -4 to -7, and -20 and Toho-1. Group III includes CTX-M-8. Group IV includes CTX-M-9, -13, -14, -16, to -19, -21, and -27 and Toho-2. Finally, group V includes CTX-M-25 and -26. We were unable to obtain a strain producing any group V enzymes.
These strains are part of an unpublished isogenic panel. The original strains producing these β-lactamases were provided to the Center for Research in Anti-Infectives and Biotechnology by the referenced authors listed in the table.
Antimicrobial susceptibility testing.
MICs of the following drugs were determined by Vitek (Vitek AMS): piperacillin (PIP), piperacillin-tazobactam (TZP), cefpodoxime (CPD), cefotaxime (CTX), and ceftazidime (CAZ). The quality control strains used for this study were E. coli ATCC 25922, E. coli ATCC 35218, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, and K. pneumoniae ATCC 700603. Throughout this study, results were interpreted using NCCLS criteria for broth dilution (30).
Screening for and confirmation of ESBLs.
The presence of ESBLs was evaluated in both the control strains and the recent clinical isolates. Screening was performed with Vitek (Vitek AMS) using 1-μg/ml CPD, CAZ, and CTX. Screening and disk confirmation tests using CTX (30 μg) and CAZ (30 μg) disks in combination with 10 μg of clavulanate (CLA) were performed and interpreted by NCCLS criteria for ESBL screening and disk confirmation tests (30). Disks for ESBL confirmation tests were obtained from Oxoid, Inc. (Nepean, Ontario, Canada). K. pneumoniae ATCC 700603 and E. coli ATCC 25922 were used as positive and negative controls, respectively.
β-Lactamase gene identification.
DNA template preparation and PCR amplification for CTX-M-β-lactamase genes were carried out on a Thermal Cycler 9600 instrument (Applied Bio-systems, Norwalk, Conn.) as previously described (23). The primers, sizes of the expected amplification product, and annealing temperatures used for PCR amplification are listed in Table 2. Magnesium chloride concentrations were 1.5 mM for all PCRs.
TABLE 2.
Primers used for amplification
| Target(s)a | Primer | Sequenceb | Product size (bp) | Annealing temp (°C) | Nucleotide positions (bp)c | GenBank accession no.d |
|---|---|---|---|---|---|---|
| CTX-M group I | CTXM1-F3 | GAC GAT GTC ACT GGC TGA GC | 499 | 55 | 416-435 | X92506 |
| CTXM1-R2 | AGC CG C CGA CGC TAA TAC A | 914-896 | ||||
| CTX-M group II | TOHO1-2F | GCG ACC TGG TTA ACT ACA ATC C | 351 | 55 | 313-334 | X92507 |
| TOHO1-1R | CGG TAG TAT TGC CCT TAA GCC | 663-643 | ||||
| CTX-M group III | CTXM825F | CGC TTT GCC ATG TGC AGC ACC | 307 | 55 | 475-495 | AF189721 |
| CTXM825R | GCT CAG TAC GAT CGA GCC | 781-764 | ||||
| CTX-M group IV | CTXM914F | GCT GGA GAA AAG CAG CGG AG | 474 | 62 | 1857-1876 | AF252622 |
| CTXM914R | GTA AGC TGA CGC AAC GTC TG | 2330-2311 |
Group I includes CTX-M-1, -3, -10 to -12, -15 (UOE-1), -22, -23, -28, -29, and -30. Group II includes CTX-M-2, -4 to -7, and -20 and Toho-1. Group III includes CTX-M-8. Group IV includes CTX-M-9, -13, -14, -16 to -19 and -21, and -27 and Toho-2. Finally, group V includes CTX-M-25 and -26.
Sequence of primer as synthesized 5′ to 3′.
Nucleotide position in base pairs for the GenBank accession number sequence.
Accession number of the sequence used for primer design.
Nucleotide sequence accession number.
The GenBank nucleotide sequence accession numbers for the sequences used in this study were as follows: CTX-M-1, X92506; CTX-M-2, X92507; CTX-M-3, AF550415; CTX-M-15, AY044436; CTX-M-4, Y14156; CTX-M-5, AF286192; CTX-M-6, AJ005044; CTX-M-7, AJ005045; CTX-M-8, AF189721; CTX-M-9, AJ416345; CTX-M-10, AF255298; CTX-M-11, AJ310929; CTX-M-12, AF305837; CTX-M-13, AF252623; CTX-M-14, AF252622; CTX-M-16, AY029068; CTX-M-17, AF454633; CTX-M-18, AF325133; CTX-M-19, AF325134; CTX-M-20, AJ416344; CTX-M-21, AJ416346; CTX-M-22, AY080894; CTX-M-23, AF488377, CTX-M-24, AY143430; CTX-M-25, AF518567; CTX-M-26, AY455830; CTX-M-27, AY156923; CTX-M-28, AJ549244; CTX-M-29, AY267213; CTX-M-30, AY292654; CTX-M-31, AJ567482; CTX-M-32, AJ557142; CTX-M-33, AY238472; Toho-1, D37830; Toho-2, AF311345; Toho-3, AB038771; and FEC-1, AB098539.
RESULTS
Molecular detection of CTX-M genes.
Currently, there are 40 gene sequences designated as blaCTX-M in the National Center for Biotechnology Information GenBank database (3). Based on percent sequence similarities, blaCTX-M genes can be clustered into five different groups (3). Nucleotide similarities between these genes were analyzed by using DNAsis for Windows 2.6 (Hitachi Software), and four primer sets were designed which would amplify family- or group-specific CTX-M genes. These groups were designated CTX-M groups I, II, III, and IV. These groups and the specific family members within the group are listed in Tables 1 and 2. The group III primer set was designed to amplify both blaCTX-M-8 and blaCTX-M-25. This was done to simplify the number of primer pairs required to identify blaCTX gene families. The forward primer of primer pair III (blaCTX-M-8) has 100% identity to blaCTX-M-8 but has three mismatches at the 5′ end for blaCTX-M-25; the reverse primer is 100% identical to blaCTX-M-8 and has a 1-base mismatch at position 5 of the 18-base primer for blaCTX-M-25. The accession numbers used for sequence alignments in this study are listed in Materials and Methods.
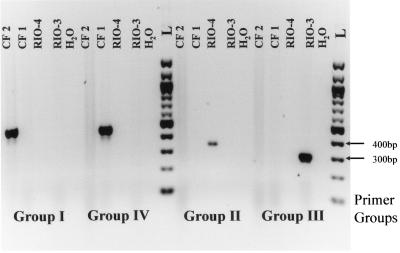
The designed primers were tested for specificity in separate PCRs using a DNA template prepared from control strains known to produce specific CTX-M β-lactamases or strains producing ESBLs other than CTX-M β-lactamases (Tables 1 and 3 and Fig. 1). PCR amplification of DNA template prepared from strains CF2, VER-1, Cfr2525/96, Eco3553/98, and EC97/38582 resulted in a single amplified product of 499 bp when CTX-M group I primers were used. No amplified product was identified with this primer set when DNA template from the rest of the control strains in Table 1 was used during PCR amplification. Group II primers amplified a single 351-bp fragment when DNA template was prepared from control strains Rio-4, 34, and CT1. All other control strain DNA templates resulted in no amplification product for this primer set. Group IV primers amplified a 474-bp product from DNA prepared from strains 785D, EC984167, CF1, Rio-6, BM4493, ILT-2, and ILT-3. This primer set was also very specific, resulting in no amplification of DNA when template was prepared from the other control strains. Use of the group III primer set resulted in amplification of DNA prepared from only one strain, Rio-3, which produced CTX-M-8. An isolate producing CTX-M-25 was requested but not obtained. Therefore, we were unable to examine the ability of the group III primer set to amplify blaCTX-M-25. A representative gel indicating the specificity of the primer pairs is shown in the Fig. 1. These data indicate a high level of specificity for these group-specific primer pairs.
TABLE 3.
Laboratory diagnosis of control strains producing known ESBLs
| Strain | Enzyme | Result by:a:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Screeningb
|
Disk confirmationc
|
PCR primer pairsd
|
||||||||
| CPD | CAZ | CTX | CAZ/CLA | CTX/CLA | CTX-M-I group | CTX-M-II group | CTX-M-III group | CTX-M-IV group | ||
| CF2 | CTX-M-1 | + | + | + | − | + | + | − | − | − |
| Rio-4 | CTX-M-2 | + | − | + | − | + | − | + | − | − |
| VER-1 | CTX-M-3 | + | + | + | − | + | + | − | − | − |
| Cfr2525/96 | CTX-M-3 | + | + | + | − | + | + | − | − | − |
| Eco3553/98 | CTX-M-15 | + | + | + | + | + | + | − | − | − |
| 34 | CTX-M-5 | + | + | + | − | + | − | + | − | − |
| Rio-3 | CTX-M-8 | + | − | + | − | + | − | − | + | − |
| 785D | CTX-M-9 | + | − | + | − | + | − | − | − | + |
| EC97/38582 | CTX-M-10 | + | − | + | − | + | + | − | − | − |
| EC984167 | CTX-M-14 | + | − | + | − | + | − | − | − | + |
| CF1 | CTX-M-14 | + | − | + | − | + | − | − | − | + |
| Rio-6 | CTX-M-16 | + | + | + | + | + | − | − | − | + |
| BM4493 | CTX-M-17 | + | + | + | + | + | − | − | − | + |
| ILT-2 | CTX-M-18 | + | + | + | − | + | − | − | − | + |
| ILT-3 | CTX-M-19 | + | + | + | + | + | − | − | − | + |
| CT1 | Toho-1 | + | − | + | − | + | − | + | − | − |
| S1 | SHV-2 | + | + | + | + | + | − | − | − | − |
| S2 | SHV-7 | + | + | + | + | + | − | − | − | − |
| T1 | TEM-3 | + | + | + | + | + | − | − | − | − |
| T4 | TEM-10 | + | + | − | + | + | − | − | − | − |
| T5 | TEM-50 | + | + | + | + | + | − | − | − | − |
−, negative; +, positive.
NCCLS guidelines for screening for ESBL-producing bacteria. CPD, CAZ, and CTX were each used at 1 μg/ml.
NCCLS guidelines for ESBL disk confirmation tests.
PCR primers for CTX-M groups (refer to Materials and Methods for details). Group I includes CTX-M-1, -3, -10 to -12, -15 (UOE-1), -22, -23, -28, -29, and -30. Group II includes CTX-M-2, -4 to -7, and -20 and Toho-1. Group III includes CTX-M-8. Group IV includes CTX-M-9, -13, -14, -16 to -19, -21, and -27 and Toho-2. Finally, group V includes CTX-M-25 and -26.
FIG. 1.
CTX-M PCR. Four primer sets representing specific groups of blaCTXM genes were evaluated for specificity with DNA prepared from strains known to produce specific CTX-M β-lacatmases. The DNA templates were prepared from CF2 producing CTX-M-1, CF1 producing CTX-M-14, Rio-4 producing CTX-M-2, and Rio-3 producing CTX-M-8. H2O represents the negative control, and L represents a 100-bp ladder (Invitrogen). Specific marker sizes in base pairs are shown on the right. Each primer pair was used in combination with both positive and negative templates. The primer groups used for each set of templates are listed below the figure and are defined in Table 2.
Clinical bacterial strains.
During the study period, 232 E. coli strains producing ESBLs were isolated from 168 patients and 11 K. pneumoniae strains were isolated from 7 patients. The overall frequencies of ESBL producers observed in this study were 1.3% for E. coli and 0.5% for K. pneumoniae. Of the 175 (168 plus 7) strains included in this study, 154 (88%) were isolated from urine cultures, 7 (4%) were isolated from blood, 5 (3%) were isolated from both the blood and urine of a single patient, 3 (2%) were isolated from wounds (purulent), 2 (1%) were isolated from sputum, 2 (1%) were isolated from both sputum and urine of a single patient, and 1 (1%) each was isolated from bronchoalveolar lavage and stool (associated with symptomatic diarrhea), respectively.
Antimicrobial susceptibility of clinical strains.
All the ESBL-producing E. coli strains were resistant to CPD and PIP, 119 (71%) were resistant to CTX, 29 (17%) were resistant to CAZ, and 7 (4%) were resistant to TZP. Among the ESBL-producing K. pneumoniae strains, all were resistant to CPD and PIP, 3 (43%) were resistant to CTX, and 1 (14%) was resistant to CAZ. All of the ESBL-producing K. pneumoniae strains were susceptible to TZP.
Identification of blaCTX-M genes in the clinical strains.
All of the ESBL-producing E. coli and Klebsiella spp. were examined by PCR for the presence of blaCTX-M genes. Of the 168 E. coli strains isolated during the study period, 24 (14%) were positive for blaCTX-M genes from the CTX-M-I group, indicating CTX-M-1-like β-lactamases. Ninety-three (55%) were positive for blaCTX-M genes from the CTX-M-IV group, indicating CTX-M-14-like β-lactamases, and the remainder (51 [31%]) were negative for blaCTX-M genes (Table 4). Of the seven K. pneumoniae strains isolated during the study period, two (29%) were positive for blaCTX-M genes from the CTX-M-IV group and the remainder (five [71%]) were negative for blaCTX-M genes (Table 4).
TABLE 4.
Laboratory diagnosis of clinical strains producing ESBLs
| Strains (n) | PCR for blaCTX-M genesa | No. (%) of strains
|
|||||
|---|---|---|---|---|---|---|---|
| Screeningb
|
Disk confirmationc
|
||||||
| CPD | CAZ | CTX | CAZ/CLA | CTX/CLA | CTX/CLA and CAZ/CLA | ||
| E. coli (51) | Negative | 51 (100) | 51 (100) | 48 (94) | 34 (67) | 49 (96) | 51 (100) |
| K. pneumoniae (5) | Negative | 5 (100) | 5 (100) | 4 (80) | 3 (60) | 5 (100) | 5 (100) |
| E. coli (24) | Positive for CTX-M-I group | 24 (100) | 24 (100) | 24 (100) | 21 (88) | 24 (100) | 24 (100) |
| E. coli (93) | Positive for CTX-M-IV group | 93 (100) | 71 (76) | 93 (100) | 3 (3) | 93 (100) | 93 (100) |
| K. pneumoniae (2) | Positive for CTX-M-IV group | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 2 (100) |
| Total (175) | 175 (100) | 151 (86) | 171 (98) | 61 (35) | 173 (99) | 175 (100) | |
PCR primers for CTX-M subgroups (see Materials and Methods for details). Group I includes CTX-M-1, -3, -10 to -12, -15 (UOE-1), -22, -23, -28, -29, and -30. Group II includes CTX-M-2, -4 to -7, and -20 and Toho-1. Group III includes CTX-M-8. Group IV includes CTX-M-9, -13, -14, -16 to -19, -21, and -27 and Toho-2. Finally, group V includes CTX-M-25 and -26.
NCCLS guidelines for screening ESBL-producing bacteria. CPD, CAZ, and CTX were each used at 1 μg/ml.
NCCLS guidelines for ESBL disk confirmation tests.
Screening and confirmation for ESBLs by NCCLS criteria.
Using the 1999 NCCLS criteria for detection of ESBLs in E. coli and Klebsiella spp., all of the control and clinical strains from the current study producing ESBLs were positive when screened with CPD (1 μg/ml) (Tables 3 and 4) (29). Because the CPD MICs for all organisms examined were greater than 4 μg/ml, the new guidelines set forth in 2003 by the NCCLS changing the CPD concentration to 4 μg/ml would not have affected the results described in this study. It is important to note that according to the references listed in Table 1, none of the control strains tested produced an ESBL other than the CTX-M β-lactamase identified. Almost every reference indicated the presence of TEM-1 in the control strains, and in the case of K. pneumoniae strains, a band with a pI of 7.6 was reported as SHV-1. The production of an AmpC β-lactamase was not indicated in any of the references describing these strains (see references in Table 1). All of the control strains expressing various ESBLs were detected with 1-μg/ml CTX except for TEM-10. In addition, 98% of the clinical strains from the current study were positive with 1-μg/ml CTX; the four exceptions were blaCTX-M-negative strains. Using CAZ at 1 μg/ml did not detect control strains producing CTX-M-2, -8, -9, -10, and -14 and Toho-1 or 14% of the clinical stains from the current study, which were all positive for blaCTX-M-14-like (Tables 3 and 4). The NCCLS disk confirmation test using CTX with and without CLA was positive for all of the control strains and 99% of the clinical strains from the present study (Table 3 and 4). The disk confirmation tests using CAZ with and without CLA were positive for the control strains producing CTX-M-15 (UOE-1), -16, -17, and -19. These β-lactamases are part of groups I and IV (Table 1 and 3). Sixty-five percent of the current clinical strains tested negative (35% positive) with CAZ with and without CLA (Table 4). These data reflect the molecular data, which indicated that only 3% of the current clinical strains with a blaCTX-M-14-like gene tested positive by the CAZ and CLA disk confirmation test (Table 4). All of the strains (control and clinical) were positive for at least one of the confirmation tests, but by using both CTX/CLA and CAZ/CLA disk confirmation tests, all strains producing an ESBL were detected (Table 4).
DISCUSSION
There has been a dramatic increase in the number of organisms reported in the literature that produce CTX-M-β-lactamases (3). This class of β-lactamases has been recognized worldwide as an important mechanism of resistance to oxyiminocephalosporins used by gram-negative pathogens (3). In most cases, organisms producing these enzymes display higher levels of resistance to CTX and ceftriaxone than CAZ (3). However, organisms producing some CTX-M variants, including CTX-M-16 and -19 and UOE-1, are resistant to CAZ (4, 33, 35).
Phenotypic differentiation of organisms producing CTX-M-β-lactamases from organisms producing other types of ESBLs can be difficult. The difficulty is due to overlapping phenotypes resulting in interference from other β-lactamases produced by the organism capable of hydrolyzing CAZ (2, 4, 35). Therefore, susceptibility testing which relies on identifying organisms that are resistant to CTX and/or ceftriaxone but susceptible to CAZ is not a reliable approach. In addition, the use of isoelectric focusing to identify β-lactamases is becoming obsolete because many isoelectric points of different β-lactamases overlap, including CTX-M-β-lactamases (3, 7). PCR amplification and sequencing of blaCTX-M genes have been used to characterize organisms producing CTX-M-β-lactamases (6, 12, 14). Recently a molecular approach for screening ESBL-positive organisms for the presence of CTX-M genes was described by Edelstein et al. (19). Consensus primers, which recognize all the known variant genes of blaCTX-Ms to date, were used to generate an amplified product of 544 bp. To identify the specific groups of blaCTX-Ms, restriction fragment length polymorphism (RFLP) analysis was employed. The technique described in this study uses four specific primer sets to detect the various groups of CTX-M-β-lactamase genes, thus negating the need for RFLP analysis. Data generated using several control strains known to produce specific CTX-M-β-lactamases validated the specificity and sensitivity of these primer sets. A control strain producing a CTX-M-25 β-lactamase (belonging to group V) was not available, and although the primers designed to amplify blaCTXM-8 (belonging to group III) contained one mismatch with respect to blaCTX-M-25 in the reverse primer and three mismatches in the 5′ portion of the forward primer, these mismatches should still allow amplification from blaCTX-M-25 given the stringency with which the PCR is modified (unpublished data on mismatched primer pairs). Even though this PCR assay involves the use of four sets of primers, a single DNA fragment is amplified for each CTX-M group. Therefore, interpretation of results is simple and can be adapted in reference laboratories for screening multiple isolates for the presence of group-specific CTX-M-β-lactamase genes (Fig. 1).
The four-primer-pair PCR-based detection system was used to screen 168 ESBL-producing E. coli strains and 7 K. pneumoniae strains for the presence of genes encoding CTX-M-β-lactamases recovered from the Calgary Health Region (CHR) during 2000 to 2002. The CHR is a fully integrated, publicly funded health system that provides health care to the residents of the cities of Calgary and Airdrie and approximately 20 nearby small towns, villages, and hamlets (overall population of 958,610 in 2001). In the CHR, Calgary Laboratory Services receives all clinical specimens submitted for bacteriologic testing, including those from all hospitals, nursing homes, physicians' offices, and community collection sites (15). The majority of ESBL-producing bacteria (68%) isolated in the CHR during 2000 to 2002 carried a CTX-M-β-lactamase gene (Table 4). To our knowledge, this is the first study that identified strains with blaCTX-M genes as the predominant type of ESBL in a well-defined North American region, although a previous study has shown the presence of CTX-Ms in Canada (28).
The limitations of molecular analyses for resistance genes result from the presence of unknown mutations which might occur in the primer target region or the evolution of gene products which have not yet been identified at the genetic level. Therefore, any negative PCR result must be evaluated with this in mind. Thirty-five percent of the 175 ESBL-producing strains in this report were negative by PCR for blaCTX-M. These data could indicate that the ESBL phenotype is due to production of ESBLs other than CTX-Ms. However, the negative PCR results in this report do not negate the possibility that modified blaCTX-Ms were present in these isolates. Due to the increased complexity of β-lactam resistance in gram-negative organisms, the key to effective surveillance is the use of both phenotypic and genotypic analyses in concert.
The detection of organisms producing ESBLs remains a contentious issue. Proficiency testing studies performed by the Centers for Disease Control and Prevention and the College of American Pathologists have raised concerns about the current capacity of many laboratories to detect organisms producing ESBLs (22, 39-41). A study recently published showed that only 8% of microbiology laboratories from rural hospitals in the United States routinely screen for ESBL-producing organisms (38). Since the majority of patients infected with a strain of ESBL-producing E. coli or Klebsiella spp. identified in our study originated from the community (32), it is conceivable to predict that ESBL-producing organisms are present in the community in North America but are not being reported.
The NCCLS guidelines for ESBL detection in E. coli and Klebsiella spp. include an initial screening with either CPD, CTX, CAZ, ceftriaxone, or aztreonam, followed by a confirmation test using both CTX and CAZ in combination with clavulanate (30). A practice exists among some clinical laboratories of using CAZ as the initial screening drug and CAZ with CLA as the confirmation test (10, 16). Data from this study indicate that 14% of ESBL-producing strains will not be detected if CAZ is used as the initial screen. However, in this study, CPD detected all of the ESBL-positive strains (Table 4). The concern raised by this study is that only 35% of ESBL-producing strains (20% of CTX-M-producers) were reported as ESBL positive when CAZ with CLA was the only confirmation test (Table 4). Therefore, a more appropriate approach for initially screening organisms for the presence of ESBLs is to use CPD followed by disk tests using both CTX with CLA and CAZ with CLA to confirm the presence of ESBLs among E. coli and Klebsiella spp.
Organisms with genes encoding CTX-M-14-like β-lactamases were responsible for an outbreak among elderly patients in the Calgary community during 2000 (32). This outbreak was only recognized after the PCR identification of CTX-M-14-like β-lactamase genes. If molecular detection assays, such as the one described in this paper are available at the time of an outbreak and performed by a reference laboratory, early recognition and the possible mechanism(s) by which resistance is spread can be identified in a timely manner. Extra efforts such as molecular procedures to identify resistance mechanisms can promote optimal patient care by early detection of antimicrobial-resistant organisms and implementation of appropriate infection control procedures.
Acknowledgments
This work was supported by a grant from the University of Calgary Dean's Starter grant (no. 75-4777).
We thank Lorraine Campbell, Wanda Wudal, Harjinder Gill, and Brenda Gallant, Calgary Laboratory Services, Calgary, Alberta, Canada, for their technical support of this study and Richard Bonnet for providing the control strains CF1, CF2, VER-1, Rio-3, Rio-4, and Rio-6; Marek Gniadkowski for strains Cfr2525/96 and Eco3553/98; Patricia Bradford for strain 34; Ferran Navaro for strain 785D; Rafael Conton for strain Ec97/38582; L Siu for strain KTC984167; Patrice Courvalin for strain BM4493; and Patrice Nordmann for strains ILT-2 and ILT-3.
REFERENCES
- 1.Baraniak, A., J. Fiett, W. Hryniewicz, P. Nordmann, and M. Gniadkowski. 2002. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum beta-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 50:393-396. [DOI] [PubMed] [Google Scholar]
- 2.Baraniak, A., J. Fiett, A. Sulikowska, W. Hryniewicz, and M. Gniadkowski. 2002. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R., C. Dutour, J. L. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, R., J. L. M. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou, G., M. Cartelle, M. Tomas, D. Canle, F. Molina, R. Moure, J. M. Eiros, and A. Guerrero. 2002. Identification and broad dissemination of the CTX-M-14 β-lactamase in different Escherichia coli strains in the northwest area of Spain. J. Clin. Microbiol. 40:4030-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, P. A., C. Urban, A. Jaiswal, N. Mariano, B. A. Rasmussen, S. J. Projan, J. J. Rahal, and K. Bush. 1995. SHV-7, a novel cefotaxime-hydrolyzing β-lactamase, identified in Escherichia coli isolates from hospitalized nursing home patients. Antimicrob. Agents Chemother. 39:899-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenwald, N. P., G. Jevons, J. M. Andrews, J. H. Xiong, P. M. Hawkey, and R. Wise. 2003. An outbreak of a CTX-M-type beta-lactamase-producing Klebsiella pneumoniae: the importance of using cefpodoxime to detect extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 51:195-196. [DOI] [PubMed] [Google Scholar]
- 11.Bush, K. 2001. New beta-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32:1085-1089. [DOI] [PubMed] [Google Scholar]
- 12.Canton, R., A. Oliver, T. M. Coque, M. del Carmen Varela, J. C. Pérez-Diaz, and F. Baquero. 2002. Epidemiology of extended-spectrum β-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao, V., T. Lambert, and P. Courvalin. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob. Agents Chemother. 46:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chanawong, A., F. H. M'Zali, J. Heritage, J.-H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Church, D. L., C. Don-Joe, and B. Unger. 2000. Effects of restructuring on the performance of microbiology laboratories in Alberta. Arch. Pathol. Lab. Med. 124:357-361. [DOI] [PubMed] [Google Scholar]
- 16.Dandekar, P. K., J. Tetreault, J. P. Quinn, C. H. Nightingale, and D. P. Nicolau. 2004. Prevalence of extended spectrum beta-lactamase producing Escherichia coli and Klebsiella isolates in a large community teaching hospital in Connecticut. Diagn. Microbiol. Infect. Dis. 49:37-39. [DOI] [PubMed] [Google Scholar]
- 17.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutour, C., R. Bonnet, H. Marchandin, M. Boyer, C. Chanal, D. Sirot, and J. Sirot. 2002. CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelstein, M., M. Pimkin, I. Palagin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrhardt, A. F., C. C. Sanders, and E. S. Moland. 1999. Use of an isogenic Escherichia coli panel to design tests for discrimination of β-lactamase functional groups of Enterobacteriaceae. Antimicrob. Agents Chemother. 43:630-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gniadkowski, M., I. Schneider, A. Pałucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hageman, J. C., S. K. Fridkin, J. M. Mohammed, C. D. Steward, R. P. Gaynes, and F. C. Tenover. 2003. Antimicrobial proficiency testing of National Nosocomial Infections Surveillance System hospital laboratories. Infect. Control Hosp. Epidemiol. 24:356-361. [DOI] [PubMed] [Google Scholar]
- 23.Hanson, N. D., K. S. Thomson, E. S. Moland, C. C. Sanders, G. Berthold, and R. G. Penn. 1999. Molecular characterization of a multiply resistant Klebsiella pneumoniae encoding ESBLs and a plasmid-mediated AmpC. J. Antimicrob. Chemother. 44:377-380. [DOI] [PubMed] [Google Scholar]
- 24.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishii, Y., A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa. 1995. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma, L., Y. Ishii, F. Y. Chang, K. Yamaguchi, M. Ho, and L. K. Siu. 2002. CTX-M-14, a plasmid-mediated CTX-M type extended-spectrum β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 46:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moland, E. S., J. A. Black, A. Hossain, N. D. Hanson, K. S. Thomson, and S. Pottumarthy. 2003. Discovery of CTX-M-like extended-spectrum β-lactamases in Escherichia coli isolates from five U.S. states. Antimicrob. Agents Chemother. 47:2382-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulvey, M. R., E. Bryce, D. Boyd, M. Ofner-Agostini, S. Christianson, A. E. Simor, S. Paton, and The Canadian Hospital Epidemiology Committee of the Canadian Nosocomial Infection Surveillance Program, Health Canada. 2004. Ambler class A extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob. Agents Chemother. 48:1204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial and susceptibility testing. Ninth informational supplement. M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 30.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial and susceptibility testing. Twelfth informational supplement. M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 31.Oliver, A., J. C. Pérez-Diaz, T. M. Coque, F. Baquero, and R. Canton. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitout, J. D., N. D. Hanson, D. L. Church, and K. B. Laupland. 2004. Population-based laboratory surveillance for Escherichia coli-producing extended-spectrum beta-lactamases: importance of community isolates with blaCTX-M genes. Clin. Infect. Dis. 38:1736-1741. [DOI] [PubMed] [Google Scholar]
- 33.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 34.Poirel, L., P. Kampfer, and P. Nordmann. 2002. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 46:4038-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirel, L., T. Naas, I. Le Thomas, A. Karim, E. Bingen, and P. Nordmann. 2001. CTX-M-type extended-spectrum β-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob. Agents Chemother. 45:3355-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabaté, M., R. Tarragó, F. Navarro, E. Miró, C. Vergés, J. Barbé, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirot, D., C. Recule, E. B. Chaibi, L. Bret, J. Croize, C. Chanal-Claris, R. Labia, and J. Sirot. 1997. A complex mutant of TEM-1 β-lactamase with mutations encountered in both IRT-4 and extended-spectrum TEM-15, produced by an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 41:1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson, K. B., M. Samore, J. Barbera, J. W. Moore, E. Hannah, P. Houck, F. C. Tenover, and J. L. Gerberding. 2003. Detection of antimicrobial resistance by small rural hospital microbiology laboratories: comparison of survey responses with current NCCLS laboratory standards. Diagn. Microbiol. Infect. Dis. 47:303-311. [DOI] [PubMed] [Google Scholar]
- 39.Steward, C. D., D. Wallace, S. K. Hubert, R. Lawton, S. K. Fridkin, R. P. Gaynes, J. E. McGowan, Jr., and F. C. Tenover. 2000. Ability of laboratories to detect emerging antimicrobial resistance in nosocomial pathogens: a survey of project ICARE laboratories. Diagn. Microbiol. Infect. Dis 38:59-67. [DOI] [PubMed] [Google Scholar]
- 40.Tenover, F. C., M. J. Mohammed, T. S. Gorton, and Z. F. Dembek. 1999. Detection and reporting of organisms producing extended-spectrum β-lactamases: survey of laboratories in Connecticut. J. Clin. Microbiol. 37:4065-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenover, F. C., M. J. Mohammed, J. Stelling, T. O'Brien, and R. Williams. 2001. Ability of laboratories to detect emerging antimicrobial resistance: proficiency testing and quality control results from the World Health Organization's external quality assurance system for antimicrobial susceptibility testing. J. Clin. Microbiol. 39:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]