Abstract
Invasive pulmonary aspergillosis (IPA) is frequent and often fatal in hematopoietic stem cell transplant patients. Diagnosis requires microbiological or histopathologic demonstration of the organism in tissues; however, cultivation of Aspergillus species from respiratory secretions has low diagnostic sensitivity. Assays to detect Aspergillus antigen or DNA in bronchoalveolar lavage (BAL) fluid could facilitate earlier diagnosis, thereby guiding optimal therapy and obviating the need for additional costly and potentially morbid diagnostic evaluation. We evaluated the performance of a galactomannan enzyme immunoassay (GM EIA; Bio-Rad) by using a range of index cutoffs to define positivity and a quantitative PCR (qPCR) assay for the detection of Aspergillus species from BAL samples of patients with proven and probable IPA (case patients; n = 49) and without IPA (control patients; n = 50). The sensitivity of the GM EIA was 61% with an index cutoff of 1.0 and 76% with an index cutoff of 0.5; the corresponding specificities were 98 and 94%, respectively. The sensitivity and specificity of qPCR assay were 67 and 100%, respectively. The sensitivity with 22 culture-negative BAL specimens from patients with IPA was 41% for GM EIA with an index cutoff of 1.0, 59% for GM EIA with an index cutoff of 0.5, and 36% for qPCR assay. GM EIA indices and DNA quantities corresponded to BAL fungal burdens, with culture-positive samples having larger amounts of antigen and DNA compared to culture-negative samples. GM EIA and qPCR assay add to the sensitivity of BAL for diagnosing IPA in high-risk patients, with excellent specificity. Adjunctive use of these tests may reduce dependence on invasive diagnostic procedures.
Invasive pulmonary aspergillosis (IPA) is a frequent cause of morbidity and mortality among hematopoietic stem cell transplant (HSCT) patients. Diagnosis currently relies on cultivation of Aspergillus species from respiratory tract secretions or visualization of hyphae in biopsied tissue. Unfortunately, the relative insensitivity of bronchoalveolar lavage (BAL) fluid culture (40 to 50%) (14) frequently necessitates invasive procedures and/or liberal antifungal “empiricism,” resulting in delayed diagnosis and potentially inappropriate antifungal therapies. With the understanding that earlier diagnosis of IPA may facilitate higher cure rates, newer diagnostic platforms that are not based on culture have been proposed and investigated. Two such approaches that have recently attracted interest are the galactomannan (GM) enzyme immunoassay (EIA) and a quantitative PCR (qPCR) assay.
One antigen-based assay is the GM double-sandwich EIA. This assay has been recently approved by the U.S. Food and Drug Administration as an adjunctive test for IPA diagnosis when applied to serum. With this approval, the index cutoff to define positivity was lowered from 1.5 to 0.5 in order to optimize sensitivity (Bio-Rad Laboratories Platelia Aspergillus ELA package insert). The utility of GM EIA in BAL fluid diagnosis has not been investigated as extensively. In small clinical studies, the sensitivity of the GM EIA applied to BAL fluid ranges from 85 to 100% with a high index cutoff (1.5 or 1.0) to define positivity; specificity has not been defined in large analyses (3, 5, 22, 24).
Diagnostic assays that detect fungal nucleic acids by PCR have been investigated for use with blood (6, 8-10, 15, 19, 20, 23), biopsied tissues (11), and BAL fluid (13, 19, 22, 23). Previous studies reported that PCR assay has reasonable sensitivity and specificity when used to test samples from patients at high risk for IPA. Sensitivity has been highly variable, ranging from 50 to 100%, primarily because of differences in assay characteristics, types of patients evaluated, and certainty of IPA diagnosis (6, 8-10, 15, 19, 23). More recent studies have evaluated the use of real-time PCR assays applied to blood and BAL fluid (6, 15, 22); results suggest utility, although no standard assay platform has been used and no large study has been performed.
The present study was performed to examine the possibility that the application, singly or together, of a commercial EIA for GM (Bio-Rad) and a qPCR assay to BAL fluid obtained from HSCT patients suspected of having IPA would enhance diagnostic accuracy for invasive aspergillosis. To optimize the performance of the assays, attention was directed toward defining appropriate cutoffs for positivity and identifying clinical variables that may affect the diagnostic yield, such as the nature of the radiographic abnormality and ongoing administration of antifungal therapy.
MATERIALS AND METHODS
Patients and samples.
Between 1993 and 2002, 443 patients at the Fred Hutchinson Cancer Research Center underwent bronchoscopy for evaluation of pulmonary disease. BAL fluid that was left over after conventional microbiologic and cytologic evaluations was stored at −70°C. A database of patients with invasive fungal infection during that time period was searched and matched with a HSCT database to identify samples from patients with IPA (case patients) or without IPA (control patients); microbiology, histopathology, and clinical criteria were obtained in separate searches (16, 17). The diagnosis of IPA was defined as either proven or probable using a modification of previously published criteria (1). Proven aspergillosis required histopathologic demonstration of the organism in sterile tissues; aspergillosis was considered probable if the organism was cultivated from BAL fluid or respiratory samples in patients with symptoms consistent with disease. The date of IPA diagnosis was defined as the first day on which infection was confirmed by culture or histopathological examination of respiratory tissue (BAL fluid, sputum, or tissue biopsy). Patients from whom BAL fluid samples had been obtained within 30 days of diagnosing probable or proven IPA were eligible for analysis. If multiple samples were available, the one taken closest to the diagnosis date was selected. Case patients were randomly selected such that both culture-positive and culture-negative BAL fluid samples were included. BAL fluid samples obtained from patients with other diagnoses (e.g., bacterial pneumonia, viral pneumonia, pneumonia caused by other fungi) were selected randomly as a control group. A single sample was randomly selected for each control patient.
The study was approved by the Institutional Review Board at the Fred Hutchinson Cancer Research Center, and informed consent was obtained from patients or their parents or guardians. Charts were reviewed to verify the diagnoses in the case and control patient groups and to assess clinical variables that may affect the performance of the assays. These factors included culture positivity of BAL fluid, receipt of mould-active antifungal therapy at the time of bronchoscopy, and the nature of any radiographic abnormality (nodules or focal infiltration versus diffuse infiltration).
BAL fluid specimens obtained from 50 case and 50 control patients were thawed and assigned randomization numbers. Five hundred microliters of thawed BAL fluid was aliquoted to separate tubes for GM EIA analyses. An additional 1.5 ml was centrifuged (3,200 × g) for 5 min, and the cellular pellet was stored for qPCR assay. BAL fluid sample volumes allowed for GM EIA analysis of samples from 49 case and 50 control patients and qPCR assay analysis of samples from 46 case and 47 control patients. Two separate investigators who were unaware of the patient group (case or control) and other diagnostic test results performed the GM EIA and qPCR assays and interpreted the results.
GM EIA.
The Platelia Aspergillus GM EIA (Bio-Rad, Edmonds, Wash.) was used to quantify GM indices in accordance with the manufacturer's instructions. BAL fluid samples were processed under a high-efficiency particulate air-filtered benchtop hood. Briefly, samples were mixed well and 300 μl of each was added to 100 μl of 4% EDTA treatment solution, boiled for 3 min, and centrifuged at 10,000 × g for 10 min. Supernatant (50 μl) was added to 50 μl of a reaction mixture containing conjugated anti-GM EB-A2 antibody, and the mixture was incubated in microtiter plates precoated with the same antibody (EB-A2) for 90 min at 37°C. Wells were washed with an automated washer (Diagnostics Pasteur, Paris, France) and incubated with 200 μl of tetramethylbenzidine solution for an additional 30 min in the dark. Reactions were stopped with 100 μl of 1.5 M sulfuric acid, and optical densities (ODs) at 450 and 620 nm were read. Positive and negative controls (provided in the kit) were included in each assay. Results were recorded as an index relative to the OD of the threshold control (GM index = OD sample/OD threshold control).
qPCR assay.
Cell pellets from centrifuged BAL fluid were digested, and the DNA was purified with the Master Pure Yeast kit (Epicentre, Madison, Wis.). A water-only digestion control was processed with every BAL fluid sample to monitor contamination. DNA was resuspended in 50 μl of water and stored frozen until assayed.
The qPCR amplified a 219-bp segment of the fungal 18S rRNA gene with broad-range primers 5′-GATACCGTYGTAGTCTTA-3′ (forward) and 5′-TGTCTGGACCTGGTGAGT-3′ (reverse). The Aspergillus 18S rRNA gene was detected with a fluorescein-labeled molecular beacon with a dabcyl quencher with the sequence 5′-FAM-CGCTGTTTCTATGATGACCCGCTCGGCACAGCG-Dab-3′. This probe detects most of the fungi in the Aspergillus genus and some Penicillium species. Each 50-μl PCR mixture contained Universal Master Mix (Applied Biosystems), 10 pmol of each primer, 200 nM Aspergillus 18S rRNA gene beacon, and 5 μl of target DNA. In each qPCR assay, test samples were run in duplicate with four no-template controls, two digestion controls, and four positive controls containing Aspergillus genomic DNA at 1, 10, 100, and 1,000 pg for generation of a standard curve. PCR was performed on an Applied Biosystems 7700 sequence detector. After incubation at 50°C for 2 min to activate uracil glycosylase, samples were heated to 95°C for 10 min to activate the Taq Gold polymerase. Forty-five cycles of PCR were then performed with a melting step at 95°C for 15 s, annealing at 55°C for 1 min, and extension at 72°C for 20 s. Fluorescence was measured in the later half of the annealing step at 55°C so that the molecular beacon could anneal to its cognate target. The change in normalized relative fluorescence was plotted over time for each reaction to generate amplification plots. Samples were interpreted as positive if both duplicates showed an increase in normalized relative fluorescence above the background and the multicomponent view demonstrated an increase in absolute fluorescence. The average quantity of Aspergillus DNA present in the positive samples was calculated on the basis of the standard curve. If duplicates were discordant, the assay was repeated. Amplification controls were performed on all of the samples to determine if PCR inhibitors were present. One thousand picograms of Aspergillus genomic DNA was added to separate PCR mixtures containing 5 μl of BAL fluid DNA, and amplification kinetics were compared to those of the 1,000-pg standard. If PCR inhibitors were detected in samples, the DNA was repurified and assayed again.
Statistical analysis.
Primary statistical analyses were performed with a single BAL fluid sample per patient. For these analyses, samples were limited to those obtained within 30 days of diagnosis from case patients. Samples that were obtained on the date closest to the day on which a diagnosis of proven or probable IPA was verified were included for case patients, and single samples were randomly selected from control patients. Given these criteria, a total of 100 BAL fluid samples from 50 control patients and 50 case patients was intended for analysis by GM EIA and qPCR assay.
Sensitivity and specificity, along with their associated 95% confidence intervals (CIs), were calculated for the GM EIA and the qPCR assay with standard binomial probabilities and normal approximations. A receiver operating characteristic (ROC) curve was drawn to assess how changing the index cutoff for GM EIA positivity affects sensitivity and 1 − specificity. Correlation of assay positivity with receipt of antifungal therapy, type of radiographic abnormality, and BAL fluid culture positivity was assessed with two-by-two tables. P values were calculated with Fisher's exact test or the Wilcoxon rank-sum test, as appropriate. Comparisons between test results were performed with McNemar's test for paired data and applied to samples for which both tests were carried out.
RESULTS
Samples from 50 case and 50 control patients were subjected to at least one diagnostic test. Both tests were performed with samples from 45 case and 47 control patients. All patients underwent bronchoscopy to evaluate pulmonary nodules or infiltrates that were detected after or while being evaluated for HSCT. Case and control patients were well matched with regard to age (means, 45.2 and 41.2 years, respectively) and underlying diseases (data not shown). The majority of the case (44 [88%] of 50) and control (36 [72%] of 50) patients had undergone an allogeneic HSCT.
Twenty-eight (56%) of 50 BAL fluid samples from case patients were culture positive for Aspergillus species. Samples from the remaining case patients (n = 22) required lung biopsy, autopsy, or culture of other respiratory samples (e.g., sputum) to establish a diagnosis. The BAL fluid samples from the 50 case patients were obtained between 16 days before and 20 days after diagnosis (median, 0 days). By definition, no BAL fluid samples from the control group were culture positive for Aspergillus species.
Assay performance.
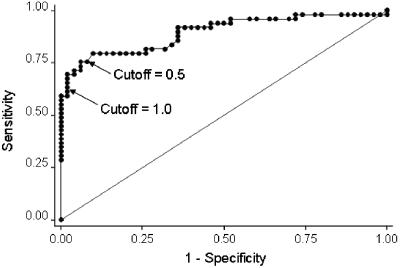
Because of limited volume in eight specimens, the GM EIA was performed with BAL fluid samples from 49 case and 50 control patients and the qPCR assay was performed with samples from 46 case and 47 control patients. The results of both tests, including calculated sensitivities and specificities, are outlined in Table 1. When the index cutoff to define GM EIA positivity was lowered from 1.0 to 0.5, the estimated sensitivity increased from 61 to 76% (P < 0.01) and the specificity decreased from 98 to 94% (P = 0.16). The ROC curve, which graphs changes in sensitivity and 1 − specificity (i.e., accuracy) as the index cutoff to define GM EIA positivity decreases, indicates that any further decrease in the cutoff below 0.5 would have compromised specificity more than it would have improved sensitivity (Fig. 1).
TABLE 1.
Performance of the diagnostic assaysa
| Test | No. of results
|
% Sensitivity (95% CI) | % Specificity (95% CI) | |||
|---|---|---|---|---|---|---|
| True positive | True negative | False positive | False negative | |||
| qPCR assay | 31 | 47 | 0 | 15 | 67 (52-81) | 100 (93-100) |
| GM EIA with index of 1.0 | 30 | 49 | 1 | 19 | 61 (46-75) | 98 (89-100) |
| GM EIA with index of 0.5 | 37 | 47 | 3 | 12 | 76 (61-87) | 94 (84-99) |
BAL fluid analysis results from 46 case and 47 control patients for qPCR and 49 case and 50 control patients for GM EIA are shown.
FIG. 1.
ROC curve demonstrating sensitivity (y axis) according to 1 − specificity (x axis) for GM EIA with a decreasing index value to define positivity (index decreases from left to right).
There were no statistically significant differences in accuracy between the qPCR test and the GM EIA. There was a trend toward higher sensitivity with the qPCR test compared to that of the GM EIA when the cutoff was 1.0 (67 versus 61%; P = 0.40), with no difference in specificity (100 versus 98%; P = 1.0). However, lowering the GM EIA cutoff to 0.5 reversed the trend in favor of the GM EIA with regard to sensitivity (76 versus 67%; P = 0.21), although the qPCR assay had better specificity (100 versus 94%; P = 0.16). Because quantitative values were not obtained for negative samples with the qPCR assay, we were not able to assess performance with alternate cutoffs or to draw ROC curves.
Both the qPCR test and the GM EIA were performed with 47 control and 45 case patient BAL fluid samples. When the results of both assays were applied to the same specimen concurrently, i.e., when a positive test was defined by a positive result of either the GM EIA with an index cutoff of 0.5 or the qPCR assay, the sensitivity was 82% (95% CI, 68 to 92) and the specificity was 96% (95% CI, 86 to 99). Interpreting the results obtained with both tests suggested an improved sensitivity compared to that of the GM EIA with an index cutoff of 0.5 alone (82 versus 76%; P = 0.08), with a nonsignificant impact on specificity (96 versus 94%; P = 1.0). Assuming a 20% prevalence of IPA in the HSCT population undergoing BAL fluid analysis, estimates of the positive and negative predictive values of the combined GM EIA and qPCR test results are 84 and 96%, respectively. Therefore, the best performance of these assays was achieved by using them in combination.
Impact of clinical variables.
For both tests, there was a trend toward better performance with BAL fluid samples obtained from patients receiving antifungal therapy (Table 2). This analysis was limited by the fact that >75% of the patients were receiving antifungal therapy at the time of bronchoscopy.
TABLE 2.
Impact of antifungal therapya
| Test | Not on antifungal therapy
|
On antifungal therapy
|
P valueb | ||||
|---|---|---|---|---|---|---|---|
| No. | Sensitivity (%) | 95% CI | No. | Sensitivity (%) | 95% CI | ||
| GM EIA with index of 0.5 | 9 | 67 | 30-95 | 35 | 80 | 63-92 | 0.40 |
| GM EIA with index of 1.0 | 9 | 44 | 14-79 | 35 | 66 | 48-81 | 0.28 |
| qPCR assay | 7 | 43 | 10-82 | 34 | 77 | 59-89 | 0.17 |
| qPCR or GM EIA with index of 0.5 | 7 | 57 | 18-90 | 33 | 91 | 76-98 | 0.06 |
Antifungal therapy data were not available for five patients.
P value from Fisher's exact test comparing sensitivities between patients receiving antifungal therapy and patients not receiving antifungal therapy.
Assay performance was evaluated on the basis of the radiographic appearance of pulmonary abnormalities. Approximately two-thirds of the patients presented with nodules or focal infiltrates, and one-third presented with bilateral infiltrates (Table 3). Although the small numbers of patients in the subgroups limited our ability to make definitive comparisons, there was a trend toward higher GM EIA and qPCR assay sensitivities with samples from patients with nodules or focal infiltrates.
TABLE 3.
Sensitivities of assays according to radiographic appearance
| Test | Nodular vs focal infiltratea
|
Bilateral infiltrate
|
P valueb | ||||
|---|---|---|---|---|---|---|---|
| No. | Sensitivity (%) | 95% CI | No. | Sensitivity (%) | 95% CI | ||
| GM EIA with index of 0.5 | 22 | 82 | 60-95 | 10 | 60 | 26-88 | 0.22 |
| GM EIA with index of 1.0 | 22 | 73 | 50-89 | 10 | 40 | 12-74 | 0.12 |
| qPCR | 22 | 73 | 50-89 | 9 | 56 | 21-86 | 0.42 |
| qPCR or GM EIA with index of 0.5 | 21 | 91 | 70-99 | 9 | 56 | 21-86 | 0.05 |
Included four patients with cavitary lesions.
P value from Fisher's exact test comparing sensitivities between patients with nodular or focal infiltration and those with bilateral infiltration.
Performance according to BAL fluid culture result.
We hypothesized that the sensitivity of the GM EIA and the qPCR assay might be higher among BAL fluid samples that were culture positive, as assay positivity and culture positivity are both likely to reflect the fungal burden. Of the BAL fluid samples that were culture positive for Aspergillus species, 89% were GM EIA (index cutoff, 0.5) positive, 96% were PCR assay positive, and 100% were either GM EIA or PCR assay positive. Accordingly, BAL fluid samples from patients with IPA that were positive by culture had higher median GM EIA indices (culture positive, 4.31; culture negative, 0.75) and DNA quantity (culture positive, 4.03 pg/ml; culture negative, 0 pg/ml). Accordingly, both tests had higher sensitivities when applied to BAL fluid samples that were also culture positive (Table 4). Of the culture-negative BAL fluid samples from patients with IPA, 59% were GM EIA (index cutoff, 0.5) positive, 36% were PCR assay positive, and 64% were either GM EIA or PCR assay positive. Therefore, almost two-third of patients who were not diagnosed by culture had a positive assay result.
TABLE 4.
Sensitivity relative to BAL fluid culture positivity for Aspergillus speciesa
| Test | BAL fluid culture positive
|
BAL fluid culture negative
|
P valueb | ||||
|---|---|---|---|---|---|---|---|
| No. | Sensitivity (%) | 95% CI | No. | Sensitivity (%) | 95% CI | ||
| GM EIA with index of 0.5 | 27 | 89 | 71-98 | 22 | 59 | 36-79 | 0.02 |
| GM EIA with index of 1.0 | 27 | 78 | 58-91 | 22 | 41 | 21-64 | 0.02 |
| qPCR | 24 | 96 | 79-100 | 22 | 36 | 17-59 | <0.001 |
| qPCR or GM EIA with index of 0.5 | 23 | 100 | 85-100 | 22 | 64 | 41-83 | 0.001 |
Among patients with proven or probable aspergillosis.
P value from Fisher's exact test comparing sensitivities between patients with BAL fluid cultures that revealed growth of Aspergillus species (positive) and those with culture-negative BAL fluid cultures.
Potential clinical impact of test results.
Eighteen culture-negative patients underwent a total of 25 additional procedures to establish a diagnosis of IPA, either a second bronchoscopy or a surgical lung biopsy (video assisted or open). Of the BAL fluid samples taken from 12 patients who subsequently underwent a second bronchoscopy, 8 were GM EIA (index cutoff, 0.5) positive, 8 were qPCR assay positive, and 11 were either GM EIA or PCR assay positive. Of the BAL fluid samples from the 13 patients who subsequently underwent an open-lung biopsy, 8 were GM EIA (index cutoff, 0.5) positive, 9 were qPCR assay positive, and 11 were either GM EIA or qPCR assay positive. Therefore, had these assays been available at the time of evaluation, 11 of 12 additional bronchoscopies and 11 of 13 lung biopsies might have been avoided. On the other hand, false-positive results obtained by GM EIA (index cutoff, 0.5) for four control patients might have led to an incorrect diagnosis.
DISCUSSION
Diagnosis of IPA remains challenging, largely because of atypical clinical presentations, coexistence with other infectious and noninfectious diseases, and a relative inability to cultivate these organisms by standard microbiological techniques. Incorporating a sensitive laboratory method that can better detect Aspergillus sp. in tissues may facilitate earlier diagnosis and, in turn, improve clinical outcomes. In this large study, we applied the Platelia Aspergillus EIA and the qPCR assay to BAL fluid samples for the diagnosis of IPA in HSCT patients. Each assay yielded a sensitivity higher than that commonly reported for BAL fluid culture while maintaining excellent specificity. Concurrent use yielded the highest sensitivity (82%), detecting Aspergillus sp. in two-thirds of the culture-negative samples from IPA patients. Use of these tests may increase the yield of bronchoscopy for the diagnosis of IPA and obviate the need for subsequent invasive procedures.
The results of this study differ somewhat from those of previous investigations. Verweij and colleagues reported that results of GM EIA applied to serum and BAL fluid correlated with possible aspergillosis in neutropenic patients but that the BAL fluid assay yielded a high rate of false positivity (24). Similarly, several investigators who applied various PCR assays to BAL fluid for the diagnosis of IPA found that although the PCR assay may have improved sensitivity, its specificity is unacceptably low, particularly for patients at lower risk of invasive disease (e.g., AIDS patients) (5, 7). The inability of the PCR assay to differentiate Aspergillus conidial colonization from true tissue invasion (7, 19) was felt to account for this relatively high rate of false positivity.
More recently, other investigators have reported higher positive predictive values for the qPCR assay (5, 13) and the Platelia Aspergillus GM EIA (2, 3, 21). Improved diagnostic utility likely stems from improved accuracy of the techniques themselves and from limiting their application to patients with a higher likelihood of disease. Given the higher incidence of IPA in HSCT patients relative to the general population, a positive assay in the context of a known pulmonary abnormality more likely indicates invasive infection than airway colonization. Although both the GM EIA and the qPCR assay may detect Penicillium species, the high specificities produced by application of these assays to BAL fluid suggest that significant colonization with Penicillium species is not common in our HSCT population.
There has recently been much discussion of the optimal index cutoff of the GM EIA applied to serum. Although the value used to define positivity has historically been ≥1.0, more recent studies suggest that using a lower index cutoff (0.7) may detect disease earlier in its course while maintaining good specificity (12). The Food and Drug Administration approved the serum-based assay for use in the United States with a cutoff index of 0.5 to define positivity, on the basis of as yet unpublished data (Bio-Rad Laboratories Platelia Aspergillus ELA package insert). In the present study, lowering the index cutoff in tests performed on BAL fluid improved sensitivity by 15% (P < 0.01) and compromised specificity by 4% (P = 0.16). The ROC curve demonstrates that lowering the index cutoff further would have considerably compromised specificity for only a modest gain in sensitivity. Optimal performance of the GM EIA was achieved by using an index cutoff of 0.5.
These findings carry important clinical implications. For those case patients who were diagnosed with a positive BAL fluid culture (i.e., culture-positive patients), application of the newer assays in real time could have yielded a diagnosis much more quickly (in hours instead of days). Also, the 64% of culture-negative patients who were positive by the GM EIA or the qPCR assay could have been diagnosed by initial bronchoscopy instead of proceeding with a lung biopsy or a second bronchoscopy. Avoiding additional invasive procedures in these patients would be especially desirable, given their predisposition to bleeding and infection. Moreover, the high specificity of both assays would assure the clinician that a positive test does, in fact, represent IPA, thereby preventing both unwarranted antifungal therapy and a failure to investigate other causes of acute pulmonary disease. Despite these encouraging data, the temptation to use GM EIA by itself to diagnose fungal infection must be tempered by the potential for misdiagnoses (false positives) and by the assay's inability to detect other fungal pathogens that might be present either alone or together with Aspergillus species. In our study, relying on the GM EIA alone (false positives with the 0.5 index cutoff) would have led to the misdiagnosis of three patients. This finding further supports a diagnostic approach that combines multiple laboratory methods (e.g., qPCR assay, GM EIA, and culture) to establish a diagnosis of disease.
Antifungal compounds may lower the fungal burden and therefore hamper the ability of an assay to detect Aspergillus species. In fact, prior human and animal studies have suggested that concurrent antifungal therapy decreases GM levels (in serum and BAL fluid), as well as PCR assay and GM EIA sensitivity (2, 3, 18, 25). However, we noted a trend toward improved sensitivity with samples from patients receiving therapy. This may be associated with therapeutic bias, as patients who received therapy in the absence of a definitive diagnosis may have had greater fungal burdens.
Our results also suggested a higher sensitivity of diagnostic tests with samples from patients with focal versus diffuse radiographic abnormalities. There are at least three scenarios that could explain these findings: (i) nodules or focal infiltrates carry a greater fungal burden, thereby exceeding the amount of cells or antigen needed to achieve the assay threshold; (ii) focal abnormalities enable the pulmonologist to perform BAL more selectively, thereby increasing the chance of collecting fluid from an involved lung; or (iii) focal abnormalities represent true IPA, whereas diffuse infiltrates may represent another process altogether. Finally, it is possible that disease presenting as focal, as opposed to diffuse, by radiography might represent subtle differences in pathogenic mechanisms and fungal burden. Prior studies with animal models have shown that such a difference may be attributed to the extent of infection or the type of immune suppression (4). In a rabbit model of pulmonary aspergillosis, equivalent inocula yielded different processes in subjects with different forms of immunosuppression: whereas neutropenic animals had focal processes with a large fungal burden, corticosteroid- or cyclosporine-treated animals had more diffuse infiltration with a smaller fungal burden (4).
With 50 case patients with IPA and 50 control patients, this study is one of the largest analyses of adjunctive testing for BAL fluid diagnosis of IPA. However, certain limitations must be considered when extrapolating these data to clinical practice. First, the current classification of IPA into possible, probable, and proven disease (rather than one distinct diagnosis) may confound the results of a study that compares case and control patient populations. We included only probable and proven case patients, as these patients are unequivocally treated for IPA in standard clinical practice. However, these definitions may not differentiate true invasive aspergillosis from Aspergillus colonization accompanying another pulmonary disease. Also, we cannot accurately determine test performance according to probable or proven IPA separately, as patients who had culture-positive BAL fluid samples (probable IPA) did not routinely undergo biopsy to confirm proven disease. Second, since these samples were collected from a population with a relatively high incidence of IPA, the results of our study should not be applied to the more general population, in which positive and negative predictive values would certainly be diminished. Third, the optimal processing of BAL fluid for the performance of these assays is unclear. Changing the quantity of the specimen or the methods of preparation may alter the sensitivity and specificity of the assays. Finally, our study was limited by its retrospective design. Since BAL was performed at various time points during the course of infection, ranging from onset of radiographic abnormalities to progression to severe disease, the timing of bronchoscopy may influence assay accuracy. Large, prospective studies are needed to define more clearly the clinical variables that affect assay performance and to determine optimal methods of specimen processing.
In summary, this large retrospective study demonstrates that incorporating the Platelia Aspergillus GM EIA and a qPCR assay into standard BAL fluid analysis may enhance bronchoscopic identification of Aspergillus species as the cause of pulmonary disease in HSCT patients. Although culture and/or histopathology of sterile tissue remain the “gold standard” indicators of invasive disease, the relatively low cost, ease of processing, and rapid results yielded by these assays may facilitate diagnoses based on bronchoscopy in high-risk patients.
Acknowledgments
This study was funded by grants from the National Institutes of Health (CA18029, CA15704, and UO1 AI54736). Bio-Rad Laboratories provided EIA kits for analysis. K. A. Marr and S. A. Balajee have served as consultants for Bio-Rad Laboratories.
REFERENCES
- 1.Ascioglu, A., J. Rex, B. DePauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Becker, M. J., S. de Marie, D. Willemse, H. A. Verbrugh, and I. A. Bakker-Woudenberg. 2000. Quantitative galactomannan detection is superior to PCR in diagnosing and monitoring invasive pulmonary aspergillosis in an experimental rat model. J. Clin. Microbiol. 38:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, M. J., E. J. Lugtenburg, J. J. Cornelissen, C. Van Der Schee, H. C. Hoogsteden, and S. De Marie. 2003. Galactomannan detection in computerized tomography-based broncho-alveolar lavage fluid and serum in haematological patients at risk for invasive pulmonary aspergillosis. Br. J. Haematol. 121:448-457. [DOI] [PubMed] [Google Scholar]
- 4.Berenguer, J., M. C. Allende, J. W. Lee, K. Garrett, C. Lyman, N. M. Ali, J. Bacher, P. A. Pizzo, and T. J. Walsh. 1995. Pathogenesis of pulmonary aspergillosis. Granulocytopenia versus cyclosporine and methylprednisolone-induced immunosuppression. Am. J. Respir. Crit. Care Med. 152:1079-1086. [DOI] [PubMed] [Google Scholar]
- 5.Buchheidt, D., C. Baust, H. Skladny, J. Ritter, T. Suedhoff, M. Baldus, W. Seifarth, C. Leib-Moesch, and R. Hehlmann. 2001. Detection of Aspergillus species in blood and bronchoalveolar lavage samples from immunocompromised patients by means of 2-step polymerase chain reaction: clinical results. Clin. Infect. Dis. 33:428-435. [DOI] [PubMed] [Google Scholar]
- 6.Costa, C., J. M. Costa, C. Desterke, F. Botterel, C. Cordonnier, and S. Bretagne. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 40:2224-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayette, M. P., D. Vaira, F. Susin, P. Boland, G. Christiaens, P. Melin, and P. De Mol. 2001. Detection of Aspergillus species DNA by PCR in bronchoalveolar lavage fluid. J. Clin. Microbiol. 39:2338-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebart, H., J. Loffler, H. Reitze, A. Engel, U. Schumacher, T. Klingebiel, P. Bader, A. Bohme, H. Martin, D. Bunjes, W. V. Kern, L. Kanz, and H. Einsele. 2000. Prospective screening by a panfungal polymerase chain reaction assay in patients at high risk for fungal infections: implications for the management of febrile neutropenia. Br. J. Haematol. 111:635-640. [DOI] [PubMed] [Google Scholar]
- 9.Hebart, H., C. Bokemeyer, J. Loffler, U. Schumacher, L. Kanz, and H. Einsele. 1997. New aspects for the diagnosis of invasive fungal disease in oncological patients. Onkologie 20:99-104. [Google Scholar]
- 10.Hebart, H., J. Loffler, C. Meisner, F. Serey, D. Schmidt, A. Bohme, H. Martin, A. Engel, D. Bunje, W. V. Kern, U. Schumacher, L. Kanz, and H. Einsele. 2000. Early detection of aspergillus infection after allogeneic stem cell transplantation by polymerase chain reaction screening. J. Infect. Dis. 181:1713-1719. [DOI] [PubMed] [Google Scholar]
- 11.Hendolin, P. H., L. Paulin, P. Koukila-Kahkola, V. J. Anttila, H. Malmberg, M. Richardson, and J. Ylikoski. 2000. Panfungal PCR and multiplex liquid hybridization for detection of fungi in tissue specimens. J. Clin. Microbiol. 38:4186-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbrecht, R., V. Letscher-Bru, C. Oprea, B. Lioure, J. Waller, F. Campos, O. Villard, K. L. Liu, S. Natarajan-Ame, P. Lutz, P. Dufour, J. P. Bergerat, and E. Candolfi. 2002. Aspergillus galactomannan detection in the diagnosis of invasive aspergillosis in cancer patients. J. Clin. Oncol. 20:1898-1906. [DOI] [PubMed] [Google Scholar]
- 13.Kawazu, M., Y. Kanda, S. Goyama, M. Takeshita, Y. Nannya, M. Niino, Y. Komeno, T. Nakamoto, M. Kurokawa, S. Tsujino, S. Ogawa, K. Aoki, S. Chiba, T. Motokura, N. Ohishi, and H. Hirai. 2003. Rapid diagnosis of invasive pulmonary aspergillosis by quantitative polymerase chain reaction using bronchial lavage fluid. Am. J. Hematol. 72:27-30. [DOI] [PubMed] [Google Scholar]
- 14.Levy, H., D. A. Horak, B. R. Tegtmeier, S. B. Yokota, and S. J. Forman. 1992. The value of bronchoalveolar lavage and bronchial washings in the diagnosis of invasive pulmonary aspergillosis. Respir. Med. 86:243-248. [DOI] [PubMed] [Google Scholar]
- 15.Lin, M., H. Lu, and W. Chen. 2001. Improving efficacy of antifungal therapy by polymerase chain-reaction-based strategy among febrile patients with neutropenia and cancer. Clin. Infect. Dis. 33:1621-1627. [DOI] [PubMed] [Google Scholar]
- 16.Marr, K. A., R. A. Carter, M. Boeckh, P. Martin, and L. Corey. 2002. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood 100:4358-4366. [DOI] [PubMed] [Google Scholar]
- 17.Marr, K., R. Carter, F. Crippa, A. Wald, and L. Corey. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34:909-917. [DOI] [PubMed] [Google Scholar]
- 18.Petraitis, V., R. Petraitiene, A. A. Sarafandi, A. M. Kelaher, C. A. Lyman, H. E. Casler, T. Sein, A. H. Groll, J. Bacher, N. A. Avila, and T. J. Walsh. 2003. Combination therapy in treatment of experimental pulmonary aspergillosis: synergistic interaction between an antifungal triazole and an echinocandin. J. Infect. Dis. 187:1834-1843. [DOI] [PubMed] [Google Scholar]
- 19.Raad, I., H. Hanna, A. Huaringa, D. Sumoza, R. Hachem, and M. Albitar. 2002. Diagnosis of invasive pulmonary aspergillosis using polymerase chain reaction-based detection of Aspergillus in BAL. Chest 121:1171-1176. [DOI] [PubMed] [Google Scholar]
- 20.Raad, I., H. Hanna, D. Sumoza, and M. Albitar. 2002. Polymerase chain reaction on blood for the diagnosis of invasive pulmonary aspergillosis in cancer patients. Cancer 94:1032-1036. [PubMed] [Google Scholar]
- 21.Salonen, J., O. P. Lehtonen, M. R. Terasjarvi, and J. Nikoskelainen. 2000. Aspergillus antigen in serum, urine and bronchoalveolar lavage specimens of neutropenic patients in relation to clinical outcome. Scand. J. Infect. Dis. 32:485-490. [DOI] [PubMed] [Google Scholar]
- 22.Sanguinetti, M., B. Posteraro, L. Pagano, G. Pagliari, L. Fianchi, L. Mele, M. La Sorda, A. Franco, and G. Fadda. 2003. Comparison of real-time PCR, conventional PCR, and galactomannan antigen detection by enzyme-linked immunosorbent assay using bronchoalveolar lavage fluid samples from hematology patients for diagnosis of invasive pulmonary aspergillosis. J. Clin. Microbiol. 41:3922-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiess, B., D. Buchheidt, C. Baust, H. Skladny, W. Seifarth, U. Zeilfelder, C. Leib-Mösch, H. Mörz, and R. Hehlmann. 2003. Development of a Light-Cycler PCR assay for detection and quantification of Aspergillus fumigatus DNA in clinical samples from neutropenic patients. J. Clin. Microbiol. 41:1811-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verweij, P. E., J. P. Latge, A. J. Rijs, W. J. Melchers, B. E. De Pauw, J. A. Hoogkamp-Korstanje, and J. F. Meis. 1995. Comparison of antigen detection and PCR assay using bronchoalveolar lavage fluid for diagnosing invasive pulmonary aspergillosis in patients receiving treatment for hematological malignancies. J. Clin. Microbiol. 33:3150-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh, T. J., V. Petraitis, R. Petraitiene, A. Field-Ridley, D. Sutton, M. Ghannoum, T. Sein, R. Schaufele, J. Peter, J. Bacher, H. Casler, D. Armstrong, A. Espinel-Ingroff, M. G. Rinaldi, and C. A. Lyman. 2003. Experimental pulmonary aspergillosis due to Aspergillus terreus: pathogenesis and treatment of an emerging fungal pathogen resistant to amphotericin B. J. Infect. Dis. 188:305-319. [DOI] [PubMed] [Google Scholar]