Abstract
A low-density, high-resolution diagnostic DNA microarray comprising 38 gene targets for 13 viral causes of meningitis and encephalitis was constructed. The array has been used for the detection of multiplex PCR-amplified viruses in cerebrospinal fluid (CSF) and non-CSF specimens. A total of 41 clinical specimens were positive for echoviruses (23 samples), herpes simplex virus type 2 (4 samples), varicella-zoster virus (4 samples), human herpesvirus 7 (1 sample), human herpesvirus 6A (1 sample) and 6B (2 samples), Epstein-Barr virus (three samples), polyomavirus JC (1 sample), and cytomegalovirus (2 samples). Probes for herpes simplex virus type 1, polyomavirus BK, and mumps and measles viruses were also included on the array. Three samples were false negative by the microarray assay due to discordant results between the multiplex PCR for all 13 viruses simultaneously and the virus-specific PCR alone. Fifteen CSF specimens were true negative. The clinical sensitivity, specificity, and negative and positive predictive values of the assay were 93, 100, 100, and 83%, respectively, when the results were compared to those of the single-virus PCR, which was used as the “gold standard.” The microarray-based virus detection assay is qualitative and provides a single-format diagnostic tool for the detection of panviral CNS infections.
Establishment of the precise etiologies of infective central nervous system (CNS) syndromes, meningitis and encephalitis, has always required the assistance of laboratory techniques. The pathogens responsible for such syndromes must be identified, as organism identification enables more precise treatment to be given, a prognosis to be made, and public health measures to be instituted in a timely and targeted manner and also allows the most cost-effective use of hospital resources. Indeed, a sensitive panviral PCR assay should be able to act as an early-warning test system for the detection of emerging viruses. For example, earlier detection of cases of West Nile fever, seen recently as a cause of encephalitis outbreaks in the United States, would allow vector control measures to begin at an earlier stage.
Despite the introduction of diagnostic PCR in routine laboratories, only a fraction of the potential pathogens can be identified in patients with suspected infective CNS syndromes (for a review, see reference 8). A recent audit at St. George's Hospital has shown that only 45% of physician-diagnosed cases of meningitis or encephalitis were confirmed by a laboratory diagnosis, comparable to figures reported by others (43%) (17). The principal reasons for this failure are the large number of potential pathogens involved and the small volume of the cerebrospinal fluid (CSF) sample, which must often be divided up for several analyses, often at different laboratories. A multiplex PCR approach has overcome the diagnostic limitations of multiple PCRs for the detection of individual neuroinvasive viruses (6, 8, 14, 18). However, even multiplex PCR allows only a few potential pathogens (about three to six) to be targeted at any one time. The amplification of a low-abundance pathogen by multiplex PCR often results in spurious bands by gel electrophoresis (5) and requires validation by hybridization to specific probes immobilized on membranes or microwell plates. These hybridization formats have limited capacities.
A technique capable of overcoming these limitations and increasing the diagnostic output of the assay is hybridization on a DNA microarray. Due to its unrestricted capacity to accommodate hundreds to thousands of individual gene probes, an array allows the simultaneous detection of potentially any amplifiable pathogen present in a specimen. As a result, this technology is ideal for the extensive parallel identification and differentiation of various pathogens and their strains. The technique is rapidly evolving from a novel research technology (10) to a practical tool for the identification of bacterial species (5, 11), the detection of environmental bacteria (24), and the genotyping of influenza viruses (13) and human immunodeficiency virus (HIV) type 1 (23). However, the microarray approach has not yet been extended to the clinical diagnosis of viral infections.
We have applied the DNA microarray technology to the identification of specific pathogens in cases of meningitis and encephalitis. We focused on developing a microarray for the detection not only of the most common neurotropic viruses in the United Kingdom (herpes simplex virus [HSV], varicella-zoster virus [VZV], and enteroviruses) (18) but also for the detection of several other viruses capable of causing CNS syndromes in very young and immunocompromised individuals. The greatest difficulties in clinical management are determining whether the etiology is bacterial or viral in an individual patient. However, we decided to concentrate on viral pathogens because we already had prospectively well characterized patients and cerebrospinal fluid (CSF) samples from patients with CNS infection for which the causative viral pathogen was confirmed by standard techniques and because the majority of infective CNS syndromes result from virus infections. Once the array was constructed, viral etiologic agent could be identified from a panel of 13 different neurotropic viral agents.
MATERIALS AND METHODS
Viruses, cells, and clinical specimens.
Cytomegalovirus (CMV; laboratory strain AD169), Epstein-Barr virus (EBV; transformed B95a cell line), and VZV and echovirus (clinical isolates) were available from the virology laboratory of St. George's Hospital. Cloned sequences of HSV type 1 (HSV-1) and HSV-2, human herpesvirus 7 (HHV-7), HHV-6A, HHV-B, and cultures of polyomaviruses BK and JC and mumps and measles viruses were kindly supplied by the Health Protection Agency (Colindale, London, United Kingdom). Coxsackie B virus was in the form of a purified PCR DNA fragment from a previous study (12). Primary human embryo lung (HEL) cells were used for virus isolation and titration and were maintained as described previously (21). The enteroviruses were cultured from CSF specimens on HEL cells and were identified by immunofluorescent microscopy with a screening set of enterovirus group- and type-specific monoclonal antibodies (Chemicon International Inc., Temecula, Calif.).
CSF samples from patients with viral meningitis diagnosed by virus culture and/or PCR were collected retrospectively over the past 2 years and were stored at −70°C. The laboratory worker who processed the specimens by PCR-microarray analysis was blinded to the clinical diagnoses and the viral agent.
Nucleic acid extraction from clinical specimens.
Every CSF specimen was divided in two, and each half was processed separately for DNA and RNA viruses. CSF volumes of 200 μl were used. If the volume was less (30 to 100 μl), it was adjusted to 200 μl with phosphate-buffered saline. These samples were supplemented with 10 μg of carrier tRNA (Sigma-Aldrich, Poole, United Kingdom). DNA was extracted by use of a QIAamp DNA blood kit (Qiagen, Crawley, United Kingdom) and RNA was extracted by use of the Trizol LS reagent (Invitrogen, Paisley, United Kingdom), according to the directions of each manufacturer. The DNA was eluted in 50 μl of nuclease-free water and was stored at −20°C for repeated use. The total RNA from each specimen was reverse transcribed, and the cDNA was stored at −70°C.
Primers and multiplex PCR amplification.
Unique pairs of gene-specific primers were designed for each viral target on the basis of a full search of the GenBank database (www.ncbi.nlm.nih.gov) with the BLAST program for known virus sequences. To ensure a nearly equal annealing efficiency, all primers were 22-mers and had similar melting temperatures (58.1 ± 1.6°C) and G+C contents (44.6% ± 3.8%). In order to prevent cross-hybridization, the target viral sequences (range, one to five per viral genome) were selected so that they represented unique regions with little or no similarity with the sequences deposited in the GenBank database. The primer sequences and their target locations within viral genomes are listed in Table 1. Three pairs of primers were designed for each virus to increase the probability of virus detection (Table 1). For enteroviral genome detection we used only one pair of primers, E1 and E2, whose sequences flank a 201-bp product within the conserved 5′ nontranslated region of the genome (4) (Table 1). In the case of CMV, three newly designed primer pairs were supplemented with two known pairs already in use; the last two primer pairs (CMV4 and CMV5 in Table 1) target the late ULIIIA region and the early-late UL21.5 region of the CMV genome, respectively, and have previously been demonstrated to have high sensitivities and specificities for the detection of CMV by PCR (3). One of three pairs of primers designed to detect both HSV-2 and HHV-6B failed to amplify the viruses when they were used with positive control material and were excluded from the study, leaving only two primer sets for the detection of these viruses.
TABLE 1.
Sequences and target locations of primers used in this study
| Virus (element)a | Sequence (5′-3′)
|
PCR fragment (Size in bp [nucleotide position within genome]) | GenBank accession no. | |
|---|---|---|---|---|
| Forward primer | Reverse primer | |||
| HHV 6B (1) | TGCTCAATGACATAACAGTCCC | GACATCTGTTCGGGGTTTAGAG | 744 (47581-48325) | NC 000898 |
| HHV-6B (2) | ATCATCACAAACATCGTCTTCG | CAGTTTAATTGGCCGAAAAGTC | 708 (26506-27214) | |
| EBV (1) | GGCCTGTCATTGAGGTAAAAAG | AGCACTACGTGCTTGAGGTACA | 483 (140018-140501) | NC 001345 |
| EBV (2) | TGATATCCACAGAAGTGCCAAC | TCAGGCAAAATAACCTGTGATG | 744 (165737-166481) | |
| EBV (3) | TGTGAACCTTATGGAGATGTGC | CACTGGAAATGCTTGGTCAATA | 443 (116271-116714) | |
| HSV-1 (1) | AAAAACGACCAAAGGAACAGAA | CTTGGCCTCCAATGAACTAAAC | 488 (41274-41762) | NC 001806 |
| HSV-1 (2) | TCCTTCTTGTCGTCTCTCTTCC | GGTGGTTTGTTCATCCTATGGT | 360 (143607-143967) | |
| HSV-1 (3) | AAAATTAAGGCGTGATCTCCAA | CGTTTGTCGTTATTGATCTGGA | 777 (116042-116819) | |
| CMV (1) | ATCACGATGAGTTTTCTCCGTT | CCTTTGAAAACCAAGCCTATTG | 518 (157758-158276) | X 17403 |
| CMV (2) | TTTAAATCACAAAGAACCGCCT | AAAACCCCGTTATGATTACACG | 602 (90840-91442) | |
| CMV (3) | ATACCTTTCGAGGACGAAAACA | ATAATGGAGCTTTGCTGATGGT | 795 (143979-144774) | |
| CMV (4) | CTATGGATCTTGAGCTTACT | TCGCGCCATCTCCGTCTGT | 258 (27120-27378) | |
| CMV (5) | CTGTCGGTGATGGTCTCTTC | CCCGACACGCGGAAAAGAAA | 234 (159681-159914) | |
| VZV (1) | GACGGCCATTTTAGTATCAAGC | TCATGTAACATTGGCACTCCTC | 707 (74311-75018) | NC 001348 |
| VZV (2) | TTCATAAACTCGTCACAATGGC | CCGAACATGAGTTACTTCCTCC | 712 (37744-38456) | |
| VZV (3) | CGGGGAGTTAAACAGAGAATTG | CAGGAGTAAATTGGAACTTGGC | 688 (28574-29262) | |
| HSV-2 (1) | TGTCGATCCGTAATTGTTTCTG | TCTATACCTCTTTCTGAGCCGC | 441 (104645-105086) | NC 001798 |
| HSV-2 (2) | CTACTATCTCATCCACCGGGAG | TGGGATATCTCACACAGACTGG | 682 (43126-43808) | |
| HHV-6A (1) | ACATGATGAGACAGCATTTTGG | GGTGGAGGTAGCAATTTTCTTG | 679 (47788-48467) | X 92436 |
| HHV-6A (2) | GGAATTGCAGTTAGGTTTCCTG | GAAAGCCACATCTACATCACCA | 318 (128271-128589) | |
| HHV-6A (3) | GTCTAGAACTCCACCACGATCC | TTGACTATGTTTCGTCTCCCCT | 298 (156151-156449) | |
| JC (1) | TATACTCATGTGGGAGGCTGTG | TACTTGAGCATCCATGCCATAC | 666 (1765-2431) | AB 048576 |
| JC (2) | GATTCCCATTCATCTGTTCCAT | TATTGCAAGGAATGGCCTAACT | 339 (4395-4734) | |
| JC (3) | GTCCTTGTTCTCCACAATCTCC | CAACTTGCCTTACCATAGAGGG | 235 (990-1225) | |
| HHV-7 (1) | CAAAGTGTGCCAAATTCGATAA | TTTTGCGTTCCTGTGATTTATG | 539 (28937-29476) | NC 001716 |
| HHV-7 (2) | CCTTACTGCCCAGTTTCATTTC | TATTTTGTGGTTGCGAAGAATG | 428 (10961-11389) | |
| HHV-7 (3) | TGTAGAATTGGCAATGTTTTCG | ATGGGCTGTTTTAGTGGAGAAA | 671 (9490-10161) | |
| BK (1) | ACTTGGGAAGAGCATTGTGATT | CAGGCTTGGTTACAATGAATGA | 396 (2840-3236) | NC 001538 |
| BK (2) | AATGCAGGGAGTGCTAATGAAT | TCAGACCCCATAGTCAAGAGT | 756 (2046-2802) | |
| BK (3) | TCACAGGAATTGCAGAGAAGAA | ACAGCCTCCCACATCAGTAGAT | 749 (1155-1904) | |
| Measles (1) | ACTGGAAATCATTTGCTGGAGT | CATGCCCTCATTTTGTAAGTCA | 351 (10549-10900) | AF 266288 |
| Measles (2) | TGACATACATCTGACAGGGGAG | TGATTTCCAGTTATCAACGCAC | 303 (10256-10559) | |
| Measles (3) | TTTGATCTCTCTTGGCTTCACA | TGGACAATCTGGTTATCACTCG | 613 (11933-12546) | |
| Mumps (1) | ATTTCTTGTCTGTGCCTGGAAT | TGATGGTCAATCTGGTTAGCAC | 610 (7747-8357) | NC 002200 |
| Mumps (2) | AATCCTCAAATGAAGCAGGTGT | TCAGCATATGTTGTGGGTCTTC | 531 (3051-3582) | |
| Mumps (3) | GTGCTAACCAGATTGACCATCA | ATATGCTTGCAGGATTGGAAGT | 404 (8336-8740) | |
| Enteroviruses | TCCTCCGGCCCCTGAATG | CACCGGATGGCCAATCCA | 201 (192-393) | ECV7337 |
JC, polyomavirus JC; BK, polyomavirus BK.
PCR was performed in triplicate with 30-μl volumes by using a set of Platinum Taq DNA polymerase reagents (Invitrogen) in the recommended proportions, except that the concentration of each primer was optimized at 2 μM. For the reverse transcription (RT) step of RT-PCR we used the Superscript II set of reagents (Invitrogen), according to the instructions of the manufacturer, and a mixture of 1 μM each eight reverse primers for RNA viruses. A single round of PCR amplification was performed in a Mastercycle Gradient thermocycler (Eppendorf, Hamburg, Germany) and entailed enzyme activation at 95°C for 2 min, followed by 45 cycles of denaturation (95°C, 1 min), annealing (60°C, 30 s), and extension (72°C, 1 min), with a final 10-min extension at 72°C. Each PCR run included two negative no-template controls that, after PCR, were combined for labeling and microarray hybridization. The viruses infecting the retrospectively analyzed CSF samples (n = 28) had previously been determined at Micropathology Ltd. (Coventry, United Kingdom) by PCR screening assays (18). Both prospective samples, i.e., from patients for whom there was not a previous diagnosis (n = 32), and retrospective samples were subjected to a multiplex PCR which incorporated 30 pairs of primers for DNA viruses or 7 pairs of primers for RNA viruses. After virus detection by our multiplex PCR and microarray assay, virus was amplified from the same specimen by virus-specific PCR with a mixture of only virus-specific primers, followed by array hybridization, in order to compare the two methods. We found that gel analysis of the multiplex PCR-amplified products was not helpful because for samples with a low abundance of the target, specific bands were difficult to detect among the multiple target-unrelated bands. Therefore, all PCR-amplified products from specimens were directly hybridized to the microarray without prior electrophoresis. The viruses detected by PCR in the prospective samples by Micropathology Ltd. were disclosed only after the multiplex PCR and microarray results were obtained at the Department of Medical Microbiology, St. George's Hospital Medical School.
Precautions to prevent contamination included spatial separation of pre- and post-PCR steps, the use of filter pipette tips, and regular treatment of surfaces and pipettes with DNA-destroying agents. Once a sample was found to be virus positive by the multiplex PCR and microarray assay, the extracted stored DNA or cDNA was reamplified with virus-specific primers and the purified PCR bands were validated by direct dideoxy sequencing with a USB Sequenase (version 2.0) kit (Amersham Biosciences).
Microarray production.
Microarrays were printed on polylysine-coated glass microscope slides with a contact microspotting robotic system (MicroGrid II; BioRobotics, Cambridge, United Kingdom) by using split pins. The average spot size was 150 to 200 μm. The printed slides were then processed by chemically blocking the unreacted lysine groups by a method previously described for use with CMT-GAPS (Corning Ltd., High Wycombe, United Kingdom)-coated slides (7). The probe elements on the array were made from gel-purified PCR products generated from the viral DNA or RNA template (see above) by using the same gene-specific primer pair sequences (Table 1) used for multiplex PCR of the clinical samples. PCR products were suspended in 50% dimethyl sulfoxide at a DNA concentration of 0.5 μg/μl. Printed slides were stored dry in lightproof protective boxes until use.
Array format.
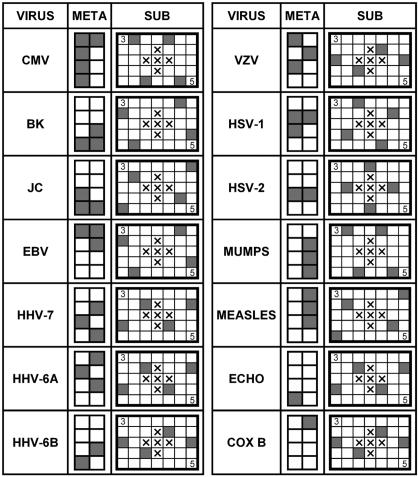
Each array contained identical sets of probes spotted in quadruplicate (for signal averaging) and arranged in a virus-specific subgrid format (Fig. 1). The subgrids were composed of probes spotted in a pattern of seven by five spots; these were combined into larger groups of eight to form a metagrid. Each subgrid had a pattern characteristic for each virus (Fig. 1). To enable positive virus identification, the individual subgrid must have the correct spot pattern and the location of the subgrid within the metagrid template must be correct. Each subgrid had “landing-light” spots (top left and bottom right) made with cyanine 3 (Cy3)- or Cy5-labeled nucleotides to mark the array orientation (Fig. 1). The center five spot locations were left blank. The slides were analyzed in an Affymetrix (Santa Clara, Calif.) 428 array scanner with the aid of ImaGene (BioDiscovery, Marina del Rey, Calif.) software (version 4.2). Two fluorescent images of the same array for alternatively labeled hybridization control (Cy5 detection at 670 nm) and sample (Cy3 detection at 570 nm) channels were first examined on their own and were then examined as a superimposed image of the two to produce a false two-color (red-green) image. For positive samples, the resulting signal formed one of the expected spot patterns depicted in Fig. 1.
FIG. 1.
Individual virus patterns on diagnostic DNA microarray. Each probe is printed on a subgrid in quadruplicate and forms a distinctive pattern, as shown by shaded squares. In turn, subgrids are arranged in larger units, metagrids, which also create a unique virus-specific pattern on a slide. The crosses in the middle of each subgrid indicate blank spaces. To mark the array orientation, each subgrid has landing-light spots, made with nucleotides labeled with Cy3 (3, top left spots) or Cy5 (5, bottom right spots). COX B, coxsackie B virus.
Target labeling and microarray hybridization.
The products from the DNA PCRs and RT-PCRs performed with the same specimen were combined and copurified with a QIAquick PCR purification kit (Qiagen). The purified PCR products were labeled with a random primer labeled with Cy3 dCTP. Each 50-μl reaction mixture contained 3 μg of random primers; 5 μl of 10× reaction buffer; 5 mM each dATP, dGTP, and dTTP; 2 mM dCTP; and 5 U of the large DNA polymerase I (Klenow) fragment (all reagents were from Invitrogen) plus 1.5 μl of fluorescently labeled Cy5 dCTP (control target) or Cy3 dCTP (sample target). The labeled deoxynucleotides were from Amersham Biosciences (Little Chalfont, United Kingdom). As a control for every experiment we used 10 ng of a mixed viral PCR product as a probe, which was suspended in 50% dimethyl sulfoxide and simultaneously labeled with Cy5 dCTP. After labeling of the product, both control and sample DNAs were pooled, copurified with a MinElute PCR purification kit (Qiagen), added to 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.3% sodium dodecyl sulfate in a 16-μl volume, denatured at 95°C for 2 min, and hybridized to the array overnight in a waterproof hybridization chamber (Corning). Posthybridization washes were performed in 1× SSC buffer with 0.06% sodium dodecyl sulfate for 2 min at 65°C and then in 0.06× SSC buffer (twice). Detailed protocols of the labeling and hybridization procedures have been published elsewhere (7).
Virus detection.
The results were considered positive for virus detection when the mean hybridization signal intensity from the quadruplicate virus-specific spotted elements (probes) on the array was three times greater than the background signal intensity in one or more of the subgrids. The background signal intensity was defined as the mean signal intensity for all the nonspecific probes on the array minus the background signal intensity for the local spot. The local background signal intensity around the nonspotted area of each spot was automatically subtracted from the spot signal intensity by image acquisition software (version 4.1; ImaGene BioDiscovery). Each experiment was performed as a cohybridization between an internal hybridization control sample (prepared from a mixture of all virus-specific PCR products spotted on the array) and the multiplex PCR-amplified test sample. Control and test samples were separately labeled with the fluorescent dye conjugate Cy5 (indicated here as computer-generated false color red) or Cy3 (indicated here as computer-generated false color green), respectively. The amount of control DNA was titrated to 10 ng so that each spot on the array gave a sufficiently intense signal for spot detection (red) while still allowing the test viral PCR products to hybridize, thus producing a green color after quantitation and computer superimposition of the signal intensity ratios from the two channels. Thus, positive hybridization signals (visualized here as green) could be determined by eye from the correct patterns of spots in both the subgrid and the metagrid locations that corresponded to the target virus.
To guard against slide-to-slide and batch-to-batch variations in signal intensities, the Cy3 and Cy5 images were scanned at the same laser power and saved as eight-bit TIFF files. The mean fluorescent signal intensity was measured for each of quadruplicate Cy3-labeled sample spots or Cy5-labeled control spots with ImaGene Biodiscovery software. Statistical analysis was performed with Axum (version 6.0) software (MathSoft, Inc.).
PCR-microarray assay sensitivity.
The sensitivity of our multiplex PCR-microarray assay was determined by using CMV and echovirus as representatives of the DNA and RNA virus groups, respectively. The CMV and echovirus stocks, both of which were adapted to HEL cells, were serially diluted 10-fold and titrated on replicate HEL cell cultures. The end points were determined by observing a clearly visible cytopathic effect 5 days postinfection in echovirus-infected replicate cell cultures or by immunoenzymatic staining of CMV-infected cultures (21). The RNA or DNA was extracted from 1 ml of cell culture supernatant, which was collected from each dilution in the virus titration experiments and clarified by centrifugation at 14,000 × g for 15 min and then processed by multiplex PCR. After amplification the products from replicate PCRs for each virus dilution were pooled, purified, and divided in two. One half was run on 6% polyacrylamide minigels, and the other half was randomly labeled and hybridized to the microarray, followed by comparison of end-point detection by either method. The CMV DNA copy number in the end-point dilution was estimated by a semiquantitative PCR with primers specific for the ULIIIA region (CMV5 in Table 1) and an internal DNA mimic standard (2). For echovirus RNA quantification, the purified 201-bp PCR DNA fragment was ligated to the T7 RNA polymerase promoter by use of the Lig'nScribe kit (Ambion, Austin, Tex.), and runoff RNA transcripts were produced with an in vitro transcription kit (Epicentre Technologies, Madison, Wis.), according to the instructions of each manufacturer. The RNA product was digested with 2 U of RNase-free DNase I (Ambion) and was quantified by measuring its optical density. A stock preparation of the transcript RNA (1.25 μg/μl) was serially diluted 10-fold in RNase-free water, and each dilution was processed by RT-PCR with a mixture of RNA virus-specific primers, followed by simultaneous gel and microarray hybridization analysis.
The clinical sensitivity, specificity, and positive and negative predictive values of the multiplex PCR were determined by comparison of the multiplex PCR results with the results of the virus-specific PCR and were calculated by using conventional formulas.
RESULTS
We have constructed a glass slide DNA microarray consisting of virus-specific PCR products from 13 neurotropic viruses commonly found in the United Kingdom in both previously healthy and immunosuppressed people. We termed the combination of virus genome amplification (by using a mixture of primers for each of the probe elements on the array) and hybridization to the array the “multiplex PCR-microarray assay.” We have applied this assay to clinical samples for both the prospective and the retrospective diagnosis of meningitis and encephalitis to investigate its diagnostic utility.
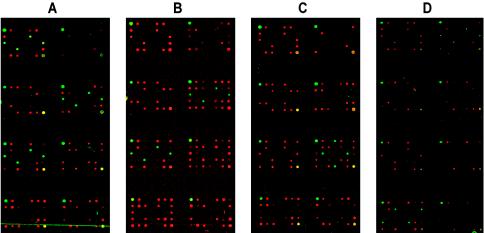
A total of 60 clinical specimens from patients with clinically suspected cases of meningitis or encephalitis were processed by the diagnostic multiplex PCR-microarray procedure. Of the 60 clinical specimens test, 41 gave patterns for a specific virus on the array, including patterns for echoviruses (23 samples), HSV-2 (4 samples), VZV (4 samples), HHV-7 (1 sample), HHV-6A (1 sample), HHV-6B (2 samples), EBV (3 samples), polyomavirus JC (1 sample), and CMV (2 samples). Three samples had false-negative results (1 sample was positive for VZV, 1 was positive for HSV-2, and 1 was positive for measles virus; Table 2), and 15 had true-negative results. The samples with true-negative results comprised 1 sample from a patient with Lyme disease, 1 sample from a patient with serologically proven primary EBV infection and Guillain-Barré syndrome, and 13 samples from patients with bacterial meningitis and other neurological conditions of an ultimately noninfective etiology. A representative example of positive detection by two-color image analysis is shown in Fig. 2A for VZV, in which the quadruplicate green spots are in the correct positions within the subgrid and in the correct three subgrids of the metagrid, as predicted from the schematic in Fig. 1. Samples with positive results with two of three probes for a separate VZV-infected CSF specimen are also shown in Fig. 2B, in which one subgrid is negative for the virus-specific probe element contained within it.
TABLE 2.
Clinical diagnoses, viruses detected, and multiplex PCR-microarray assay resultsa
| Specimen origin, clinical diagnosis | Virus detected | No. of specimens tested | No. of targets detected/ no. of probes arrayed
|
|
|---|---|---|---|---|
| Multiplex PCR for all viruses arrayed | Virus-specific PCR | |||
| CSF, multifocal leukoencephalopathy in HIV-infected patient | Polyomavirus JC | 1 | 1/3 | 2/3 |
| CSF, CNS lymphoma in HIV-infected patient | EBV | 2 | 1/3 (2)b | 1/3 |
| 1 | 2/3 | 2/3 | ||
| CSF, febrile fits | HHV-6A | 1 | 1/3 | 1/3 |
| CSF, viral meningitis | HHV-6B | 2 | 1/2 (2) | 1/2 |
| CSF, febrile fits | HHV-7 | 1 | 1/3 | 1/3 |
| Vesicular fluid, chickenpox | VZV | 1 | 2/3 | NTc |
| CSF, chickenpox | VZV | 1 | 2/3 | 2/3 |
| CSF, zoster | VZV | 1 | 2/3 | 2/3 |
| CSF, renal transplant | VZV | 1 | 2/3 | NT |
| CSF, leukemia | VZV | 1 | 0/3d | 1/3 |
| CSF, primary genital herpes | HSV-2 | 3 | 1/2 (3) | 1/3 |
| CSF, recurrent genital herpes | HSV-2 | 2 | 0/2 (1) | 1/2 |
| 1/2 (1) | ||||
| CSF, encephalitis in an HIV-infected patient | CMV | 1 | 1/5 | NT |
| Amniotic fluid (congenital infection) | CMV | 1 | 5/5 | NT |
| CSF, viral meningitis | Echovirus | 23 | 1/1 | NT |
Measles virus RNA was not detected in a saliva swab specimen from a patient with a typical infection, and the reference laboratory confirmed measles by PCR in assays with the the same swab specimen (data not shown). The data for two specimens with true-negative results and one specimen with false-negative results are not shown.
Values in parentheses indicate the number of samples that gave the same results.
NT, not tested.
Italics indicate CSF samples that gave false-negative results.
FIG. 2.
Identification of viral pathogens by DNA microarray-hybridization assay-specific patterns by using computer-assisted false two-color imaging. Each virus (except enteroviruses) was probed for more than one target, and at least one target per virus was detected on the array. Virus detection patterns are illustrated for VZV (VZV-infected culture [A] and a CSF specimen [B]); HSV-2 in CSF (C); and enterovirus in CSF (D) showing cross-hybridization between the echovirus- and coxsackie B virus-specific probe elements (top right-hand and bottom left-hand subgrids). The key to reading the array results is given in Fig. 1. The internal control red spots are not seen equally strongly in each panel. This is due to a software-simulated reduction in red-spot intensities in order to enhance the virus-specific green hybridization signal (e.g., panels C and D) and is for presentation purposes only.
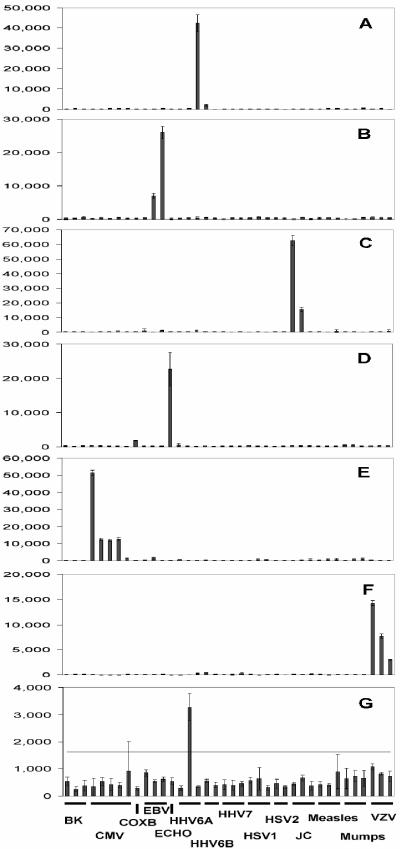
Quantitative analysis of the mean fluorescence signal intensities of quadruplicate virus-specific elements on the array for hybridizations with seven separate clinical samples are presented in Fig. 3 as representative results of this study. The signal intensities for virus-specific hybridizations varied between different samples, as shown by the range of values on the y axes of Fig. 3. We defined the signal cutoff above which a signal was regarded as positive as being greater than a value of three times the mean intensity for all nonspecific elements. The threefold negative cutoff is indicated as a horizontal line in Fig. 3G. In Fig. 3A to F, the threefold cutoff value is not shown, as it is too close to zero on the scale used. The means, standard deviations, and threefold negative control values for each panel are given in the legend to Fig. 3. The weakest signal was obtained for a sample containing HHV-6A (Fig. 3G), for which the maximum signal intensity for the positive element (one from a possible three) was 3,275 and the negative cutoff value was 1,627. The high error bar for CMV-specific probe element 5 in Fig. 3G was due to a nonspecific signal in a single spot and was disregarded. For the other samples (Fig. 3A to F), the positive signal exceeded the negative cutoff values by 10- to 50-fold, thus indicating clear-cut positive results for accurate virus detection. Positive values ranged from 1,410 (CMV-specific probe 5; Fig. 3E) to 62,603 (polyomavirus JC virus probe 1; Fig. 3C) with threefold cutoff values ranging from 247 (VZV; Fig. 3F) to 1,627 (HHV-6A; Fig. 3G), with the mean value for the seven arrays in Fig. 3 being 879.
FIG. 3.
Representative bar charts of the mean (bar) and standard deviations (error bars) of the relative fluorescent signal intensities of quadruplicate virus-specific elements on the array for hybridizations for seven separate clinical samples. (A) HHV-6A; (B) EBV; (C) polyomavirus JC; (D) echovirus (ECHO); (E) CMV; (F) VZV; (G) HHV-6A. Two, three, or five virus-specific elements are indicated along the x axis and are described in detail in Table 1. The threefold negative cutoff (three times the nonspecific signal minus the local background signal) is indicated as a horizontal line in panel G. For panels A to F, the threefold cutoff value is not shown, as it is too close to zero on the scale used. The mean, standard deviation, and threefold cutoff values for each panel are as follows: (A) 147, 94, and 440; (B) 371, 207, and 1,114; (C) 341, 267, and 1,024; (D) 236, 279, and 709; (E) 332, 302, and 499; (F) 82, 103, and 499; (G) 542, 200, and 1,627. COXB, coxsackie B virus.
We obtained positive virus identifications for 41 of 44 infected specimens, with at least one of the viral probes (probe 2, 3, or 5) on the array showing positive hybridization signals (Table 2). The clinical sensitivity of our combined multiplex PCR-microarray assay, determined by comparison with the virus-specific detection rate by PCR with the same specimens, was 93%. The specificity of the assay was calculated to be 100%, and its positive and negative predictive values were 100 and 83%, respectively.
In comparison to PCR amplification only with primers specific for a particular virus followed by microarray hybridization (referred to as virus-specific PCR in Table 2), the multiplex PCR-microarray assay failed to hybridize to any probe on the array for only two samples. These were positive, however, with one of three probes (VZV) or one of two probes (HSV-2) after virus-specific PCR. For two other samples (one containing polyomavirus JC and one containing HSV-2; Table 2), the multiplex PCR-microarray assay detected one less target than the virus-specific PCR did. The virus in one specimen (a throat swab specimen) found to be positive for measles virus at the Health Protection Agency reference laboratories by a single-gene nested PCR assay was not detected by the PCR-microarray assay or virus-specific PCR assay, presumably because of very low viral titers. Thus, the performance of our multiplex PCR-microarray assay was close to that of the standard diagnostic PCR tests used by most clinical microbiology laboratories.
The analytical sensitivity of our assay was estimated for a DNA virus (CMV) and an RNA virus (echovirus) by correlating the PCR end-point signals on the gel and on the microarray by using serial dilutions of viral stocks (see Materials and Methods). The sensitivity of CMV ULIIIA DNA band detection by our polyacrylamide minigel system is 2.5 ng of DNA (2); that compared well with the reported detection limit of 1 ng of randomly labeled DNA detectable by microarray hybridization (24). In terms of infectivity or viral copy numbers, the calculated sensitivity of our multiplex PCR-microarray assay for CMV was 1.6 infectious CMV units, or 46 CMV DNA copies. For the echovirus, the values were 0.01 50% tissue culture infective doses or 28 RNA copies (125 ag), which are comparable to the sensitivities attained by nonnested PCR techniques for these two viruses reported by others (1, 15, 19).
DISCUSSION
In multiplex PCR, problems with primer pair compatibility have previously been reported to result in variable suppression effects on some of the primer pairs, leading to false-negative results (8, 16). However, others have demonstrated detection rates by PCRs with multiplex primers comparable to those achieved by PCRs with single primer pairs (6, 14, 18). Such effects are thought to result from variations in primer binding and amplification efficiencies, the differential accessibilities of the targets within genomes due to their secondary structures, and/or differences in G+C contents (8, 22). To minimize any potential effect of PCR suppression or selection on the diagnostic utility of our multiplex PCR-microarray assay, we introduced into the PCR and microarray design multiple primer sets specific for each virus (except enteroviruses; see Materials and Methods), assuming that at least one primer pair per virus would overcome any interference effect. We found that neither the multiplex PCR for all viruses nor the virus-specific PCR could amplify all possible targets at the same time and also that the targets amplified were not always identical in different samples containing the same virus.
The phenomenon of PCR selection is best illustrated in our study by several examples. In the first, three of five VZV-infected samples were positive with two of three viral probes, namely, probe 1 and probe 2 (VZV1 and VZV2, respectively, in Table 1; Fig. 2B); one sample was positive for probes 1 and 3 (regardless of whether the mixture of all viral primers or only pooled VZV-specific PCR primers was used); and the fifth sample gave no signal on the array after multiplex PCR amplification (the results are summarized in Table 2). However, when this VZV array-negative sample was reamplified with only three pairs of VZV-specific primers, the positive signal on the array was due exclusively to target 3 (VZV3 in Table 1). Another example of PCR selection was seen with five HSV-2-infected specimens, all of which were positive by assays with one of two probes (Table 2). However, three specimens were positive for probe 2 (HSV-2 element 2 in Table 1; Fig. 2C) and two specimens were positive in assays with probe 1 (the results are summarized in Table 2). Similar effects were observed for CMV, as shown in Fig. 3E with four of five CMV-specific probe elements showing strong positive signal intensities of 51,477, 12,376, 11,767, and 12,691, respectively. A weak signal was seen with CMV element 5, with a signal intensity of 1,410, but the signal was still above the threefold cutoff value of 995. The variability in viral targets detected among specimens suggests that, apart from the amplification superiority of some primer pairs, some specimen-related factor(s) might affect the PCR. Indeed, two CMV-infected specimens of different origins showed only a 10-fold difference in viral loads but gave remarkably different scores for the viral targets detected by the multiplex PCR-microarray assay (Table 2)
Although PCR suppression-selection was occurring, its impact on the diagnostic utility of the multiplex PCR-microarray assay was minimized through the inclusion of multiple probes on the array for each virus such that a positive result with any one probe was sufficient for a correct identification. For both retrospective and prospective specimens, the virus detected by the multiplex PCR-microarray assay was verified by virus-specific PCR in the Department of Medical Microbiology, St. George's Hospital, and was independently confirmed by commercial PCR screening assays (18).
The majority of meningitis cases in this study (56%) were caused by enteroviruses. This was not unexpected, as their prevalence over other viral causes of meningitis is well established (for a review, see reference 20). Also, our specimens were collected over a 2-year period, inclusive of seasonal countrywide outbreaks of enterovirus infection. For 11 of 23 enterovirus-positive specimens we observed a cross-hybridization pattern between the echovirus and coxsackie B virus targets due to their known sequence similarity (see Fig. 2D and Fig. 3D, which show a weak signal intensity of 1,786 with the single coxsackie B virus-specific probe and a signal intensity of 22,659 with the echovirus-specific probe). Indeed, when the coxsackie B1 virus sequence was used to query other enterovirus sequences with the BLASTN program for sequence homology comparison, the sequence variability among the top 50 hits was 7 to 10%; these included strains of coxsackie B virus, echovirus, and other enteroviruses. Given that PCR product-based DNA microarrays cannot distinguish between DNA targets sharing more than 80% sequence identity (9, 24), the cross-hybridization between coxsackie B virus and echovirus targets was not unexpected. However, the echovirus target cross-reacted with the closely related coxsackie B virus-specific probe for only half of the samples, with a mean intensity of signal of 65% (range, 28 to 89%). This might be due to a combination of factors, such as viral load, amplification efficiency, and the degree of sequence diversity, that affect the cross-hybridization patterns of enteroviruses on the array.
The impacts of various factors on microarray scanner-generated digital images have been reviewed at length by Li et al. (13). They concluded that judging images of closely related influenza viruses just by eye can be misleading. Indeed, we observed differences in spot sizes on our arrays due to batch-to-batch or beginning-to-end printing variations. To ascertain whether the signal intensities vary from slide to slide or batch to batch, we analyzed the most prevalent echovirus target, which was detected by using 4 slides from batch 1, 15 slides from batch 2, and 3 slides from batch 3. On the basis of the average intensity of four echovirus spots per slide, the mean variations in echovirus signal intensities were 10, 7.7, and 4.6% for slide batches 1, 2, and 3, respectively. Therefore, good intra- and interbatch array reproducibilities, coupled with the lack of cross-hybridization between unrelated viruses, make the visual signal readout from the computer-assisted two-color superimposition image very clear. Of practical consideration is the ability of the multiplex PCR-microarray assay to provide a consistently robust qualitative detection of at least one target per viral genome. We found no difficulties in distinguishing by eye true-positive from true-negative results (Fig. 2). Although we showed that there was a 100% concordance for positive virus detection results and that the theoretical limits of detection were as low as 48 genome copies for CMV and 28 genome copies for echovirus, it should not be assumed that negative results indicate the absence of virus. Issues of assay sensitivity, low viral genome copy number, and sample-specific inhibition affect the interpretation of negativity. The sensitivities for the detection of virus in clinical samples are likely to be lower than those calculated here for tissue culture samples.
There is a need for multicomponent panel-type diagnostic assays that allow the differentiation and identification of the pathogens responsible for disease syndromes with many potential causes, such as CNS or respiratory infections. The capacity of microarrays to accommodate high probe element densities and the potential use of several multiplex pools of primer pairs permit the development of pan-pathogen-specific probe arrays that will encompass other, primarily bacterial, causes of CNS infection, in addition to viruses. One limitation of the present array is that strain-specific sequence variations cannot be detected, and this would be of particular relevance to RNA viruses. The primers used in this study were not designed to target strain-specific nucleotide polymorphisms. Such limitations could be overcome by the use of strain-specific primer sets that represent known polymorphisms or by choosing nonvariant target sequences for primer design. Such modifications to PCR-microarray assays are tractable and would provide great diagnostic and clinical utility for certain viral infections. Single-format pan-pathogen-specific microarrays will contribute significantly to the speed and accuracy of laboratory diagnosis. This will have a beneficial impact on clinical care and hospital economics. This study has demonstrated the simplicity, accessibility, and applicability of such an approach in the context of CNS infections and is the first time that the diagnostic power of a microarray linked to multiplex PCR amplification of multiple pathogens has been demonstrated in a clinical laboratory.
Acknowledgments
We thank Li Jin, Wendy Knowles, and Bernard Cohen of the Health Protection Agency for providing live and cloned viruses.
This work was supported by the NHS R&D Support Fund from St. George's Healthcare Trust. R.A.S. and J.H. were supported by a grant from The Wellcome Trust Functional Genomics Resources Initiative for a microbial pathogen microarray facility in the Department of Medical Microbiology, St. George's Hospital.
REFERENCES
- 1.Allen, R. D., P. E. Pellet, J. A. Stewart, and M. Koopmans. 1995. Non-radioactive PCR-enzyme-linked immunosorbent assay method for detection of human cytomegalovirus DNA. J. Clin. Microbiol. 33:725-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boriskin, Y. S. 2001. Quantitative PCR for human cytomegalovirus DNA: analysis using NucleoScan 2001. New Microbiol. 24:1-9. [PubMed] [Google Scholar]
- 3.Boriskin, Y. S., K. Fuller, R. L. Powles, I. B. Vipond, P. S. Rice, J. C. Booth, E. O. Caul, and P. D. Butcher. 2002. Early detection of cytomegalovirus infection in bone marrow transplant patients by reverse transcription-PCR for CMV spliced late gene UL21.5: a two site evaluation. J. Clin. Virol. 24:13-23. [DOI] [PubMed] [Google Scholar]
- 4.Chapman, N. M., S. Tracy, C. J. Gauntt, and U. Fortmueller. 1990. Molecular detection and identification of enterovirus using enzymatic amplification and nucleic acid hybridisation. J. Clin. Microbiol. 28:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corne, J. M., S. Green, G. Sanderson, E. O. Caul, and S. L. Johnston. 1999. A multiplex RT-PCR for the detection of parainfluenza viruses 1-3 in clinical samples. J. Virol. Methods 82:9-18. [DOI] [PubMed] [Google Scholar]
- 7.Dorell, N., J. A. Mangan, K. G. Lang, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elnifro, E. M., A. M. Ashshi, R. J. Cooper, and P. E. Klapper. 2000. Multiplex PCR: optimisation and application in diagnostic virology. Clin. Microbiol. Rev. 13:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evertsz, E. M., J. Au-Young, M. V. Ruvolo, A. C. Lim, and M. A. Reynolds. 2001. Hybridisation cross-reactivity within homologous gene families on glass cDNA microarrays. BioTechniques 31:1182-1192. [DOI] [PubMed] [Google Scholar]
- 10.Freeman, W. M., D. J. Robertson, and K. E. Vrana. 2000. Fundamentals of DNA hybridization arrays for gene expression analysis. BioTechniques 29:1042-1055. [DOI] [PubMed] [Google Scholar]
- 11.Hamels, S., J. L. Gala, S. Dufour, P. Vannuffel, N. Zammatteo, and J. Remacle. 2001. Consensus PCR and microarray for diagnosis of the genus Staphylococcus, species, and methicillin resistance. BioTechniques 31:1364-1366. [DOI] [PubMed] [Google Scholar]
- 12.Jeffery, S., P. J. Keeling, A. Lukaszyk, Y. S. Boriskin, J. C. Booth, J. Hodgson, M. J. Davies, and W. J. McKenna. 1997. Molecular evaluation of enteroviruses in the pathogenesis of idiopathic dilated cardiomyopathy. Clin. Cardiol. 20:857-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markoulatos, P., A. Georgopoulou, N. Siafakas, E. Plakokefalos, G. Tzanakaki, and J. Kourea-Kremastinou. 2001. Laboratory diagnosis of common herpesvirus infections of the central nervous system by a multiplex PCR assay. J. Clin. Microbiol. 39:4426-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muir, P., A. Ras, P. E. Klapper, G. M. Cleator, K. Korn, C. Aepinus, A. Fomsgaard, P. Palmer, A. Samuelsson, A. Tenorio, B. Weissbrich, and A. M. van Loon. 1999. Multicenter quality assessment of PCR methods for detection of enteroviruses. J. Clin. Microbiol. 37:1409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onodera, K., J. d'Offay, and U. Melcher. 2002. Nylon membrane-immobilized PCR for detection of bovine viruses. BioTechniques 32:74-78. [DOI] [PubMed] [Google Scholar]
- 17.Oostenbrink, R., K. G. Moons, C. C. Theunissen, G. Derksen-Lubsen, D. E. Grobbee, and H. A. Moll. 2001. Signs of meningeal irritation at the emergency department: how often bacterial meningitis? Pediatr. Emerg. Care 17:161-164. [DOI] [PubMed] [Google Scholar]
- 18.Read, S. J., and J. B. Kurtz. 1999. Laboratory diagnosis of common viral infections of the central nervous system by using a single multiplex PCR screening assay. J. Clin. Microbiol. 37:1352-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers, C. D., and L. A. Burgoyne. 2000. Reverse transcription of an RNA genome from databasing paper (FTA(R)). Biotechnol. Appl. Biochem. 31:219-224. [DOI] [PubMed] [Google Scholar]
- 20.Rotbart, H. A. 2000. Viral meningitis. Semin. Neurol. 20:277-292. [DOI] [PubMed] [Google Scholar]
- 21.Steel, H. M., J. C. Booth, Y. S. Tryhorn, and H. Stern. 1988. A simple immunoalkaline phosphatase method for the rapid diagnosis of cytomegalovirus infection. Serodiagn. Immunother. Infect. Dis. 2:193-200. [Google Scholar]
- 22.Wagner, A., N. Blackstone, P. Cartwright, M. Dick, B. Misof, P. Snow, G. P. Wagner, J. Bartels, M. Murtha, and J. Pendleton. 1994. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst. Biol. 43:250-261. [Google Scholar]
- 23.Wilson, J. W., P. Bean, T. Robins, F. Graziano, and D. H. Persing. 2000. Comparative evaluation of three human immunodeficiency virus genotyping systems: the HIV-GenotypR method, the HIV PRT GeneChip assay, and the HIV-1 RT line probe assay. J. Clin. Microbiol. 38:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]