Abstract
Recent molecular studies have shown Mycobacterium porcinum, recovered from cases of lymphadenitis in swine, to have complete 16S rDNA sequence identity and >70% DNA-DNA homology with human isolates within the M. fortuitum third biovariant complex. We identified 67 clinical and two environmental isolates of the M. fortuitum third biovariant sorbitol-negative group, of which 48 (70%) had the same PCR restriction enzyme analysis (PRA) profile as the hsp65 gene of M. porcinum (ATCC 33776T) and were studied in more detail. Most U.S. patient isolates were from Texas (44%), Florida (19%), or other southern coastal states (15%). Clinical infections included wound infections (62%), central catheter infections and/or bacteremia (16%), and possible pneumonitis (18%). Sequencing of the 16S rRNA gene (1,463 bp) showed 100% identity with M. porcinum ATCC 33776T. Sequencing of 441 bp of the hsp65 gene showed four sequevars that differed by 2 to 3 bp from the porcine strains. Clinical isolates were positive for arylsulfatase activity at 3 days, nitrate, iron uptake, d-mannitol, i-myo-inositol, and catalase at 68°C. They were negative for l-rhamnose and d-glucitol (sorbitol). Clinical isolates were susceptible to ciprofloxacin, sulfamethoxazole, and linezolid and susceptible or intermediate to cefoxitin, clarithromycin, imipenem, and amikacin. M. porcinum ATCC 33776T gave similar results except for being nitrate negative. These studies showed almost complete phenotypic and molecular identity between clinical isolates of the M. fortuitum third biovariant d-sorbitol-negative group and porcine strains of M. porcinum and confirmed that they belong to the same species. Identification of M. porcinum presently requires hsp65 gene PRA or 16S rRNA or hsp65 gene sequencing.
The rapidly growing mycobacterial species Mycobacterium porcinum was described in 1983 by Tsukamura et al. as a causative agent of submandibular lymphadenitis in swine (23). Tsukamura et al. characterized these strains as being similar to M. fortuitum but differing from this species by being d-mannitol and i-myo-inositol positive, nitrate negative, and succinamidase positive and utilizing benzoate as a sole source of carbon in the presence of ammoniacal nitrogen. Until recently, no isolation of this species had been reported since that report.
Bönicke studied isolates previously identified as M. fortuitum and placed them into three subgroups (1). Subgroup A was negative for d-mannitol and i-myo-inositol and now is recognized as M. fortuitum. Subgroup B was positive for d-mannitol but negative for i-myo-inositol and is now recognized as M. peregrinum. Subgroup C was positive for mannitol and inositol, and its taxonomic status has taken much longer to establish and is still evolving. A study by Wallace et al. in 1991 showed this subgroup to consist of two major groups. One group was d-sorbitol positive, had low semiquantitative catalase, and was pipemidic acid resistant, and MICs of cefoxitin (27) and clarithromycin (4) were high for members of the group. β-Lactamase patterns on polyacrylamide gels following isoelectric focusing (IEF) had previously showed these isolates to have primarily a single enzyme pattern (28). These isolates were named the M. fortuitum third biovariant complex d-sorbitol-positive group, and ATCC 49403 was chosen as the reference strain (27).
Subsequent 16S rRNA gene sequencing showed these isolates to have a unique hypervariable A region (11, 19) and an 880-bp sequence of ATCC 49403 to differ by 6 to 15 bp from previously recognized species within the M. fortuitum group (11). To date, three species are recognized within this d-sorbitol-positive group based on 16S rDNA complete sequencing and DNA-DNA homology studies: M. mageritense (7, 26), M. houstonense (of which ATCC 49403 is the proposed type strain), and M. brisbanense (16).
The other major subgroup identified by Wallace et al. (27) within Bonicke's subgroup C (1) was d-sorbitol negative, had a high (height of column of bubbles, >100 mm) semiquantitative catalase, was pipemidic acid susceptible, and was susceptible or intermediate to cefoxitin and clarithromycin (4). This group was named the M. fortuitum third biovariant complex d-sorbitol-negative group (27). Subsequent 16S rRNA gene sequencing showed all studied isolates to have an identical but unique hypervariable A region (19). However, IEF studies of the β-lactamase from this group showed a surprising multitude of enzyme patterns, suggesting that multiple taxa or species might be present (32). The first species within this group to be identified was M. septicum (17). Only one strain of M. septicum has been reported (17), and the majority of isolates of this group were still to be identified to the species level.
This quickly changed when recent molecular studies showed complete sequence identity between the 16S rRNA gene of M. porcinum and some of the unnamed isolates of the M. fortuitum third biovariant d-sorbitol-negative group (16, 25). Schinsky et al. (16) found four human strains of the third biovariant d-sorbitol-negative group to have 100% sequence identity over 1,401 bp with the M. porcinum type strain, ATCC 33776. This latter strain had 91% homology with DNA-DNA comparison to ATCC BAA-328, which is a typical clinical strain of the third biovariant d-sorbitol-negative group.
The original description of M. porcinum in 1983 by Tsukamura et al. (23) appears very similar to that of isolates within Bonicke's subgroup C (1) that presently contain the M. fortuitum third biovariant, but phenotypic and molecular comparison of these two groups was not done when M. porcinum was first described (23). Some phenotypic and drug susceptibility studies on 13 clinical strains of M. porcinum were recently reported (16). We compared the type strain of M. porcinum and select clinical isolates of the third biovariant d-sorbitol-negative group, including the 13 clinical isolates in the recent taxonomy study (16), by a number of phenotypic and molecular methods to provide additional comparative data for these two taxa. (This work was presented in part as an abstract to the American Society for Microbiology meeting in Washington, D.C., in May 2003.)
MATERIALS AND METHODS
Mycobacterial isolates.
Clinical isolates of the M. fortuitum third biovariant sorbitol-negative group submitted to the Mycobacteria/Nocardia Laboratory at The University of Texas Health Center at Tyler for identification or susceptibility testing and the type strain of M. porcinum (ATCC 33776) were subjected to PCR restriction enzyme analysis (PRA) of the Telenti sequence of the hsp65 gene (20, 22). Two clinical isolates had previously been submitted to the American Type Culture Collection (ATCC; Manassas, Va.) and numbered as ATCC 49939 and ATCC BAA-328. Information on clinical histories, specimen type, and geographic location was obtained at the time of isolate submission. Chart reviews and informed consent were not obtained. Most clinical information was obtained before current Health Information Patient Protection Agency guidelines were in place. Care was taken not to utilize any specific patient identifiers and to protect patient confidentiality.
Sixty-seven clinical and two environmental isolates of the M. fortuitum third biovariant d-sorbitol-negative group were subjected to PRA. Of these, 46 clinical and two environmental isolates (48 isolates or 70%) had a PRA pattern identical to that of M. porcinum ATCC 33776T and were studied for clinical site and geographical source. Among the 45 patient isolates for which culture sites were known, 8 (18%) were respiratory; 7 (16%) were from central catheter-related infections and/or bacteremia and included five known cases of bacteremia; 28 (62%) were from postsurgical (7 cases), posttraumatic (12 cases), or unknown types (9 cases) of wound infections; 1 (2%) was from an infected lymph node; and 1 (2%) was from urine. Seven cases involved known associated osteomyelitis (Table 1). Two environmental isolates were recovered from tap water as part of an investigation of catheter-related infections in a bone marrow transplant unit in Texas (8). When treatment of the patients' infections was based on susceptibility results, with appropriate surgical intervention, all 12 cases with detailed follow-up resulted in clinical resolution.
TABLE 1.
Clinical and geographic locations of the 46 clinical and 2 reference isolates of the selected PRA group of the sorbitol-negative third biovariant complex
| Isolate type and name | Source | Geographic location | Clinical disease |
|---|---|---|---|
| Respiratory | |||
| Mf-91 | Sputum | Florida | Unknown |
| Mf-131 | Lung biopsy | Georgia | Pneumonia |
| Mf-239 | Sputum | Texas | Unknown |
| Mf-487 | Sputum | Texas | Pneumonia |
| Mf-607 | Sputum | Australia | Unknown |
| Mf-2075 | Sputum | Virginia | Unknown |
| Mf-2084 | Sputum | Texas | Bronchiectasis |
| Mf-2108 | Sputum | Illinois | Cystic bronchiectasis |
| Catheter or blood related | |||
| Mf-115 | Blood | Florida | Catheter sepsis |
| Mf-533 | Abdominal wall drainage | Texas | Peritoneal catheter exit site infection |
| Mf-537 | Blood | Texas | Catheter sepsis |
| Mf-1581 | Blood | Texas | Catheter sepsis |
| Mf-1600 | Not specified | Iowa | Catheter sepsis |
| Mf-1847 | Catheter site | Washington | Infected catheter site |
| Mf-2190 | Blood | North Carolina | Hemodialysis related |
| Wound related | |||
| Mf-114 | Leg amputation wound | Texas | Surgical wound infection |
| Mf-147 | Open ankle fracture | Texas | Cellulitis, osteomyelitis, inguinal lymphadenitis |
| Mf-182 | Traumatic knee wound | Texas | Septic arthritis |
| Mf-205 | Surgical abdominal wound | Texas | Surgical wound infection, abdominal wall abscess |
| Mf-388 | Traumatic foot wound | Texas | Cellulitis |
| Mf-443 | Surgical chest wound | Texas | Surgical wound infection |
| Mf-450 | Traumatic leg lesion | Florida | Cellulitis |
| Mf-485 | Open ankle fracture | South Carolina | Cellulitis, osteomyelitis |
| Mf-611 | Sinus, left leg | Australia | Cellulitis |
| ATCC 49939 (Mf-661) | Surgical back wound | Louisiana | Surgical wound infection |
| Mf-809 | Traumatic thigh wound | Texas | Cellulitis |
| Mf-1363 | Abdominal wound | Missouri | Unknown |
| Mf-1411 | Leg wound | Florida | Unknown |
| Mf-1472 | Spinal cord stimulator pocket | Florida | Unknown |
| Mf-1476 | Arm bone tissue | Oklahoma | Osteomyelitis, wound infection |
| Mf-1533 | Leg tissue | Arkansas | Unknown |
| Mf-1561 | Traumatic arm wound | Florida | Osteomyelitis, cellulitis |
| Mf-1568 | Base of toe | Florida | Unknown |
| Mf-1691 | Breast | Texas | Infected implant |
| Mf-1719 | Breast needle aspiration | Alabama | Cellulitis |
| Mf-1746 | Traumatic foot wound | Florida | Cellulitis |
| Mf-1824 | Hand tissue | Florida | Cystic lesion |
| Mf-1965 | Surgical face wound | Texas | Surgical wound infection |
| Mf-1966 | Open ankle fracture | North Carolina | Cellulitis, osteomyelitis |
| Mf-1997 | Traumatic foot wound | Texas | Osteomyelitis, cellulitis |
| Mf-2083 | Breast tissue | Texas | Unknown |
| Mf-2106 | Abscess, unknown site | Unknown | Unknown |
| Mf-2121 | Arm bone tissue | North Carolina | Osteomyelitis, cellulitis |
| Lymph node related | |||
| ATCC BAA-328 (Mf-771) | Inguinal node | Texas | Lymphadenitis |
| Environmental | |||
| Mf-1615 | Water softener | Texas | None |
| Mf-1616 | Water softener | Texas | None |
| Other | |||
| Mf-2100 | Urine | Texas | Unknown |
| Unknown | |||
| Mf-2107 | Unknown | Unknown | Unknown |
The 46 isolates for which geographical information was provided were from 16 states and Queensland, Australia. Multiple isolates were obtained from Texas (19 isolates or 41%), Florida (9 isolates or 20%), North Carolina (3 isolates), and Australia (2 isolates). All but eight isolates from the United States (36 of 44 isolates or 82%) were from southern coastal states.
The ATCC reference strains included M. porcinum ATCC 33776T and ATCC 33775, M. peregrinum type I ATCC 14467T, M. senegalense ATCC 35796T and ATCC 13781, M. neworleansense ATCC 49404T, M. houstonense ATCC 49403T, and M. septicum ATCC 700731T.
Growth and biochemical characteristics.
Isolates were examined for colony morphology on Mueller-Hinton agar, pigmentation after 1 and 2 weeks (early and late), growth within 7 days on Trypticase soy agar and Middlebrook 7H11 agar at 30°C, and carbohydrate utilization of d-mannitol, i-myo-inositol, and d-glucitol (sorbitol) (2, 10, 18, 24, 26, 31). Selected isolates were tested for growth at 35 and 45°C; arylsulfatase activity at 3 days; catalase activity at 68°C; semiquantitative catalase activity; nitrate reductase activity; iron uptake; growth on MacConkey agar; utilization of l-rhamnose, d-trehalose, citrate, benzoate, and acetamide as sole carbon sources (2, 10, 18, 24, 26, 31); and IEF patterns of β-lactamase by using polyacrylamide gels (32). Additionally, most isolates were tested for inhibition by commercial disks of polymyxin B, cephalothin, and kanamycin using agar disk diffusion on Mueller-Hinton agar (Table 2) (26). Results for some isolates have previously been published as part of earlier taxonomic studies (27, 32).
TABLE 2.
Laboratory features of clinical and reference isolates of the selected PRA group of the M. fortuitum d-sorbitol-negative third biovariant complex and the ATCC type strain of M. porcinum
| Isolate group and feature | M. porcinum ATCC 33776T | Third biovariant complex isolates
|
||
|---|---|---|---|---|
| ATCC BAA-328 | ATCC 49939 | Clinical isolates (no. positive/no. tested) (%) | ||
| All isolates | ||||
| Pigment (early/late) | −/− | −/− | −/− | 0/46 (0) |
| Rough colony morphology | + | + | + | 36/46 (78) |
| Growth at: | ||||
| <7 days | + | + | + | 46/46 (100) |
| 30°C | + | + | + | 46/46 (100) |
| 45°Ca (14 days) | + | + | + | 14/46 (30) |
| Acetamide | + | + | + | 46/46 (100) |
| Carbohydrate utilization | ||||
| d-Mannitol | + | + | + | 46/46 (100) |
| i-myo-Inositol | + | + | + | 46/46 (100) |
| d-Glucitol (sorbitol) | − | − | − | 0/46 (0) |
| l-Rhamnose | − | − | − | 0/46 (0) |
| d-Trehalose | + | − | + | 35/46 (76) |
| Citrate | − | − | − | 0/46 (0) |
| Disk inhibition | ||||
| Polymyxin B (diameter of inhibition, >6 mm)a | + | + | + | 46/46 (100) |
| Kanamycin (diameter of inhibition, ≥20 mm)a | + | − | + | 17/46 (37) |
| Random isolates | ||||
| Arylsulfatase activity (3 days) | + | + | + | 31/31 (100) |
| Semiquantitative catalase (≥100 mm)a | −b | NDc | − | 17/17 (100) |
| Catalase (68°C)a | + | ND | + | 14/19 (74) |
| Nitrate reductiona | − | ND | + | 19/19 (100) |
| Iron uptakea | + | ND | + | 18/19 (95) |
| MacConkey agar | ND | ND | + | 17/17 (100) |
| Disk inhibition | ||||
| Cephalothin (diameter of inhibition, >6 mm)a | − | − | − | 0/29 (0) |
First reported data for clinical isolates and for most tests for the ATCC type strain.
For M. porcinum ATCC 33776T, the semiquantitative catalase test outcome was weakly positive (the column of bubbles was 65 mm high), a result we considered negative.
ND, not determined.
HPLC.
Mycolic acids were prepared, esterified, and then subjected to fluorescence detection high-performance liquid chromatography (FL-HPLC) as previously described (2, 26, 31). HPLC reference strains included M. fortuitum ATCC 6841T, M. peregrinum ATCC 14467T, M. intracellulare ATCC 13950T, and M. neworleansense ATCC 49404T.
Susceptibility testing.
Susceptibility testing for 11 antimicrobial agents was performed using the broth microdilution method (3, 21) and the recently approved NCCLS standard M24-A (13). Drugs tested were amikacin, tobramycin, cefoxitin, imipenem, doxycycline, ciprofloxacin, gatifloxacin, levofloxacin, clarithromycin, linezolid, and sulfamethoxazole. Control strains included Staphylococcus aureus ATCC 29213 and M. peregrinum ATCC 700686 (13) (Table 3). Thirty-nine of the 48 isolates (including the two ATCC reference strains) recovered from all time periods were tested against eight of the drugs (the remaining nine isolates had been submitted only for identification, and drug susceptibilities had not been determined), while a smaller number of only recent isolates were tested against three drugs only recently made available (levofloxacin, gatifloxacin, and linezolid).
TABLE 3.
Susceptibility results for isolates of the selected PRA group of the M. fortuitum sorbitol-negative third biovariant and M. porcinum ATCC 33776T with the same PRA patterna
| Drug | MIC for M. porcinum ATCC 33776T (μg/ml) | MICs for third biovariant isolates (μg/ml)
|
No. of clinical third biovariant isolates tested | |||||
|---|---|---|---|---|---|---|---|---|
| ATCC 49939 | ATCC BAA-328 | Clinical isolates
|
||||||
| Range | MIC50 | MIC90 | Mode | |||||
| Group I (isolates from all time periods) | ||||||||
| Amikacin | ≤2 | 4, 2 | 4 | ≤1-64 | 4 | >32 | 32 | 39 |
| Cefoxitin* | 32 | 32 | 32 | 8-64 | 32 | 32 | 32 | 39 |
| Ciprofloxacin | 0.5 | 0.5, 1 | 0.125 | 0.25-1 | 0.5 | 1 | 0.5 | 39 |
| Clarithromycin* | 2 | 1 | 1 | 0.25-4 | 4 | 4 | 4 | 39 |
| Doxycycline | 4 | 8, 16 | >32 | ≤0.12-128 | 16 | >32 | 8, 32 | 39 |
| Imipenem | 8 | 4 | ≤0.5 | 1-8 | 4 | 8 | 4 | 39 |
| Sulfamethoxazole | ≤4 | ≤1, 2 | ≤1 | ≤2-8 | 2 | 8 | ≤1 | 39 |
| Tobramycin* | 2 | 4 | 32 | ≤1->32 | 16 | 32 | 4 | 39 |
| Group II (later isolates only) | ||||||||
| Gatifloxacin* | ND | ND | ND | ≤0.12-0.25 | ≤0.12 | ≤0.12 | ≤0.12 | 16 |
| Levofloxacin* | ND | ND | ND | ≤0.25-1 | 0.5 | 1 | 0.25, 0.5 | 27 |
| Linezolid* | 4 | ND | ND | 1-8 | 4 | 8 | 4 | 26 |
*, First reported data for clinical isolates and for most drugs for the ATCC reference strains. ND, not determined. MIC50, concentration inhibitory for 50% of strains. MIC90, concentration inhibitory for 90% of strains.
PRA.
Genomic DNA from all isolates was subjected to PCR amplification of the 441-bp Telenti sequence of the hsp65 gene (20, 22). The PCR product was digested with BstEII and HaeIII, and the DNA restriction fragments were separated using 3% metaphor agarose gels. Selected strains were also cut with BsaHI. Fragment sizes (in base pairs) were estimated using a computerized Bio Image System (Millipore, Bedford, Mass.).
16S rRNA gene sequencing.
Selected isolates of the third biovariant complex sorbitol-negative group and the M. porcinum type strain underwent sequencing of the first 500 bp using the MicroSeq 500 16S rDNA Bacterial Sequencing Kit (Applied Biosystems, Foster City, Calif.) at Mayo Clinic (9, 14). More complete 16S rRNA gene sequencing was also performed on select isolates using an in-house sequencing protocol (25). This included bases 27 to 1490 (total, 1,463 bases) spanning both hypervariable regions A and B.
hsp65 partial gene sequencing.
Selected isolates of the third biovariant complex sorbitol-negative group, M. porcinum ATCC 33776T, and M. porcinum ATCC 33775 were chosen for sequencing of the 441-bp Telenti fragment of the hsp65 gene (25). Sequencing of the PCR product obtained using the previously described protocol (22) was performed using the same PCR primers (TB11 and TB12) in a forward and reverse sequencing reaction according to the manufacturer's instructions (Applied Biosystems). Sequence editing and phylogenetic analyses by ClustalV (neighbor-joining method) were performed using the Seqman and Megalign components of Lasergene 5 (DNASTAR).
Nucleotide sequence accession numbers.
Examples of the five sequevars of hsp65 seen with M. porcinum have been deposited in GenBank as sequevar 1, accession number AY496137 (ATCC 33776T); sequevar 2, accession number AY496138 (ATCC BAA-328); sequevar 3, accession number AY496139 (Mf-114); sequevar 4, accession number AY496140 (ATCC 49939); and sequevar 5, accession number AY496141 (Mf-205). The hsp65 sequence for M. neworleansense ATCC 49404T was submitted to GenBank as AY496143, and the sequence for M. septicum ATCC 700731T was submitted as AY496142.
RESULTS
Growth and biochemical characteristics.
All 48 isolates were negative for early and late pigmentation, grew at 30°C, grew in <7 days, and were susceptible to polymyxin B. They were typical of members of the M. fortuitum third biovariant sorbitol-negative group in that all isolates were positive for d-mannitol and i-myo-inositol carbohydrate utilization tests but negative for d-sorbitol (Table 2, All isolates section). Testing of a small number of isolates was done in 13 additional tests (Table 2, Random isolates section). This showed isolates also to be typical members of the M. fortuitum complex by virtue of having a positive arylsulfatase reaction at 3 days, growing on MacConkey agar without crystal violet, and being positive for nitrate reduction, iron uptake, and acetamide utilization (Table 2). The isolates were also negative for citrate, l-rhamnose, and d-trehalose.
These results matched those for M. porcinum ATCC 33776T except the latter was nitrate negative as previously described (23). These are the first reported growth and biochemical data for a number of features (Table 2) for M. porcinum clinical isolates and for most of these tests for the M. porcinum ATCC type strain. Some growth and biochemical characteristics for 13 of these strains have been previously reported but were tested in a different laboratory (16).
β-Lactamase.
β-Lactamase IEF patterns showed 9 of 14 (64%) isolates to exhibit β-lactamase pattern 2 and 12 of 14 (86%) isolates to exhibit pattern 2 or 4. All isolates with pattern 2 or 4 that were subjected to PRA belonged to this group. None of the isolates matched β-lactamase IEF pattern 3, which was shown to be present in the M. fortuitum third biovariant d-sorbitol-negative group in previous studies (28, 32).
FL-HPLC.
Four clinical isolates of the selected PRA group of M. fortuitum third biovariant d-sorbitol-negative isolates were compared to M. porcinum ATCC 33776T, M. neworleansense ATCC 49404T, M. intracellulare 13950T, M. fortuitum ATCC 6841T, and M. peregrinum ATCC 14467T. The isolates of the selected PRA group and M. porcinum yielded mycolic acid chromatograms that were typical of Mycobacterium species. All produced an indistinguishable pattern of two closely clustered sets of peaks (Fig. 1). This pattern was indistinguishable from the pattern produced by M. fortuitum, M. peregrinum, and M. neworleansense. It was separable from the pattern of the control strain of M. intracellulare (Fig. 1) as well as those of M. mageritense and members of the M. smegmatis group as reported previously (2, 26).
FIG. 1.
Results of FL-HPLC with two strains of M. porcinum and three reference strains. The patterns of the last four isolates (B to D) are considered indistinguishable. (A) M. intracellulare ATCC 13950T; (B) M. porcinum ATCC 33776T; (C) M. porcinum ATCC 49939; (D) M. peregrinum type I ATCC 14467T; (E) M. fortuitum ATCC 6841T.
Susceptibility testing.
Drug susceptibility tests using current NCCLS mycobacterial breakpoints (where available) showed the clinical isolates to be susceptible to ciprofloxacin, gatifloxacin, levofloxacin, sulfamethoxazole, and linezolid and intermediate or susceptible to cefoxitin, clarithromycin, imipenem, and amikacin (13). The isolates were intermediate or resistant to doxycycline except for 1 (2.6%) of 39 strains tested which was susceptible. The eight drugs tested against all 39 clinical isolates and the two reference isolates are shown in the Group I section of Table 3, while the three drugs tested against only a small number of isolates are shown in the Group II section of Table 3. The M. porcinum type strain, ATCC 33776, gave the same susceptibility pattern (Table 3). Susceptibilities to amikacin, ciprofloxacin, imipenem, doxycycline, and sulfamethoxazole for 13 of these isolates (Mf-91, Mf-131, Mf-147, Mf-239, Mf-450, Mf-485, Mf-487, Mf-533, Mf-607, Mf-611, Mf-661, Mf-771, and Mf-809) (determined in another laboratory) were published previously as part of an earlier taxonomy study (16). These are the first reported susceptibility data for seven antimicrobial agents for M. porcinum (Table 3), including the ATCC type strain, 33776.
PRA.
By PRA of the hsp65 gene, M. porcinum ATCC 33776T and 49 of the 70 isolates of the third biovariant d-sorbitol-negative group exhibited the same restriction fragment length polymorphism pattern. They gave fragments of 235 and 210 bp with BstEII and 140, 125, and 100 bp with HaeIII. This pattern was unique from those of other currently recognized members of the M. fortuitum group (Fig. 2), with the exception of M. septicum. Enzyme restriction using BsaHI, however, distinguished the third biovariant isolates and the M. porcinum reference strain (260-, 100-, and 80-bp fragments) from M. septicum (260- and 80-bp fragments). Twenty-four of the clinical isolates were subjected to BsaHI digestion, and all gave the 260-, 100-, and 80-bp fragments of M. porcinum. PRA results of two ATCC isolates and two clinical isolates (ATCC 49939, ATCC BAA-328, Mf-131, and Mf-487) and M. porcinum ATCC 33776T have been reported previously (16).
FIG. 2.
PRA schema of the 441-bp Telenti fragment of the hsp65 gene for separating members of the M. fortuitum group. An ATCC reference strain, where applicable, is included.
16S rRNA gene sequencing.
Seventeen (37%) of the 46 clinical isolates (Mf-91, Mf-114, Mf-115, Mf-131, Mf-147, Mf-205, Mf-388, Mf-533, Mf-771, Mf-1568, Mf-1615, Mf-1847, Mf-1965, Mf-2106, Mf-2107, Mf-2108, and ATCC 49939) among the third biovariant d-sorbitol-negative group of isolates with the M. porcinum PRA pattern underwent partial 16S rRNA gene sequencing, and all exhibited 16S rRNA gene sequences identical to that of M. porcinum ATCC 33776T over the first 500 bp beginning at the 5′ end. This region of the gene also shows 100% identity with M. neworleansense ATCC 49404T (presently not included in the MicroSeq database), and therefore 12 clinical isolates (Mf-91, Mf-114, Mf-115, Mf-131, Mf-147, Mf-205, Mf-388, Mf-661, Mf-771 Mf-1568, Mf-1615, and Mf-2190) and ATCC 49939 were subjected to near-complete sequencing (1,463 bp) of the 16S rRNA gene and compared with the sequences previously determined for M. neworleansense ATCC 49404T and M. porcinum ATCC 33776T and ATCC 33775 (25). Ten of the 12 clinical third biovariant isolates and the reference isolates of M. porcinum had 100% sequence identity to each other, while one clinical strain (Mf-2190) differed by 1 bp from the other strains at Escherichia coli base-pair position 187. One isolate revealed a 1-bp variation from M. neworleansense at E. coli base-pair position 1135.
hsp65 gene partial sequencing.
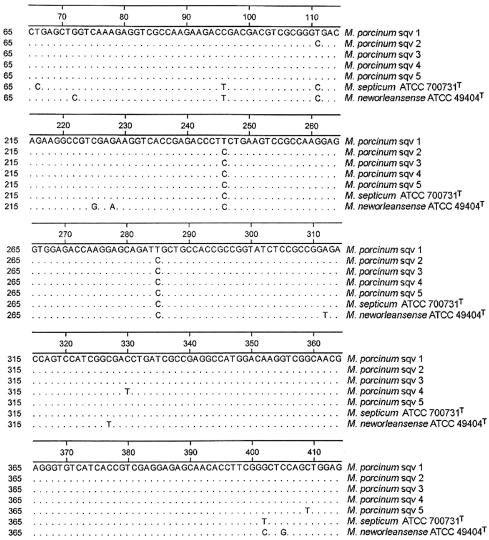
Twelve of the clinical M. fortuitum third biovariant d-sorbitol-negative isolates and both reference strains of M. porcinum were subjected to sequencing of the 441-bp Telenti fragment (hsp65 gene). Four distinct sequence variants (sequevars) were observed among the clinical isolates, varying from each other by 1 to 2 bases. Both ATCC strains of M. porcinum were identical to each other and showed 2- to 3-bp variations from the clinical strains. The four clinical sequevars all contained a T→C substitution at positions 246 and 285 that was not present in the two porcine strains. None of these variations were in regions affected by restriction enzyme sites for BstEII, HaeIII, or BsaHI. The hsp65 sequences of M. septicum ATCC 700731T (with a PRA profile identical to that of M. porcinum as determined by use of BstEII and HaeIII) and M. neworleansense ATCC 49404T (identical in the first 500 bp to the 16S rRNA gene sequence from M. porcinum) were also determined for comparison (Fig. 3). M. neworleansense differed by 11 bp, and M. septicum differed by 6 bp from the M. porcinum reference strains. Interestingly, both of these species also had the T→C substitution at positions 246 and 285.
FIG. 3.
Alignment of the partial hsp65 gene of the various sequevars of the M. porcinum group and the closely related M. septicum ATCC 700731T and M. neworleansense ATCC 49404T. Base positions represent numbering for the 441-bp Telenti fragment. Regions not shown contain no variations. M. porcinum sequevar (sqv) 1, M. porcinum ATCC 33776T and M. porcinum ATCC 33775; M. porcinum sqv 2, clinical strains Mf-2190, Mf-131, Mf-147, Mf-388, Mf-115, ATCC BAA-328 (Mf-771), Mf-1568, and Mf-91; M. porcinum sqv 3, Mf-1615 and Mf-114; M. porcinum sqv 4, ATCC 49939; M. porcinum sqv 5, Mf-205.
DISCUSSION
These studies identified only a few minor phenotypic, drug susceptibility-related, or molecular differences between the human PRA group of the M. fortuitum third biovariant complex d-sorbitol-negative group and the porcine reference strains of M. porcinum. The original 10 strains of M. porcinum, characterized by Tsukamura et al., were found to be nitrate negative (23). The M. porcinum type strain was negative for nitrate reduction on repeat testing in the present study, while all third biovariant d-sorbitol-negative isolates tested, including the two ATCC strains, were positive for nitrate reduction (20 of 20 isolates; Tsukamura also tested 12 of these isolates in his laboratory, and all were positive). The type strain of M. porcinum had a low semiquantitative catalase (65 mm), while 17 of the 17 third biovariant isolates had a high (>100 mm) semiquantitative catalase. A small number of the third biovariant isolates were tested for benzoate as a sole carbon source in Tsukamura's laboratory, with 4 of 12 (33%) being positive compared to 100% positivity of the original published strains of M. porcinum (data not presented) (23). Other studies including drug susceptibilities, FL-HPLC, PRA, and carbohydrate utilization gave comparable results for the two groups.
The 16S rRNA gene sequence of the M. fortuitum third biovariant d-sorbitol-negative strain ATCC 49404 was first reported by Kirschner et al. in 1992 (11). This group subsequently sequenced hypervariable region A of the 16S rRNA gene of three additional third biovariant d-sorbitol-negative strains (ATCC 49935, ATCC 49937, and ATCC 49939) and found them identical to ATCC 49404 (19).
ATCC 49404 was found to have a PRA pattern of the hsp65 gene that differed from those of the other third biovariant d-sorbitol-negative strains (20), although Turenne et al. recently showed M. porcinum ATCC 33776T to differ from this strain by only 1 bp over the entire 1,400-bp 16S rRNA gene sequence (25). Schinsky et al. showed ATCC 49404 to have DNA-DNA homologies of <50% with all other taxa within the third biovariant group including M. porcinum and named this isolate M. neworleansense (16). No other third biovariant d-sorbitol-negative clinical strains were sequenced in the study by Turenne et al. (25). That group did complete sequences of other M. fortuitum group members, including M. porcinum ATCC 33776T, and showed that the latter differed by 8 bp from its closest sequences. These were M. farcinogenes (a nonpigmented slow grower originally reported by Chamoiseau) (5, 15) and the proposed type strain of M. houstonense, ATCC 49403 (formerly of the M. fortuitum third biovariant d-sorbitol-positive group) (11, 16). Some of those differences are present in the first 500 bp of the 16S rRNA gene, and this sequence is presently different from those of all other recognized species.
Of the isolates of the M. fortuitum third biovariant d-sorbitol-negative group whose hsp65 PRA patterns matched that of M. porcinum ATCC 33776T, 4 isolates in the Schinsky et al. study (16) and 17 in the present study gave partial 16S rDNA gene sequences that matched that of the M. porcinum type strain. Eleven clinical isolates with almost complete 16S rRNA gene sequencing also showed 100% identity. This suggests that hsp65 PRA will accurately identify isolates of M. porcinum.
The phenotype, drug susceptibility, and hsp65 gene sequence similarity, the 16S rDNA identity, and the high degree of overall DNA homology (presented in the study by Schinsky et al.) (16) provide convincing evidence that the nonpigmented pig pathogen described in 1983 by Tsukamura et al. (23) is the same species as the present human pathogen previously grouped within the M. fortuitum third biovariant d-sorbitol-negative group (27). We would agree with calling these isolates M. porcinum, with three reference strains (one porcine, two human) presently catalogued in the ATCC.
The sources and clinical disease associated with these isolates of M. porcinum clearly define them as human pathogens. Their role in causing chronic lung infection is not well established, but there is little question of their ability to cause posttraumatic or postsurgical wound infections, osteomyelitis, and catheter-related infections. These findings are similar to what has been reported for all members of the M. fortuitum third biovariant group (27). As has been noted with several types of surgical wound infections, the M. porcinum clinical isolates submitted to us are concentrated among southern coastal states (6, 12, 29, 30).
This study identified two environmental isolates of M. porcinum (Mf-1615 and Mf-1616), the first of these to be reported. Both were recovered from tap water (8). This reservoir could explain the frequent association of this species with catheter-related infections, since catheter exposure to tap water probably occurs frequently.
M. porcinum isolates are phenotypically separable from isolates of the three recognized or proposed species of the M. fortuitum third biovariant d-sorbitol-positive group using l-rhamnose (M. mageritense), d-sorbitol (M. mageritense, M. houstonense, and M. brisbanense), and clarithromycin susceptibility (same three species) (Table 4). It is not phenotypically separable from any of the other four presently recognized species or species of the M. fortuitum third biovariant d-sorbitol-negative group (M. septicum, M. neworleansense, M. concordense, and M. boenickei) (Table 4). All four species have the same antibiotic susceptibility pattern, including being intermediate or susceptible to cefoxitin and clarithromycin. This comment is qualified by the observation that only a single strain each of M. septicum, M. neworleansense, and M. concordense has been reported and characterized to date (16, 17).
TABLE 4.
Useful tests for separating M. porcinum from other members of the M. fortuitum groupd
| Species (strain) (reference) | Antibiotic resistance
|
Carbohydrate utilization
|
Low catalase (<100 mm) | Unique PRA hsp65 gene | Unique 16S r-DNA
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clari (MIC, >4 μg/ml) | Fox (MIC, ≥128 μg/ml) | d-man | i-myo-inos | d-sorb | l-rham | Hyper-variable A region | First 500 bp (5′ end) | Entire sequence | |||
| M. fortuitum (ATCC 6841T) (4) | Var | − | − | − | − | − | − | + | + | + | + |
| M. neworleansense (ATCC 49404T) (16, 27) | − | − | + | + | − | − | − | − | −a | − | + |
| M. mageritense (ATCC 700351T) (26) | + | − | + | + | + | + | + | + | + | + | + |
| M. porcinum (ATCC 33776T, BAA-328, ATCC 49939) | − | − | + | + | − | − | − | + | −a | − | + |
| M. septicum (ATCC 700731T) (17) | − | − | + | + | − | − | − | + | −a,b | −b | −b |
| M. houstonense (ATCC 49403T) (4, 16, 27) | + | + | + | + | + | − | + | − | −c | −c | −c |
| M. boenickei (ATCC 49935T) (16) | − | − | + | + | − | − | − | + | −a | NK | + |
| M. peregrinum type I (ATCC 14467T) (4) | − | − | + | − | − | − | − | + | −b | −b | −b |
| M. senegalense (ATCC 35796T) | − | − | + | − | − | − | − | + | −c | −c | + |
M. porcinum and other members of the M. fortuitum third biovariant sorbitol-negative group, other than M. septicum, have the same hypervariable A sequence.
M. septicum and M. peregrinum type I have the same sequence over the entire 1,400-bp 16S rRNA gene sequence.
M. senegalense, M. houstonense, and M. farcinogenes have the same sequence over the first 500 bp. M. houstonense and M. farcinogenes are also identical over the entire 1,400 bp.
An ATCC reference strain, where applicable, is included. Abbreviations: Var, variable; Clari, clarithromycin; Fox, cefoxitin; man, mannitol; inos, inositol; sorb, sorbitol (D-glucitol); rham, rhamnose; +, ≥90% positive for indicated characteristic; −, ≤10% negative for indicated characteristic; NK, not known.
The best biochemical screening test for selection of M. fortuitum among isolates belonging to the M. fortuitum group would appear to be d-mannitol. d-Mannitol-negative isolates are presumptive M. fortuitum, while d-mannitol-positive isolates are not M. fortuitum but require additional testing to define the species. Sequencing of the hypervariable A region of the 16S rRNA gene or even the first 500 bp results in a great deal of overlap for numerous taxa (25). This region identifies M. porcinum and M. neworleansense as distinct from the rest of the M. fortuitum complex but identical to each other. A single additional sequencing reaction encompassing the V7 region of the 16S rRNA gene can differentiate between these two species due to a 1-bp variation at position 1135 and could therefore provide accurate species identification. These species are better distinguished by their hsp65 sequences, showing 11 mismatches or a 97.5% sequence identity, contributing to clearly distinct PRA patterns. The best single molecular test for identifying species within the M. fortuitum group would appear to be hsp65 PRA, as it clearly separated the sorbitol-positive species and M. boenickei from M. porcinum within the sorbitol-negative group.
Acknowledgments
We thank Stacie Yarborough, Maria McGlasson, Amanda Burnett, Brian Campbell, and Cole Fiser and the staff in the mycobacteriology laboratory at the Texas Department of Health, Austin, Texas, for their laboratory assistance and Joanne Woodring for preparation of the manuscript.
Footnotes
This paper is dedicated to the three coauthors Vella Silcox, Zeta Blacklock, and Michio Tsukamura, early investigators who contributed greatly to the present study but who retired before the present taxonomic status of isolates of the third biovariant complex was established. Both Zeta Blacklock and Michio Tsukamura passed away prior to submission of this study.
REFERENCES
- 1.Bönicke, R. 1966. The occurrence of atypical mycobacteria in the environment of man and animal. Bull. Int. Union Tuberc. Lung Dis. 37:361-368. [Google Scholar]
- 2.Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, M. C. Menendez, B. Rodriguez-Salgado, K. C. Jost, S. H. Chiu, G. O. Onyi, E. C. Böttger, and R. J. Wallace, Jr. 1999. Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49:1493-1511. [DOI] [PubMed] [Google Scholar]
- 3.Brown, B. A., J. M. Swenson, and R. J. Wallace, Jr. 1992. Broth microdilution MIC test for rapidly growing mycobacteria, p. 5.11.1. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook. American Society for Microbiology, Washington, D.C.
- 4.Brown, B. A., R. J. Wallace, Jr., G. Onyi, V. DeRosas, and R. J. Wallace III. 1992. Activities of four macrolides including clarithromycin against Mycobacterium fortuitum, Mycobacterium chelonae, and Mycobacterium chelonae-like organisms. Antimicrob. Agents Chemother. 36:180-184. [DOI] [PMC free article] [PubMed]
- 5.Chamoiseau, G. 1979. Etiology of farcy in African bovines: nomenclature of the causal organisms Mycobacterium farcinogenes Chamoiseau and Mycobacterium senegalense (Chamoiseau) comb. nov. Int. J. Syst. Bacteriol. 29:407-410. [Google Scholar]
- 6.Clegg, H. W., M. T. Foster, W. E. Sanders, Jr., and W. B. Baine. 1983. Infection due to organisms of the Mycobacterium fortuitum complex after augmentation mammaplasty: clinical and epidemiologic features. J. Infect. Dis. 147:427-433. [DOI] [PubMed] [Google Scholar]
- 7.Domenech, P., M. S. Jimenez, M. C. Menendez, T. J. Bull, S. Samper, A. Manrique, and M. J. Garcia. 1997. Mycobacterium mageritense sp. nov. Int. J. Syst. Bacteriol. 47:535-540. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, S. L., R. M. Plemmons, D. P. Dooley, C. E. Davis, M. C. Garces, B. A. Brown, D. Grubber, and R. J. Wallace. 1999. Identification and control of an outbreak of rapidly growing mycobacterial infections in a bone marrow transplant unit, abstr. 469, p. 121. 37th Annu. Meet. Infect. Dis. Soc. Am., Philadelphia, Pa.
- 9.Hall, L., K. A. Doerr, S. L. Wohlfiel, and G. D. Roberts. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control, U.S. Department of Health and Human Services, Atlanta, Ga.
- 11.Kirschner, P., M. Kiekenbeck, D. Meissner, J. Wolters, and E. C. Böttger. 1992. Genetic heterogeneity within Mycobacterium fortuitum complex species: genotypic criteria for identification. J. Clin. Microbiol. 30:2772-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuritsky, J. N., M. G. Bullen, C. V. Broome, R. C. Good, and R. J. Wallace, Jr. 1983. Sternal wound infections and endocarditis due to organisms of the Mycobacterium fortuitum complex. Ann. Intern. Med. 98:938-939. [DOI] [PubMed] [Google Scholar]
- 13.NCCLS. 2003. Susceptibility testing of mycobacteria, nocardia and other aerobic actinomycetes. Approved standard. NCCLS document M24-A. NCCLS, Wayne, Pa. [PubMed]
- 14.Patel, J. B., D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridell, M., and M. Goodfellow. 1983. Numerical classification of Mycobacterium farcinogenes, Mycobacterium senegalense and related taxa. J. Gen. Microbiol. 129:599-611. [DOI] [PubMed] [Google Scholar]
- 16.Schinsky, M. F., R. E. Morey, A. G. Steigerwalt, M. P. Douglas, R. W. Wilson, M. M. Floyd, W. R. Butler, M. I. Daneshvar, B. A. Brown-Elliott, R. J. Wallace, Jr., M. M. McNeil, D. J. Brenner, and J. M. Brown. 2004. Taxonomic variation in the Mycobacterium fortuitum third biovariant complex: description of Mycobacterium boenickei sp. nov., Mycobacterium houstonense sp. nov., Mycobacterium neworleansense sp. nov. and Mycobacterium brisbanense sp. nov. and recognition of Mycobacterium porcinum from human clinical isolates. Int. J. Syst. Evol. Microbiol. 54:1653-1667. [DOI] [PubMed] [Google Scholar]
- 17.Schinsky, M. F., M. M. McNeil, A. M. Whitney, A. G. Steigerwalt, B. A. Lasker, M. M. Floyd, G. G. Hogg, D. J. Brenner, and J. M. Brown. 2000. Mycobacterium septicum sp. nov. a new rapidly growing species associated with catheter-related bacteraemia. Int. J. Syst. Evol. Microbiol. 50:575-581. [DOI] [PubMed] [Google Scholar]
- 18.Silcox, V. A., R. C. Good, and M. M. Floyd. 1981. Identification of clinically significant Mycobacterium fortuitum complex isolates. J. Clin. Microbiol. 14:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Springer, B., E. C. Böttger, P. Kirschner, and R. J. Wallace, Jr. 1995. Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int. J. Syst. Bacteriol. 45:262-267. [DOI] [PubMed] [Google Scholar]
- 20.Steingrube, V. A., J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr. 1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swenson, J. M., C. Thornsberry, and V. A. Silcox. 1982. Rapidly growing mycobacteria: testing of susceptibility to 34 antimicrobial agents by broth microdilution. Antimicrob. Agents Chemother. 22:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukamura, M., H. Nemoto, and H. Yugi. 1983. Mycobacterium porcinum sp. nov., a porcine pathogen. Int. J. Syst. Bacteriol. 33:162-165. [Google Scholar]
- 24.Tsukamura, M. 1984. Identification of mycobacteria. Mycobacteriosis Research Laboratory of the National Chubu Hospital, Obu, Aichi, Japan.
- 25.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace, R. J., Jr., B. A. Brown-Elliott, L. Hall, G. Roberts, R. W. Wilson, L. B. Mann, C. J. Crist, S. H. Chiu, R. Dunlap, M. J. Garcia, J. T. Bagwell, and K. C. Jost, Jr. 2002. Clinical and laboratory features of Mycobacterium mageritense. J. Clin. Microbiol. 40:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace, R. J., Jr., B. A. Brown, V. A. Silcox, M. Tsukamura, D. R. Nash, L. C. Steele, V. A. Steingrube, J. Smith, G. Sumter, Y. Zhang, and Z. Blacklock. 1991. Clinical disease, drug susceptibility, and biochemical patterns of the unnamed third biovariant complex of Mycobacterium fortuitum. J. Infect. Dis. 163:598-603. [DOI] [PubMed] [Google Scholar]
- 28.Wallace, R. J., Jr., D. R. Nash, T. Udou, V. A. Steingrube, L. C. Steele, J. M. Swenson, and V. A. Silcox. 1985. Isoelectric focusing of beta-lactamases in Mycobacterium fortuitum. Am. Rev. Respir. Dis. 132:1093-1097. [DOI] [PubMed] [Google Scholar]
- 29.Wallace, R. J., Jr., L. C. Steele, A. Labidi, and V. A. Silcox. 1989. Heterogeneity among isolates of rapidly growing mycobacteria responsible for infections following augmentation mammaplasty despite case clustering in Texas and other southern coastal states. J. Infect. Dis. 160:281-288. [DOI] [PubMed] [Google Scholar]
- 30.Wallace, R. J., Jr., J. M. Musser, S. I. Hull, V. A. Silcox, L. C. Steele, G. D. Forrester, A. Labidi, and R. K. Selander. 1989. Diversity and sources of rapidly growing mycobacteria associated with infections following cardiac bypass surgery. J. Infect. Dis. 159:708-716. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, R. W., V. A. Steingrube, E. C. Böttger, B. Springer, B. A. Brown-Elliott, V. Vincent, K. C. Jost, Jr., Y. Zhang, M. J. Garcia, S. H. Chiu, G. O. Onyi, H. Rossmoore, D. R. Nash, and R. J. Wallace, Jr. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks, and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 51:1751-1764. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y., R. J. Wallace, Jr., V. A. Steingrube, B. A. Brown, D. R. Nash, A. Silcox, and M. Tsukamura. 1992. Isoelectric focusing patterns of β-lactamases in the rapidly growing mycobacteria. Tuber. Lung Dis. 73:337-344. [DOI] [PubMed] [Google Scholar]