Abstract
The pancreas is comprised of epithelial cells that are required for food digestion and blood glucose regulation. Cells of the pancreas microenvironment, including endothelial, neuronal, and mesenchymal cells were shown to regulate cell differentiation and proliferation in the embryonic pancreas. In the adult, the function and mass of insulin-producing cells were shown to depend on cells in their microenvironment, including pericyte, immune, endothelial, and neuronal cells. Lastly, changes in the pancreas microenvironment were shown to regulate pancreas tumorigenesis. However, the cues underlying these processes are not fully defined. Therefore, characterizing the different cell types that comprise the pancreas microenvironment and profiling their gene expression are crucial to delineate the tissue development and function under normal and diseased states. Here, we describe a method that allows for the isolation of mesenchymal cells from the pancreas of embryonic, neonatal, and adult mice. This method utilizes the enzymatic digestion of mouse pancreatic tissue and the subsequent fluorescence-activated cell sorting (FACS) or flow-cytometric analysis of labeled cells. Cells can be labeled by either immunostaining for surface markers or by the expression of fluorescent proteins. Cell isolation can facilitate the characterization of genes and proteins expressed in cells of the pancreas mesenchyme. This protocol was successful in isolating and culturing highly enriched mesenchymal cell populations from the embryonic, neonatal, and adult mouse pancreas.
Keywords: Developmental Biology, Issue 119, pancreas, pancreas microenvironment, pancreas development, pancreatic cancer, pancreatic mesenchyme, pancreatic ductal adenocarcinoma
Introduction
Energy homeostasis and food digestion in mammals depend on proper pancreatic function. The adult pancreas is comprised of two main cellular compartments: the exocrine and the endocrine. Exocrine cells, including the acinar cells that produce and secrete digestive enzymes and the duct cells that transport these enzymes to the gut, encompass more than 80% of the total pancreatic mass1. Endocrine cells, which include insulin-producing beta cells and glucagon-producing alpha cells, are organized in the islets of Langerhans that are embedded in the exocrine tissue and secrete hormones to regulate blood glucose levels2.
Pancreatic cells acquire their differentiated fate through a highly-regulated, multistep process3. Evidence suggests that extrinsic cues provided by neuronal, endothelial, and mesenchymal cells guide pancreatic cell differentiation and proliferation in the embryo3,4,5. One example is the requirement of the aorta for the specification of early pancreatic precursors6. Later in development, endothelial cells were shown to play a central role in the development of both pancreatic endocrine and exocrine cells and to promote beta-cell differentiation4,6,7. Mesenchymal cells were shown to support the survival and expansion of common pancreatic progenitors, mainly through the secretion of the growth factor Fgf108,9. We have further shown that these cells support the proliferation of endocrine and exocrine precursors, as well as of differentiated cells (including acinar and beta cells) in the embryonic pancreas5. Recently, mesenchymal cells were further shown to regulate endocrine cells differentiation10.
In the adult, beta-cell function and mass were shown to depend on cells in their microenvironment, including neuronal, immune, and endothelial cells, as well as pericytes11,12,13. During injury, endothelial cells were shown to recruit immune cells to the pancreas to promote beta-cell replication13. Endothelial cells were further shown to produce extracellular matrix (ECM) components to support insulin expression and beta-cell function14. We recently demonstrated the requirement of islet pericytes for beta-cell function11. Lastly, cells of the pancreatic stroma were shown to regulate the progression of pancreatic ductal adenocarcinoma (PDAC)15,16. However, the identity of extrinsic cues that guide pancreas development, function, and tumorigenesis are largely unknown.
Identifying cues provided by cells of the pancreas microenvironment requires characterizing the genes and proteins expressed by these cells. This depends on isolating these cells from the pancreas in order to perform gene expression and proteomic analyses and/or on establishing cell lines. Here, we propose a method to isolate mesenchymal cells of the pancreas microenvironment by utilizing tissue enzymatic digestion and fluorescence-activated cell sorting (FACS) of either immunofluorescently-labeled cells or cells expressing fluorescent proteins. This protocol was successfully performed to isolate and analyze yellow fluorescent protein (YFP)-expressing mesenchymal cells of the embryonic, neonatal, and adult pancreas5,17.
Protocol
Experiments were conducted according to protocols approved by the Committee on Animal Research at Tel Aviv University.
1. Isolation of Pancreatic Tissue from Mice
- For adult mice:
- Prepare the digestion buffer: 0.4 mg/mL collagenase P and 0.1 ng/mL DNase I dissolved in Hanks' Balanced Salt Solution (HBSS). Freshly prepare 5 mL of buffer per mouse in 15-mL conical tubes and keep them on ice until use.
- Euthanize the mice according to institutional guideline.
- Dissect each mouse to remove the pancreas (for instruction, please see Reference 18); place it in a culture dish containing HBSS. It is possible to perform this stage under a stereomicroscope, and to increase digestion efficiency, cut the tissue into 2 - 4 pieces.
- Place the pancreatic tissue into a tube containing digestive buffer (prepared in step 1.1.1). Keep the tubes on ice until all mice are dissected and pancreatic tissues collected.
- Repeat steps 1.1.2 - 1.1.4 with the remaining mice.
- For embryos and neonatal mice:
- Prepare the digestion buffer: 0.4 mg/mL collagenase P and 0.1 ng/mL DNase I dissolved in HBSS. For embryos and pups at postnatal day 7 (p7) or younger, freshly prepare 1.5 - 2 mL of buffer per mouse in a 15-mL conical tube. For p7 or older pups, prepare 3 - 4 mL of buffer per mouse in a 15-mL conical tube. Keep on ice until use.
- Euthanize the mice according to institutional guideline. For embryos, remove all embryos from their mother's uterus and place them in a 10-mm culture dish containing PBS. Dissect one embryo at a time.
- Lay the mouse on its back, with its head pointing away from the investigator. Using fine forceps, pull up the abdominal skin and open at the midline to expose the abdominal cavity up to the mouse diaphragm18.
- Using fine forceps, detach the liver from the back abdominal wall. With a continuous motion toward the mouse tail, scoop out the internal organs (including the liver, stomach, spleen, intestines, kidney, and pancreas) and place them in a culture dish containing HBSS.
- Under a stereomicroscope, remove the liver and kidneys to expose the pancreas (Figure 1). Using fine forceps, detach the pancreas from the stomach, duodenum, and finally, from the spleen.
- Place the pancreatic tissue into a tube containing digestive buffer (prepared in step 1.2.1). For p14 or older pups, cut the tissue into 2 pieces to increase the digestion efficiency. Keep the tubes on ice until all mice are dissected and pancreatic tissues collected.
- Repeat steps 1.2.2 - 1.2.6 with the remaining mice.
2. Pancreas Digestion
Incubate the tubes containing the pancreatic tissue (in digestion buffer) in a heating block at 37 °C for 30 min with agitation at 700 rpm. After 15 min, manually shake the tubes by inverting them 3 - 4 times and place them back in the heating block. NOTE: If using a heating block without agitation capabilities, invert the tubes every 5 - 10 min. Make sure that the tissue is properly digested. If not, cut the tissue into smaller pieces before incubation or increase the agitation.
To stop the tissue digestion, place the tubes on ice and add 10 mL of cold HBSS to each tube. Centrifuge the tubes at 4 °C and 300 x g for 5 min and aspirate the supernatant.
- Re-suspension:
- For adult mice and p14 or older pups, re-suspend the pellet with 6 mL of PBS and strain it through a 70-µm cell strainer placed on top of a new 15-mL conical collection polypropylene tube. To ensure maximal cell recovery, wash the original tube with an additional 6 mL of PBS and strain it through the cell strainer onto the collection tube.
- For embryos and pups younger than p14, re-suspend the pellet with 2 mL of PBS and strain it through a 35-µm cell strainer placed on top of a 5-mL round-bottom polystyrene collection tube (FACS tube). To ensure maximal cell recovery, wash the original tube with an additional 2 mL of PBS and strain it through the cell strainer onto the collection tube.
Centrifuge the tubes at 4 °C and 300 x g for 5 min and aspirate the supernatant. Remove the liquid carefully, as cells are loosely attached to the polystyrene tube.
3. Preparing Labeled Single Cells
Buffer preparations: For cell isolation by FACS, prepare sorting buffer by mixing PBS (without calcium chloride and magnesium chloride), 5% fetal bovine serum (FBS), and 5 mM EDTA. For flow cytometry analysis, prepare analysis buffer by supplementing sorting buffer with 0.05% sodium azide. Both buffers may be stored at 4-8 °C for up to 2 months.
Re-suspend cells in sorting or analysis buffer. Re-suspend each pancreas isolated from embryos and pups younger than p7 in 1 mL, from pups between p8 and one month old in 1.5 mL, and from mice older than one month in 3 mL. Strain the cells through a 35-µm cell strainer placed on top of a round-bottom polystyrene tube (FACS tube).
For staining controls, take out 50- to 100-µL aliquots from the samples made in the previous step (not more than 5% of the total sample volume) and place them into new FACS tubes; include a tube for each fluorophore used, including DAPI and fluorescent proteins, as well as for an unstained control. NOTE: When using transgenic mice with cells expressing a fluorescent protein, or when cell number is limited, include a non-transgenic tissue as a staining control. If cells expressing a fluorescent protein are used without further staining, proceed to step 3.8.
Spin-down the cells at 300 x g for 5 min at 4 °C and aspirate the supernatant.
For blocking, re-suspend the cells with blocking solution (100 µL sorting or analysis buffer supplemented with 1 µL of goat IgG) and incubate for 30 min on ice.
To stain cell surface markers, prepare a mixture that includes all desired fluorophore-conjugated antibodies in a volume of 100 µL per sample . Prepare 2x antibody dilutions in sorting or analysis buffer (i.e., dilute the antibody to obtain double the required concentration; if a final dilution of 1:200 is desired, dilute the antibody 1:100). Without washing the blocking solution, add 100 µL of the mixture to the sample cells to achieve a final staining volume of 200 µL. Incubate for 30-60 min on ice and in the dark.
To set the analysis or sorting parameters (see steps 4.3 and 5.2), prepare additional tubes that contain only one of the antibodies used (staining control). Prepare 2x antibody dilutions in sorting or analysis buffer (as described in step 3.6). Make sure to include a single staining control dilution for each fluorophore used, as well as an unstained control. Without washing the blocking solution, add 100 µL of antibody mixes to the cells (prepared in step 3.3) to achieve a final staining volume of 200 µL. Incubate for 30 - 60 min on ice and in the dark.
Wash by filling each tube with analysis or sorting buffer to a maximal volume of 4 mL. Optionally, re-strain the cells through a 35-µm cell strainer (as described in step 3.2).
Centrifuge at 300 x g for 5 min at 4 °C and remove the supernatant carefully.
Re-suspend the samples in analysis or sorting buffer. Re-suspend the cells isolated from embryos in 500 µL, from pups younger than one month in 1 mL, from 1 - 3 months old in 2 mL, and from mice older than 3 months in 3 mL. Staining controls may be re-suspended in 300 µL of buffer.
Add 200 ng/mL of DAPI to re-suspended cells to identify the dead cells. Make sure to include a tube containing unstained cells without DAPI for cytometer settings. Proceed to cell sorting (step 4) or analysis (step 5).
4. Cell Sorting
- Preparations:
- For cell sorting prior to RNA extraction, coat a 1.5-mL collecting tube with RNase inhibitor immediately before sorting. To this end, add 1 mL of sorting buffer, containing 0.01 U/mL RNase inhibitor, to a sterile 1.5-mL collection tube. After 5 min, vortex the tube and remove the liquid.
- For cell sorting prior to cell culture, add 3 mL of sterile culturing media (Dulbecco's Modified Eagle Medium (DMEM) supplemented with 20% FBS, 1% L-glutamine, and 1% penicillin-streptomycin) into a sterile 15-mL collection tube.
Before loading a tube into a FACS sorter, vortex it briefly to re-suspend the cells. Keep the remaining tubes on ice.
Start by analyzing the staining controls to determine the sorting parameters (e.g., voltage and compensation) and sorting gates (e.g., total cell population, live DAPI-negative cells, and cell populations to be sorted).
Once the sorting parameters and gates are set up, load the samples and initiate cell sorting into the collection tubes. NOTE: Sorting conditions are highly dependent on the instrument. We use a nozzle width of 100 µm, a pressure of 23.1 psi, and a maximal sorting speed of 5.
Proceed to RNA extraction or the culturing of sorted cells. NOTE: For RNA extraction, centrifuge the cells at 2,000 x g for 5 min and remove the excess liquid before continuing with a standard extraction protocol. For culturing cells, if the cells were sorted under non-sterile conditions, wash them twice by filling the tube with culturing medium and centrifuging it at 300 x g for 7 min before culturing in order to minimize their contamination.
5. Cell Analysis by Flow Cytometry
Before loading each tube into the cytometer, vortex it briefly to re-suspend the cells. Keep the remaining tubes on ice.
Start by analyzing the unstained and single-stained samples in order to determine the analysis parameters (e.g., voltage and compensation).
Once the analysis parameters are set up, load each sample, including the staining control, and record the results. Analyze the obtained results using flow cytometry analysis software.
Representative Results
The pancreatic mesenchyme is required during development and adulthood. The method described here allows the isolation of mesenchymal cells from the embryonic, neonatal, and adult pancreas. Mesenchymal cells, but no other cell types, express yellow fluorescent protein (YFP) in the pancreas of Nkx3.2-Cre;R26R-YFP mice5,11,17,19. During development, Nkx3.2 (also known as BapX1) is expressed by embryonic pancreatic, stomach, and gut mesenchyme, as well as in a subset of skeletal somites19,20,21. This gene was expressed in the pancreatic mesenchyme from e9.5 until e11.5, allowing gene expression under the control of Nkx3.2-Cre from e9.55,19,20. Based on this labeling, cells can be purified from bulk pancreatic tissue using flow cytometry. Figure 2 shows a flow-cytometry analysis of single cells from embryonic, neonatal, and adult pancreatic tissues, isolated as described here. Whereas non-transgenic pancreatic tissues did not contain fluorescent cells, Nkx3.2-Cre;R26R-YFP pancreatic tissue from all analyzed ages contains a distinct YFP-labeled cell population (Figure 2; marked with gates).
Following the method described here, cells expressing fluorescent proteins can be either purified or analyzed by flow cytometry, with or without additional immunostaining. For example, after sorting based on fluorescent labeling, mesenchymal cells can be cultured to establish a cell line (for at least five passages), as shown in Figure 3A. Note the fibrocytic morphology of the cultured cells, typical to mesenchymal cells. In addition, this system was used to analyze gene expression by sorted cells. To this end, RNA was extracted from sorted mesenchymal cells to synthesize cDNA, and gene expression levels were analyzed by qPCR. Such analysis revealed that sorted cells express the pan-mesenchymal marker vimentin (Figure 3B). Lastly, surface marker expression by pancreatic cells can be analyzed by flow-cytometry. For example, we isolated cells from the pancreatic tissue of Nkx3.2-Cre;R26R-YFP adult mice using the method described here and stained them with the cell surface glycoprotein CD9, which was reported to be expressed by fibroblasts22. As shown in Figure 3C, all fluorescently-labeled cells in the Nkx3.2-Cre;R26R-YFP pancreas express CD9.
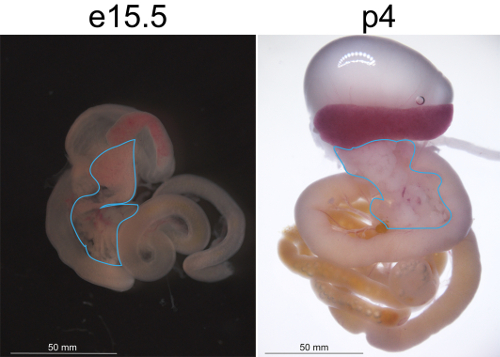 Figure 1: Embryonic and neonatal pancreatic tissue. Isolated whole gastrointestinal tract, including the stomach, spleen, intestine, and pancreas, of an e15.5 embryo (A) and a p4 pup (B). The pancreatic tissue is demarcated with a blue line. Please click here to view a larger version of this figure.
Figure 1: Embryonic and neonatal pancreatic tissue. Isolated whole gastrointestinal tract, including the stomach, spleen, intestine, and pancreas, of an e15.5 embryo (A) and a p4 pup (B). The pancreatic tissue is demarcated with a blue line. Please click here to view a larger version of this figure.
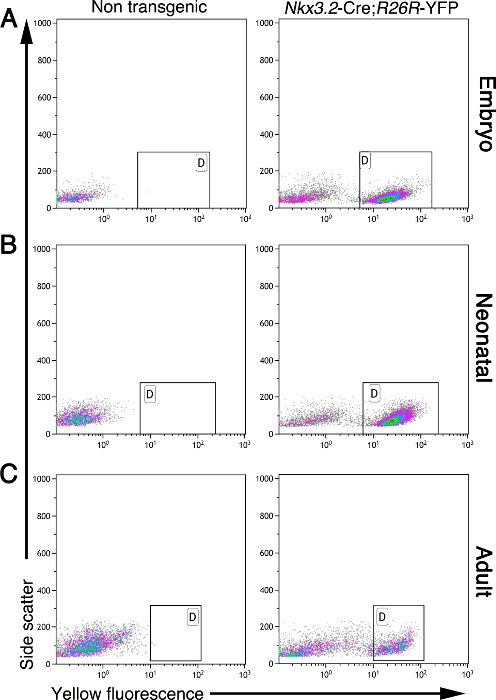 Figure 2: Mesenchymal cells are fluorescently labeled in Nkx3.2-Cre;R26R-YFP pancreatic tissue. Flow-cytometry analysis of pancreatic cells isolated from non-transgenic (left panels) and Nkx3.2-Cre;R26R-YFP transgenic (right panels) mice at various ages: embryonic (A), neonatal (B), and adult (C). Cells were analyzed for side scatter (y-axis) and yellow fluorescence (x-axis). Gates (marked with "D") indicated the presence of a YFP+ cell population in transgenic mice but not in non-transgenic controls. Please click here to view a larger version of this figure.
Figure 2: Mesenchymal cells are fluorescently labeled in Nkx3.2-Cre;R26R-YFP pancreatic tissue. Flow-cytometry analysis of pancreatic cells isolated from non-transgenic (left panels) and Nkx3.2-Cre;R26R-YFP transgenic (right panels) mice at various ages: embryonic (A), neonatal (B), and adult (C). Cells were analyzed for side scatter (y-axis) and yellow fluorescence (x-axis). Gates (marked with "D") indicated the presence of a YFP+ cell population in transgenic mice but not in non-transgenic controls. Please click here to view a larger version of this figure.
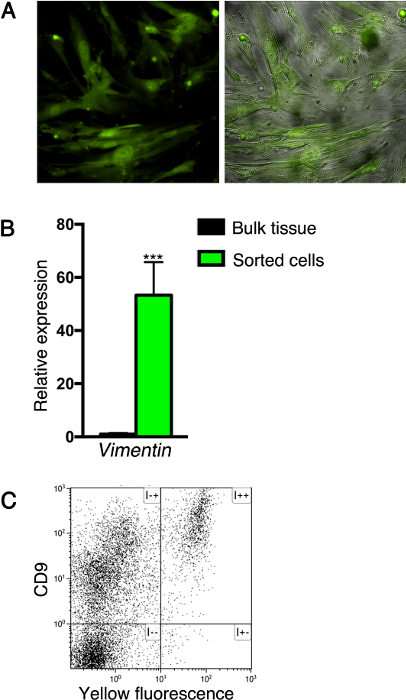 Figure 3: Analyses of isolated pancreatic cells. (A) Cells sorted from the pancreatic tissue of Nkx3.2-Cre;R26R-YFP neonatal mice (as described in Figure 2B) were cultured to establish a cell line. Cultured cells were imaged for fluorescence (green; right and left panels) and phase contrast (gray; right panel). (B) Bar diagram showing Vimentin1 (Vim1) expression levels by YFP+-sorted cells from the pancreatic tissue of Nkx3.2-Cre;R26R-YFP adult mice (as described in Figure 1C; green) as compared to unsorted pancreatic tissue (black). RNA was extracted and gene expression was analyzed by qPCR; expression was normalized to Cyclophilin. N = 4. ***P < 0.001. Data represent the mean ± SD. (C) Flow-cytometry analysis of dispersed pancreatic cells from Nkx3.2-Cre;R26R-YFP adult mice immuno-stained with allophycocyanin (APC)-conjugated anti-CD9 antibody and analyzed for APC (y-axis) and yellow fluorescence (x-axis). Please click here to view a larger version of this figure.
Figure 3: Analyses of isolated pancreatic cells. (A) Cells sorted from the pancreatic tissue of Nkx3.2-Cre;R26R-YFP neonatal mice (as described in Figure 2B) were cultured to establish a cell line. Cultured cells were imaged for fluorescence (green; right and left panels) and phase contrast (gray; right panel). (B) Bar diagram showing Vimentin1 (Vim1) expression levels by YFP+-sorted cells from the pancreatic tissue of Nkx3.2-Cre;R26R-YFP adult mice (as described in Figure 1C; green) as compared to unsorted pancreatic tissue (black). RNA was extracted and gene expression was analyzed by qPCR; expression was normalized to Cyclophilin. N = 4. ***P < 0.001. Data represent the mean ± SD. (C) Flow-cytometry analysis of dispersed pancreatic cells from Nkx3.2-Cre;R26R-YFP adult mice immuno-stained with allophycocyanin (APC)-conjugated anti-CD9 antibody and analyzed for APC (y-axis) and yellow fluorescence (x-axis). Please click here to view a larger version of this figure.
Discussion
Here, we describe a method to isolate and analyze cells of the pancreatic microenvironment. This method can be used to isolate mesenchymal cells from embryonic and adult pancreatic tissue. In addition, we successfully used this protocol to isolate endothelial cells from the adult and neonatal pancreas5,17. However, it may not be suitable for obtaining a reproducible single-cell suspension of pancreatic epithelial cells (alternative protocols are described in References 18, 23, and 24). Using this method, fluorescently-labeled cells, either expressing fluorescent proteins or immunostained for surface markers, can be purified by FACS or analyzed by flow cytometry. RNA can be extracted from purified cells to profile their gene expression pattern. Alternatively, purified cells can be cultured to establish a cell line for subsequent proteomic analysis. This method will enable the characterization of factors expressed by the pancreas microenvironment, which govern its organogenesis, physiology, and pathophysiology.
The pancreatic mesenchyme supports tissue organogenesis by promoting the proliferation of precursors and differentiated cells5,9. These cells were shown to support the expansion of human embryonic stem cell (hESC)-derived pancreatic progenitors17,25,26. Therefore, delineating the identity of embryonic mesenchymal factors would facilitate current efforts to generate insulin-producing beta cells from hESCs and induced pluripotent stem cells (iPSCs) as a potential cure to diabetes. Mouse genetic studies allowed the identification of growth factors, such as Fgf10, that are produced by the mesenchyme to promote pancreatic epithelium expansion during the early stages of pancreas development3,9. With the aim of identifying additional factors expressed in the embryonic mesenchyme, we isolated these cells using laser-captured microdissection, extracted their RNA, and performed gene expression analysis26. However, in addition to being labor-intense, this method relies on identifying cells based on their morphological features, which restricts its use to developmental stages prior to the branching of the epithelium into the surrounding mesenchyme (i.e., e12.5). To characterize mesenchymal cells at later developmental stages, we employed the method described here5,17.
We used this method to analyze surface marker expression by neonatal pancreatic mesenchyme5. In addition, mesenchymal cells were isolated from embryonic and neonatal pancreatic tissue of Nkx3.2-Cre;R26-EYFP mice, based on their fluorescent labeling in this mouse line, and were cultured to establish cell lines17. The proteomic analysis of these cells allowed for the identification of factors secreted by the pancreatic mesenchyme with the ability to promote hESC-derived pancreatic progenitors17. We further used this cell isolation method to purify mesenchymal cells from adult pancreatic tissues for RNA extraction and gene expression analysis17. Therefore, this method can be used to identify genes and proteins expressed by the pancreatic mesenchyme, with the ability to support pancreatic cell development.
Pancreatic mesenchymal cells were further shown to play a role in pancreas tumorigenesis. PDAC is characterized by the formation of a fibroblast-rich desmoplastic stroma comprised of fibroblasts, immune cells, and ECM27. While the stroma was thought to promote the development of many types of cancer, it was shown to restrain PDAC progression15,16,28. This suggests that components of the pancreatic stroma secrete factors that inhibit tumorigenesis. Furthermore, changes in stroma cellular composition as well as in cell phenotype can underlie their effect on epithelial cells15,16,28. The method described here can therefore assist in characterizing the different cell types that make up a PDAC stroma as compared to healthy pancreatic tissue. It would further allow the purification of the different stromal cell types to characterize potential changes in their gene expression profiles during PDAC progression. However, due to changes in pancreatic ECM composition during tumorigenesis27, adjustments of the tissue digestion parameters, such as the inclusion of additional collagenase types or increasing the incubation time, may be required.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors thank Adi Sasson for the technical assistance and Helen Guez for the critical reading of the manuscript. This work was supported by European Research Council starting grant no. 336204.
References
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren P-O, Caicedo A. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes GK. Developmental biology of the pancreas: a comprehensive review. Developmental biology. 2009;326(1):4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Villasenor A, Cleaver O. Crosstalk between the developing pancreas and its blood vessels: An evolving dialog. Seminars in cell & developmental biology. 2012. pp. 1–8. [DOI] [PMC free article] [PubMed]
- Landsman L, Nijagal A, et al. Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS Biology. 2011;9(9):e1001143. doi: 10.1371/journal.pbio.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294(5542):564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Pierreux CE, Cordi S, et al. Epithelial: Endothelial cross-talk regulates exocrine differentiation in developing pancreas. Developmental biology. 2010;347(1):216–227. doi: 10.1016/j.ydbio.2010.08.024. [DOI] [PubMed] [Google Scholar]
- Golosow N, Grobstein C. Epitheliomesenchymal interaction in pancreatic morphogenesis. Developmental biology. 1962;4:242–255. doi: 10.1016/0012-1606(62)90042-8. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, et al. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128(24):5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- Larsen BM, Hrycaj SM, Newman M, Li Y, Wellik DM. Mesenchymal Hox6 function is required for mouse pancreatic endocrine cell differentiation. Development. 2015;142(22):3859–3868. doi: 10.1242/dev.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasson A, Rachi E, et al. Islet pericytes are required for beta-cell maturity. Diabetes. 2016;65(10):3008–3014. doi: 10.2337/db16-0365. [DOI] [PubMed] [Google Scholar]
- Stanley SA, Kelly L, et al. Bidirectional electromagnetic control of the hypothalamus regulates feeding and metabolism. Nature. 2016;531(7596):647–650. doi: 10.1038/nature17183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissova M, Aamodt K, et al. Islet microenvironment, modulated by vascular endothelial growth factor-A signaling, promotes β cell regeneration. Cell metabolism. 2014;19(3):498–511. doi: 10.1016/j.cmet.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova G, Jabs N, et al. The vascular basement membrane: a niche for insulin gene expression and Beta cell proliferation. Developmental cell. 2006;10(3):397–405. doi: 10.1016/j.devcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Rhim AD, Oberstein PE, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer cell. 2014;25(6):735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özdemir BC, Pentcheva-Hoang T, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer cell. 2014;25(6):719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ HA, Landsman L, et al. Dynamic Proteomic Analysis of Pancreatic Mesenchyme Reveals Novel Factors That Enhance Human Embryonic Stem Cell to Pancreatic Cell Differentiation. Stem cells international. 2016;2016:6183562. doi: 10.1155/2016/6183562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout J, Pommier RM, et al. Isolation and Culture of Mouse Primary Pancreatic Acinar Cells. Journal of Visualized Experiments. 2013. p. e50514. [DOI] [PMC free article] [PubMed]
- Verzi MP, Stanfel MN, et al. Role of the homeodomain transcription factor Bapx1 in mouse distal stomach development. Gastroenterology. 2009;136(5):1701–1710. doi: 10.1053/j.gastro.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribioli C, Frasch M, Lufkin T. Bapx1: an evolutionary conserved homologue of the Drosophila bagpipe homeobox gene is expressed in splanchnic mesoderm and the embryonic skeleton. Mech Dev. 1997;65(1-2):145–162. doi: 10.1016/s0925-4773(97)00067-1. [DOI] [PubMed] [Google Scholar]
- Hecksher-Sorensen J, Watson R, et al. The splanchnic mesodermal plate directs spleen and pancreatic laterality, and is regulated by Bapx1/Nkx3.2. Development. 2004;131(19):4665–4675. doi: 10.1242/dev.01364. [DOI] [PubMed] [Google Scholar]
- Walmsley GG, Rinkevich Y, Hu MS. Live Fibroblast Harvest Reveals Surface Marker Shift In Vitro. Tissue Engineering Part C: Methods. 2014. [DOI] [PMC free article] [PubMed]
- Morris JP, Greer R, et al. Dicer regulates differentiation and viability during mouse pancreatic cancer initiation. PloS one. 2014;9(5):95486. doi: 10.1371/journal.pone.0095486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatsuka T, Matsuoka T-A, et al. Chronological analysis with fluorescent timer reveals unique features of newly generated β-cells. Diabetes. 2014;63(10):3388–3393. doi: 10.2337/db13-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon JB, Borowiak M, Melton DA. Self-renewal of embryonic-stem-cell-derived progenitors by organ-matched mesenchyme. Nature. 2012;491(7426):765–768. doi: 10.1038/nature11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Landsman L, Li N, Hebrok M. Factors expressed by murine embryonic pancreatic mesenchyme enhance generation of insulin-producing cells from hESCs. Diabetes. 2013;62(5):1581–1592. doi: 10.2337/db12-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz Y, Erez N. An inflammatory vicious cycle: Fibroblasts and immune cell recruitment in cancer. Experimental cell research. 2013;319(11):1596–1603. doi: 10.1016/j.yexcr.2013.03.022. [DOI] [PubMed] [Google Scholar]


