Abstract
Finding the best technique to identify BRAF mutations with a high sensitivity and specificity is mandatory for accurate patient selection for target therapy. BRAF mutation frequency ranges from 40 to 60% depending on melanoma clinical characteristics and detection technique used.
Intertumoral heterogeneity could lead to misinterpretation of BRAF mutational status; this is especially important if testing is performed on primary specimens, when metastatic lesions are unavailable.
Aim of this study was to identify the best combination of methods for detecting BRAF mutations (among peptide nucleic acid – PNA-clamping real-time PCR, immunohistochemistry and capillary sequencing) and investigate BRAF mutation heterogeneity in a series of 100 primary melanomas and a subset of 25 matched metastatic samples.
Overall, we obtained a BRAF mutation frequency of 62%, based on the combination of at least two techniques. Concordance between mutation status in primary and metastatic tumor was good but not complete (67%), when agreement of at least two techniques were considered. Next generation sequencing was used to quantify the threshold of detected mutant alleles in discordant samples. Combining different methods excludes that the observed heterogeneity is technique-based. We propose an algorithm for BRAF mutation testing based on agreement between immunohistochemistry and PNA; a third molecular method could be added in case of discordance of the results. Testing the primary tumor when the metastatic sample is unavailable is a good option if at least two methods of detection are used, however the presence of intertumoral heterogeneity or the occurrence of additional primaries should be carefully considered.
Keywords: BRAF, primary melanoma, PNA-clamping, immunohistochemistry, heterogeneity
INTRODUCTION
The discovery of BRAF mutations in melanoma and the development of specific BRAF inhibitors has led to significant advances in treatment of metastatic melanoma, which has now been further improved by the use of new immunologic therapy [1]. Melanoma patients with metastatic or unresectable disease, according to recent NCCN guidelines [2], should be tested for BRAF V600 mutations to evaluate therapy options. However, there is no consensus on the best BRAF testing method, except for the need for a certified laboratory. In Italy, national guidelines from the Italian association of medical oncology and Pathology (AIOM-SIAPEC) point to standardization of different testing methods with internal controls, fast turnaround time of results, and the additional value of CE-IVD techniques [3].
BRAF mutation rate in melanoma ranges from 40 to 60% of cases [4–7]. This difference, besides reflecting clinical characteristics of the cohort studied, could also be due to the different methods used: a wide range of detection techniques is available, including classical capillary sequencing, pyrosequencing, next generation sequencing techniques (NGS), several real-time PCR (RT-PCR) based methods and immunohistochemistry (IHC) antibodies [8–14]; some of these methods have obtained CE-IVD certification.
Since the approval of BRAF inhibitors for the treatment of metastatic melanoma, the importance of BRAF heterogeneity in melanoma samples has gained much attention [15]. Tumour heterogeneity and the evolution of branching tumour subclones are a challenge even for multi-targeted therapies and intermittent regimens [16]. Currently, general advice is to test metastatic melanoma for the presence of BRAF mutation in the most recent metastatic sample. In some instances, biopsy from metastasis is unavailable, so checking mutational status in the primary tumor has been proposed as an alternative [17]. The consistency in BRAF mutation detection among primary and metastatic tumors is however still debated [18–20].
In addition, the clinical significance of finding BRAF mutation using a high sensitivity technique in a low number of cells in the primary tumor is still unclear, especially when not confirmed in the metastatic sample. Indeed, a different cell clone, driven by other mutations, could be positively selected and responsible for progression and the development of the metastasis [21].
Currently, data on primary melanoma are scarce, as well as data on matched primary and metastatic melanomas tested with several high sensitivity techniques [21–24]. Generally, IHC staining shows good consistency between primary and metastatic samples, although limited by a lower sensitivity compared to recent molecular techniques, which have, conversely, the disadvantage of relying on DNA quantity and quality [25].
Overall, while recent data applying IHC point to intra and inter tumor homogeneity of BRAF-mutated expression, the issue of genetic heterogeneity is still debated [23].
Aim of this study was to identify the best combination of methods for detecting BRAF mutations and investigate BRAF mutation heterogeneity within 100 primary melanomas and between primary melanoma and a subset of 25 matched metastatic samples. Furthermore, we aim to propose a reliable and efficient scheme for diagnostic mutation testing.
RESULTS
Cohort characteristics
Our cohort included 94 primary cutaneous melanomas, 4 mucosal melanomas and 2 uveal melanomas. Median follow-up for survivors was 74 months. During this period, 22 patients relapsed and 19 patients died, of which 13 deaths were melanoma-related. Twelve patients developed multiple melanomas. A summary of patients characteristics is shown in Table 1. In addition 25 metastatic samples, from 15 out of the 22 patients who relapsed during follow-up, were analyzed. Of these, 19 samples were metastatic lymph nodes, and 6 were distant metastases. In 5 patients, lymph nodes were excised at the same time as the primary.
Table 1. Clinical and pathological features of the 100 primary melanomas in the study cohort.
| Number of Patients(n=100) | |
|---|---|
| Gender and age | |
| Male | 45 |
| Female | 55 |
| Median Age | 52 years |
| Primary tumor site | |
| Trunk | 50 |
| Upper limb | 16 |
| Lower limb | 24 |
| Head and Neck | 4 |
| Uveal | 2 |
| Mucosal | 4 |
| Breslow thickness | |
| <1mm | 38 |
| 1-2mm | 23 |
| 2-4mm | 17 |
| >4mm | 16 |
| Unclassified* | 6 |
| Median tumor thickness | 1,7 |
| Ulceration | |
| Yes | 23 |
| No | 77 |
| Histological subtype | |
| SSM | 59 |
| Nodular | 20 |
| Acral-lentiginous | 2 |
| Other | 19 |
| Tumor stage | |
| I-II | 86 |
| III-IV | 14 |
| Prognostic features | |
| Median follow-up | 74 months (range 47 Q1 -158 Q3) |
| Alive | 81 |
| Dead | 19 |
| Melanoma-related deaths | 13 |
| Multiple primary melanomas | 12 |
| Relapse | 22 |
The unclassified cases correspond to 2 uveal and 4 mucosal melanomas, not classified using Breslow. (SSM: superficial spreading melanoma)
BRAF mutation analysis, using three different techniques, reveals a high percentage of V600 mutations in primary melanomas
We analyzed 100 FFPE samples from primary melanoma with three different techniques to determine the mutational status of BRAF and test which could be the most sensitive method to identify mutations in the BRAF V600 codon (Supplementary Table 1 and Figure 1). We chose PNA clamping real-time PCR (here named PNA) and capillary sequencing, to compare the sensitivity of the IHC antibody specific for the BRAF V600E mutated protein to molecular techniques.
Figure 1. A combination of IHC and molecular techniques reveals a high percentage of V600 mutations.
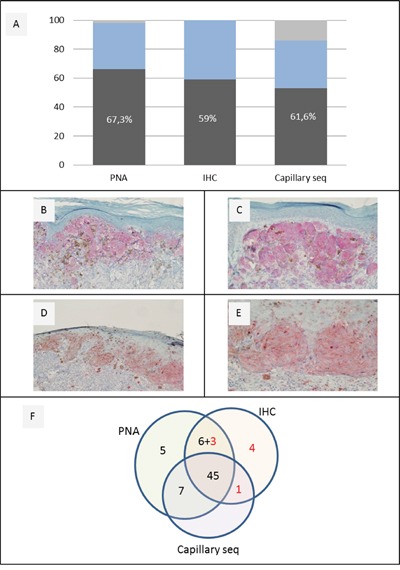
A. BRAF V600 different mutation rate and yield in 100 primary melanomas tested by PNA, IHC and capillary sequencing. B-E. representative IHC analysis of BRAF positive cases [Magnification 20x (B,D); 40x (C,E)]. F. Comparison among 100 primary melanomas, which gave a positive successful result with at least one method. Black Numbers indicate BRAF V600 positive cases obtained with each single technique; red numbers show those additional BRAF V600 positive cases obtained with one or two techniques only.
PNA identified 66 BRAF V600 mutated samples out of 98 (67,3%) and 32 wild type (33%); two samples gave invalid results (Figure 1A).
IHC was available for every primary melanoma sample, identifying 59 mutated samples with positive staining (59%) and 41 with negative staining. Among the positive samples, 52 showed an homogeneous staining pattern, while 7 showed an heterogeneous pattern (% of stained cells <95%), in a percentage variable between 10 and 80% (Figure 1A-1E).
Classical capillary sequencing was successful in 86 samples out 100, identifying 53 mutated samples (61,6%); 51 (96%) presented the canonical BRAF V600E mutation (c.1799 T>A), one (2%) presented the same amino acidic change V600E due to the less frequent substitution c.1799_1800delTGinsAA, (also called V600E2) and one (2%) the BRAF V600K mutation (c.1798_1799delGTinsAA).
Overall, BRAF mutation rate in 100 primary melanoma samples was 67,3% when analyzed by PNA, and 59% when the V600E mutation was assessed using the VE1 specific antibody for IHC staining. The classical capillary sequencing technique is sensitive (61,6%) but shows poor performance as 14 cases were not amplified. (Figure 1A). At least one of these highly sensitive methods identified a positive result in 71% of cases.
Sanger sequencing, PNA analysis and IHC provided the same results in 63 cases (46 positive and 21 negative for BRAF mutation). BRAF mutation rate was 62% if we consider the results obtained with at least two methods. Overall, agreement among the 3 methods was substantial (Light's k= 0.701, z=3.95, p=7.77e-05).
Interestingly, two samples invalid at PNA analysis were found to be positive by IHC (and one of them also at capillary sequencing) (Figure 1F and Supplementary Table 1).
Characterization of BRAF mutational status in matched primary and metastatic melanomas reveals a good consistency
We analyzed 25 samples of metastatic melanomas, coupled with their primary tumors, deriving from 15 of the 22 patients who relapsed, using the same three methods mentioned above for the primary samples: PNA, IHC and capillary sequencing (Table 2 and Table 3)
Table 2. BRAF mutational status in matched primary and metastatic melanomas reveals a good consistency.
| Case n. | Primary melanoma | M1 | M2 | M3 | Intrapatient Concordance | Intrapatient Overall Concordance | |
|---|---|---|---|---|---|---|---|
| 4 | PNA | V600 | wt | +/− | |||
| IHC | weak | weak | +/+ | +/− | |||
| Capillary seq | V600E | wt | +/− | ||||
| Cell %/Localization | 20/Upper limb | 70/LN | |||||
| 13 | PNA | V600 | wt | wt | +/− | ||
| IHC | weak | negative | negative | +/− | +/− | ||
| Capillary seq | V600E | wt | wt | +/− | |||
| Cell %/Localization | 50/Lower limb | 50/LN | 50/LN | ||||
| 18 | PNA | V600 | V600 | V600 | n.a. | +/+ | |
| IHC | weak | moderate | strong | moderate | +/+ | +/+ | |
| Capillary seq | WT | V600E | V600E- | n.a. | -/+ | ||
| Cell %/Localization | 80/Lower limb | 80/Skin | 70/Skin | 70/LN | |||
| 25 | PNA | V600 | n.a. | wt | n.a. | +/− | |
| IHC | moderate | moderate | moderate | moderate | +/+ | +/− | |
| Capillary seq | V600E- | wt | wt | n.a. | +/− | ||
| Cell %/Localization | 80/Lower limb | 70/Skin | 40/LN | 30/LN | |||
| 39 | PNA | V600 | V600 | V600 | +/+ | ||
| IHC | weak | moderate | strong | +/+ | +/+ | ||
| Capillary seq | WT | V600E | V600E | -/+ | |||
| Cell %/Localization | 50/Trunk | 80/LN | 80/Skin | ||||
| 46 | PNA | wt | wt | −/− | |||
| IHC | negative | negative | −/− | −/− | |||
| Capillary seq | n.a. | wt | n.a. | ||||
| Cell %/Localization | 80/Trunk | 80/LN | |||||
| 55 | PNA | V600 | wt | +/− | |||
| IHC | negative | negative | −/− | +/− | |||
| Capillary seq | V600E | wt | +/− | ||||
| Cell %/Localization | 10/Upper limb | 80/LN | |||||
| 67 | PNA | V600 | V600 | +/+ | |||
| IHC | weak | weak | +/+ | +/+ | |||
| Capillary seq | n.a. | V600K | n.a. | ||||
| Cell %/Localization | 60/Trunk | 80/LN | |||||
| 70 | PNA | V600 | V600 | +/+ | |||
| IHC | weak | strong | +/+ | +/+ | |||
| Capillary seq | V600E | V600E | +/+ | ||||
| Cell %/Localization | 70/Trunk | 40/LN | |||||
| 72 | PNA | V600 | V600 | V600 | +/+ | ||
| IHC | moderate | strong | strong | +/+ | +/+ | ||
| Capillary seq | V600E2 | V600E2 | V600E2 | +/+ | |||
| Cell %/Localization | 80/Trunk | 60/LN | 60/LN | ||||
| 75 | PNA | V600 | wt | wt | +/− | ||
| IHC | negative | negative | negative | −/− | +/− | ||
| Capillary seq | V600E | wt | wt | +/− | |||
| Cell %/Localization | 40/Head-Neck | 90/LN | 80/LN | ||||
| 76 | PNA | V600 | V600 | +/+ | |||
| IHC | moderate | strong | +/+ | +/+ | |||
| Capillary seq | V600E | n.a. | n.a. | ||||
| Cell %/Localization | 70/Upper limb | 90/LN | |||||
| 77 | PNA | WT | wt | wt | wt | −/− | |
| IHC | negative | weak | weak | weak | -/+ | −/− | |
| Capillary seq | WT | wt | wt | wt | −/− | ||
| Cell %/Localization | 80/Upper limb | 70/LN | 60/LN | 80/Mucosal | |||
| 98 | PNA | wt | wt | −/− | |||
| IHC | negative | negative | −/− | −/− | |||
| Capillary seq | wt | wt (k601k) | −/− | ||||
| Cell %/Localization | 80/Mucosal | 90/Mucosal | |||||
| 100 | PNA | wt | wt | −/− | |||
| IHC | negative | negative | −/− | −/− | |||
| Capillary seq | wt | wt | −/− | ||||
| Cell %/Localization | n.a,/Trunk | n.a./LN |
Intrapatient overall concordance means that at least two techniques gave the same results in matched samples (M1, M2, M3: progressive matched metastatic samples; LN: lymph-node).
Table 3. Concordance between primary and metastatic samples as observed by three different methods and with the combination of at least two different techniques.
| BRAF mutation | Concordance (%) | ||||
|---|---|---|---|---|---|
| Primary/Metastasis | Concordant | Discordant | |||
| +/+ | −/− | +/− | -/+ | ||
| IHC | 8 | 5 | 1 | 1 | 87 |
| PNA* | 6 | 4 | 5 | 0 | 67 |
| Sanger* | 2 | 3 | 5 | 2 | 42 |
| At least two methods | 6 | 4 | 5 | 0 | 67 |
when different metastatic samples were considered for each case, partial concordance was considered as discordance (n.a.: not applicable; +/+: both primary and metastatic samples BRAF positive; −/−: both primary and metastatic samples BRAF negative; +/−: primary sample BRAF positive and metastatic sample(s) BRAF negative; -/+: primary sample BRAF negative and metastatic sample(s) BRAF positive).
First, we considered the concordance of the results obtained with the three techniques, as we did for the primary samples: only 5/15 (33%) cases showed concordant results with all the three techniques; considering the results obtained combining the two most sensitive techniques, (IHC and PNA), we observed concordance in 67% of cases (10/15); if we considered at least two techniques we had concordant results in 100% (Table 2). Due to the small sample size (N < 30), inter-rater agreement for metastatic samples was not calculated in order to avoid false results.
Overall there was a high consistency between the data relative to BRAF mutation observed in primary and matched metastatic melanoma. Ten cases showed the same results between the primary and the metastatic sample, with the combination of the three methods (67% of concordance, considering a result obtained with at least two methods). The five discordant cases showed a BRAF mutation in the primary, which was not confirmed in the metastatic sample. The contrary was never observed (i.e. negative primary tumor showing BRAF mutation in the matched metastasis). Interestingly, 4 of the 5 discordant cases displayed metastatic lymph nodes which were not excised synchronously with the primary tumor.
When analyzing concordance per single method, we were able to verify that IHC showed results with the highest concordance (in 13/15 cases, 87%), followed by PNA (10/15 cases) with a concordance rate of 67% (the same observed with the combination of two methods). Instead, the concordance with capillary sequencing was very low (5/12, 42% of cases) (Table 3).
NGS excludes that inconsistency between primary and metastatic sample is due to a low threshold of mutated alleles as detected in the primary tumor
NGS with a custom designed hot spot panel including BRAF gene was used to analyze 3 of the 5 primary/metastases discordant cases (n. 4, 13 and 25) in order to confirm our observations and to quantify the threshold of mutated alleles that could be identified through the other methods.
We confirmed the positive BRAF mutational status obtained with our previous results (Supplementary Table 1, Table 2), in primary melanoma at high rates (30, 47 and 41 % of mutated alleles, respectively) and lack of BRAF mutation in the metastatic samples (Figure 2A, 2C, 2D). Interestingly, in case 4 we observed a BRAF V600E mutation in the primary melanoma, undetectable in the paired metastasis, which instead revealed a NRAS Q61R mutation (Figure 2A, 2B). A possible explanation was that when we checked the history of this patient we found out that he was one of the 12 patients who developed multiple primary melanomas, before the development of metastatic disease (including the analyzed metastasis). Therefore, this non-consistency may be explained if the metastasis that we found did not originate from the same primary melanoma that we analyzed, but from the second primary melanoma (never tested because not available in our archives, but with a higher metastatic potential because of its Breslow level) that could have been NRAS-positive and BRAF-negative. For the other non-consistent pairs, no other primary melanomas were reported.
Figure 2. NGS results of 4 samples from three different patients.

A. BRAF NGS of Case 4 primary melanoma, which shows a mutant allele percentage of 30%. B. Case 4 melanoma metastasis, positive for NRAS Q61R mutation (mutant allele 74%); the same sample was BRAF wild type (not shown). C-D. BRAF results for other two primary melanomas (case 13 and 25), whose metastases were BRAF wild type; primary tissues presented 47% and 41% of BRAF mutant alleles respectively.
DISCUSSION
Mutations in the BRAF oncogene are of increasing relevance in clinical oncology, especially after the approval of the anti-BRAF molecules in the therapy of BRAF positive metastatic melanomas. Therefore, finding the best technique to identify BRAF mutations with a high sensitivity and specificity is mandatory of accurate patient selection.
Here we tested BRAF mutations in a series of 100 primary melanomas with different techniques. Aim of our study was also to investigate intertumoral heterogeneity on the basis of the combination of different techniques used to detect BRAF mutations. Therefore, we analyzed 100 samples of primary melanomas using a combination of immunohistochemical analyses and real-time polymerase chain reaction (PNA in our cohort), in addition to classical capillary sequencing. IHC was more effective than capillary sequencing for detecting the V600E mutation. In a diagnostic setting it is the cheapest and the least time consuming technique (as it does not depend on DNA extraction) and can work on small melanoma samples.
CE-IVD PNA proved to be the most sensitive method in our cohort, as well as highly specific: it was also able to detect all mutations at codon 600, including the V600K mutation (as confirmed by capillary sequencing), while IHC antibody is marketed only for V600E identification. Due to its high sensitivity, PNA identified 67.3% of BRAF mutated samples, which is, to our knowledge, one of the highest percentages of BRAF mutations reported in the literature [4, 5, 7]. To our knowledge PNAs have been described one time only, very recently, for BRAF detection in melanomas; in addition they have been efficiently applied to BRAF testing for colorectal cancer and papillary thyroid carcinoma (PTC) [31, 37–40].
Two issues may justify this high percentage of mutated samples: the high sensitivity of PNA (compared to IHC and most to Sanger Sequencing) and the high prevalence, in our population, of melanomas in sites not chronically sun exposed (CSE) as trunk and limbs (Table 1), which are known to be associated with BRAF mutation [41].
The use of three different methods was useful in identifying the method with higher sensitivity and concordance compared to the others. Data obtained with IHC and PNA were quite consistent, while data obtained with capillary sequencing confirmed the difficulty in identifying a mutated allele when the percentage of tumor cells was below a threshold (which is generally considered to be 50%), in addition to the difficulty of working with little DNA, as in small primary melanoma, limiting PCR amplification prior to sequencing.
We conclude that PNA testing in primary melanoma is highly specific and reliable in detecting even a small fraction of mutated allele, undetectable with other methods (Figure 1F) making PNA, which is CE-IVD approved, applicable in clinical practice. Limitations in the choice of this technique could be its higher cost if compared to IHC [31] and dependence on DNA extraction.
While BRAF V600E mutation appears to play a critical role in tumor initiation, its expression during tumor progression remains controversial. Various authors claim that BRAF V600E heterogeneity present in melanoma, could be due to tumor sampling, the detection method used, or secondary to biological differences [18, 21, 22, 24, 43, 44]. Since coexistence of BRAF wild-type and BRAF mutant tumor cells within the same patient has important implications for clinical decisions, more sophisticated tools are needed to characterize patients’ cancers and guide their treatment.
With the aim to evaluate the issue of possible intertumoral heterogeneity in our cohort, we used the same three-methods approach, mentioned above, on 25 matched primary and metastatic samples from 15 patients in our cohort. In several papers, relying mainly on IHC, BRAF expression is assumed to be homogeneous in the majority of the lesions from the same patients [17, 23]. In our cohort, a consistency of nearly 90% was found using IHC, which supports this observation (Table 3). The addition of molecular techniques further clarifies that the observed heterogeneity is not due to the single techniques used. We observed that 10 (67%) cases showed consistency between primary and metastases for BRAF mutational status (6 mutated and 4 wild type), when assessed with at least two methods. In 5 cases there was no consistency, all the cases had a BRAF mutation in the primary which was not identified in the subsequent metastases. This result partially supports recent large studies in which, in the majority of cases, the primary and metastatic lesions had the same BRAF genotype [20, 45]. Indeed, we observed a good but not complete consistency between primary and metastatic samples from the same patients (10/15) while different metastatic lesions from the same patients revealed a concordant BRAF status.
The finding that BRAF mutations in primary melanomas is not always identified in the correspondent metastases can also partly explain the high percentage of BRAF mutations observed in our cohort of primary tumors (62% with at least two methods) compared to the literature, in which only metastatic tissue is evaluated. In addition, the natural history of the disease and the occurrence of multiple primary melanoma in the same patients should always be considered.
Overall, despite our 15 patients analyzed represent the 68% of the cases which relapsed, our results are limited by the small numbers and the mutation testing of only for BRAF gene.
While it is still unclear what is the selective pressure that induces migration of a BRAF-wild-type subclone instead of an expected more aggressive BRAF-mutant subclone, an approach based on NGS targeting multigene panels could help in clarifying this issue. Finally a recent study using NGS identifies very low heterogeneity in matched primary and lymph node metastatic samples, especially in synchronous metastases, while heterogeneity is more frequent in metachronous lesions [46]. Our results, using the concordance between at least two methods, confirm this finding, since our discordant cases displayed metastatic lymph-nodes which were not excised synchronously with the primary tumor.
In conclusion, the intertumoral (primary and metastatic) heterogeneity described here is not attributable to technical issues when molecular methods are used, since it was observed with a combination of different technique and partially confirmed by a fourth, quantitative, technique as NGS.
A combination of IHC, used as a screening tool, and parallel formal molecular mutation testing seems to provide the highest reliability for identifying the majority of BRAF mutated cases with the highest sensitivity and specificity [47, 48]. The use of IHC alone, without parallel molecular testing should be limited to samples of small size or with low tumor cell content for optimal DNA extraction. Conversely, the use of very sensitive molecular techniques, which could detect a minor BRAF-mutated subclone in a predominantly wild-type tumor, may not be clinically relevant as BRAF inhibitors may have opposite signaling effects in cells with mutated or wild-type BRAF [23].
The limits of this approach are in the comparison of techniques with very different sensitivity (IHC vs PNA) and specificity with the capability to identify different cellular structure (proteins for IHC; DNA for molecular techniques). In addition, when a second molecular technique is necessary, the increase in turnaround time and costs is another possible limit to this type of approach.
In clinical practice we propose the analysis of the most recent and clinically relevant sample (metastatic when available) with IHC and at least one molecular method (in our case we propose PNA). If BRAF mutation is identified in PNA, but not with IHC a third analysis with another method (in our case capillary sequencing or, for a quantification, NGS or pyrosequencing) is recommended in order to obtain a concordance of at least two methods, or identification of another BRAF V600 mutation which is undetectable with VE1 antibody (Figure 3). Testing the primary tumor when the metastatic sample is unavailable is a good option if at least two methods of detection are used, however the presence of intertumoral heterogeneity or the occurrence of additional primaries should be carefully considered.
Figure 3. Diagnostic algorithm proposed for BRAF testing in metastatic melanoma.

Report is based on the results obtained with two concordant methods. Applicable also to primary tumors when needed.
Interestingly, a recent study suggests that the staining pattern (homogeneous vs. heterogeneous) might be associated with differences in therapeutic response to BRAF inhibitors [19]. This is a field which will need further investigation in the near future and parallel IHC testing, which is currently being developed with antibodies directed to other BRAF or NRAS mutations, is an advantageous opportunity.
MATERIALS AND METHODS
Case selection
A total of 137 melanoma samples were included. 100 primary melanomas were selected based on formalin-fixed, paraffin embedded (FFPE) tissue availability at the Pathology department, IRCCS AOU San Martino IST, Genoa.
All melanoma patients had been referred to our center for germline testing either for diagnostic (familial and multiple melanoma cases) or research purposes (sporadic cases from an ongoing case-control study [26–28]). All the patients signed an informed consent under local IRB approved protocols for both germline testing and other research purposes dealing with the archived melanoma tissues in the pathology department. A local database collecting information from the pathology report, tumor cell content, in the examined section, and follow-up was designed. The database is regularly updated with the oncologists’ and local registries database, and includes detailed information on relapse, survival, time and cause of death.
A total of 25 metastatic samples, from 15 patients, were available and tested with the same techniques used for the primary tumors. Twelve metastatic samples, already tested for diagnostic purposes by molecular techniques, were used as positive and negative controls for IHC assessment (4 BRAF positive e 8 negative).
DNA extraction
All diagnoses were performed by two expert dermatopathologists (FC and MG), and subsequently reviewed for tumor content, neoplastic cell morphology (epithelioid vs spindled) and immunohistochemistry evaluation (LM and FG).
The tumor area was identified, and marked by the pathologists, on hematoxylin and eosin stained slides. Marked tumor tissue was manually dissected from two to six slides (8-micron thick) of FFPE tissue for each case.
Genomic DNA was extracted from the sections using the QIAmp DNA FFPE tissue kit (Qiagen, Valencia, CA, USA) or the Genomic DNA FFPE One-Step Kit for Diatech MagCore® HF16Plus extractor (RBC Bioscience, New Taipei City, Taiwan), in both cases according to the manufacturer's instructions. Quality and concentration of the DNA samples were examined by SPECTROstar Nano (BMG Labtech, Offenburg, Germany).
Capillary sequencing
Capillary sequencing was performed as previously described [29–31]. Briefly, we amplified tumor DNA in a 15 μl PCR containing approximately 40 ng of purified genomic DNA in a mixture of 1.5 μl of Buffer (10x), 1.5 μl dNTP (2 mM), 0.08 μl Qiagen HotStart Taq (5U/μl) and 2 μl forward and reverse primer (5 mM). After denaturation at 95°C for 15 minutes, 35 amplification cycles at 95°C for 40s, 60°C for 30s, 72°C for 50s followed by elongation at 72°C for 10 minutes were performed.
Sequencing was conducted with Big Dye Terminator sequencing kit v1.1 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions, and sequencing reactions were electrophoresed on an ABI 3130xl genetic analyzer (Applied Biosystems, Carlsbad, CA, USA). Sequencing reactions were repeated at least twice by independent PCR, with forward and reverse primers, and the sample was scored as being mutated when the mutation was observed both times. BRAF primers for amplification of exon 15, used for both PCR and Capillary sequencing were previously described [31].
PNA clamping quantitative PCR analysis
BRAF V600 codon mutational status was tested using the PNAClamp™ BRAF Mutation Detection Kit (Panagene, Daejeon, Korea), a CE-IVD certified technique based on peptide nucleic acid (PNA)-mediated real-time PCR clamping technology. PNA is a synthetic DNA analog in which the phosphodiester backbone is replaced by a peptide-like repeat formed by (2-aminoethyl)-glycine units. The technique is based on the principle that PNA inhibits wild type by hybridizing normal sequences, and therefore mutant DNA is preferentially amplified.
The reaction was performed according to the manufacturer's instructions, with slight modifications, as previously described [31]. The threshold cycle (Ct) was automatically calculated from the PCR amplification plots where fluorescence was plotted against the number of cycles. Delta-Ct values were calculated as the Ct values of the samples minus those of the controls. The higher delta-Ct values showed that the mutant was efficiently amplified. A cut-off value of 2.0 was used to determine the presence of mutant DNA.
Immunohistochemistry
Immunohistochemistry was performed using the BRAF V600E mutation-specific antibody (Springer-Bio, clone VE1, 1:50 dilution). Four micron-thick tissue sections were freshly cut [32], dried, deparaffinised and rehydrated. Endogenous peroxidase was blocked with 5% H2O2 for 10 minutes. Immunoreactions were performed using the automated BenchMark XT immunostainer® (Ventana Medical Systems, Arizona, USA). Standard heat-based antigen retrieval was performed for 30 minutes. The ultraVIEW Universal Alkaline Phosphatase Red Detection Kit (Ventana Medical Systems, Arizona, USA), was used. After immunostaining, slides were counterstained with haematoxylin and coverslipped. All reactions were carried out on the same day on consecutive runs and positive and negative controls were added for each run. Positive and negative controls used were chosen from metastatic samples already characterized for BRAF molecular status for diagnostic purposes (both Capillary sequencing and PNA clamping real-time PCR, as performed by the manufacturer).
All immunostained slides were simultaneously scored by two pathologists (LM and FG) blinded to the BRAF status; disagreement was resolved by consensus. IHC was considered positive when BRAF V600E cytoplasmic protein expression was scored according to the 3 categories score used by Tetzlaff et al [33]. Briefly, negative staining was defined as either absence of any cytoplasmic labeling or nuclear staining only in melanoma cells; positive staining was distinguished in either positive homogeneous (staining in > 95% of cells) or positive heterogeneous (staining in < 95% of cells) based on percentage of cytoplasmic staining in melanoma cells; intensity of staining was also scored as weak, moderate or strong.
Next generation sequencing
Re-analysis of primary and metastatic samples showing discordant results with the other techniques from 3 selected patients was performed by targeted next generation sequencing (NGS) as previously described [31]. Briefly, 10 nanograms of DNA were amplified with an in-house, customized Ion AmpliSeq primer tool, comprising 32 amplicons of 19 genes, targeting 79 hotspot mutations, including the hotspot exon 15 of the BRAF gene. PCR products were ligated to adapters and enriched for target regions using the Ion AmpliSeq™ Library Kit 2.0 according to the manufacturer's instructions (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The samples were quality checked for proper amplicon length and quantity on the 2100 Bioanalyzer Instruments (Agilent Technologies, Santa Clara, California, USA), then sequenced on the Ion Personal Genome Machine™ (PGM™) System. Data were analyzed using the Torrent Suite Software, version 5.0.4.0 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and visualized in the integrative genome viewer (IGV); annotation was performed by Ion Reporter™ Software (Thermo Fisher Scientific, Waltham, Massachusetts, USA).
Inter-rater agreement
To measure agreement between 3 techniques (Sanger sequencing, PNA and IHC) in determining BRAF mutational status, we computed the Light's Kappa coefficient [34]. The analysis was performed using the IRR package [35] within the R computational environment (R Foundation for Statistical Computing, Vienna, Austria) [36].
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Abbreviations
- PNA
peptide nucleic acid –PNA-clamping real-time PCR
- IHC
immunohistochemistry
- NGS
next generation sequencing
- FFPE
formalin-fixed, paraffin embedded
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest. FS received lecture fees from Bristol-Myers Squibb, GlaxoSmithKline, Novartis, MSD and Roche. PQ served as advisor to Bristol-Myers Squibb, Roche-Genentech, MSD, GlaxoSmithKline and Novartis, and received lecture fees from Bristol-Myers Squibb, GlaxoSmithKline, Novartis, MSD and Roche.
GRANT SUPPORT
This study was supported by AIRC IG15460 to PG, Genoa Atheneum FRA to WB and Italian Ministry of Health 5×1000 per la Ricerca corrente to PG, GBS and PQ.
REFERENCES
- 1.Reddy SM, Reuben A, Wargo JA. Influences of BRAF Inhibitors on the Immune Microenvironment and the Rationale for Combined Molecular and Immune Targeted Therapy. Curr Oncol Rep. 2016;18:42. doi: 10.1007/s11912-016-0531-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coit DG, Thompson JA, Algazi A, Andtbacka R, Bichakjian CK, Carson WE, Daniels GA, DiMaio D, Fields RC, Fleming MD, Gastman B, Gonzalez R, Guild V, et al. NCCN Guidelines Insights: Melanoma, Version 3.2016. J Natl Compr Canc Netw. 2016;14:945–58. doi: 10.6004/jnccn.2016.0101. http://www.ncbi.nlm.nih.gov/pubmed/27059193. [DOI] [PubMed] [Google Scholar]
- 3.Pinto C, Normanno N, Ascierto P, Testori A, M Del Vecchio, Chiarion-Sileni V, Maio M, Queirolo P, Clemente C, Taddei GL, Barberis M, Botti G, Collina G, et al. Raccomandazioni per la determinazione dello stato mutazionale di BRAF nel melanoma. AIOM-SIAPEC;
- 4.Hodis E, Watson IR, Kryukov G V, Arold ST, Imielinski M, Theurillat J-P, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, et al. A Landscape of Driver Mutations in Melanoma. Cell. 2012;20:251–26306. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colomba E, Hélias-Rodzewicz Z, Von Deimling A, Marin C, Terrones N, Pechaud D, Surel S, Côté J-FF, Peschaud F, Capper D, Blons H, Zimmermann U, Clerici T, et al. Detection of BRAF p.V600E mutations in melanomas: Comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. J Mol Diagnostics. 2013;15:94–100. doi: 10.1016/j.jmoldx.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Colombino M, Sini M, Lissia A, De Giorgi V, Stanganelli I, Ayala F, Massi D, Rubino C, Manca A, Paliogiannis P, Rossari S, Magi S, Mazzoni L, et al. Discrepant alterations in main candidate genes among multiple primary melanomas. J Transl Med. 2014;12:117. doi: 10.1186/1479-5876-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R TCGA Akbani, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, Arachchi H, Arora A, Auman JT, Ayala B, Baboud J, Balasundaram M, et al. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ihle MA, Fassunke J, König K, Grünewald I, Schlaak M, Kreuzberg N, Tietze L, Schildhaus H-U, Büttner R, Merkelbach-Bruse S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAFmutations. BMC Cancer. 2014;14:13. doi: 10.1186/1471-2407-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spagnolo F, Ghiorzo P, Queirolo P. Overcoming resistance to BRAF inhibition in BRAF-mutated metastatic melanoma. Oncotarget. 2014;5:10206–21. doi: 10.18632/oncotarget.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spagnolo F, Ghiorzo P, Orgiano L, Pastorino L, Picasso V, Tornari E, Ottaviano V, Queirolo P. BRAF-mutant melanoma: Treatment approaches, resistance mechanisms, and diagnostic strategies. Onco Targets Ther. 2015;8:157–68. doi: 10.2147/OTT.S39096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiefer A-I, Parlow L, Gabler L, Mesteri I, Koperek O, von Deimling A, Streubel B, Preusser M, Lehmann A, Kellner U, Pauwels P, Lambin S, Dietel M, et al. Multicenter Evaluation of a Novel Automated Rapid Detection System of BRAF Status in Formalin-Fixed, Paraffin-Embedded Tissues. J Mol Diagn. 2016;18:370–7. doi: 10.1016/j.jmoldx.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Anwar MAF, Murad F, Dawson E, ZY Abd Elmageed, Tsumagari K, Kandil E. Immunohistochemistry as a reliable method for detection of BRAF-V600E mutation in melanoma: a systematic review and meta-analysis of current published literature. J Surg Res. 2016;203:407–15. doi: 10.1016/j.jss.2016.04.029. [DOI] [PubMed] [Google Scholar]
- 13.Pinto R, De Summa S, Strippoli S, Pilato B, Azzariti A, Guida G, Guida M, Tommasi S. The next generation of metastatic melanoma: Uncovering the genetic variants for anti-BRAF therapy response. Oncotarget. 2016. [DOI] [PMC free article] [PubMed]
- 14.Eigentler T, Assi Z, Hassel JC, Heinzerling L, Starz H, Berneburg M, Bauer J, Garbe C. Which melanoma patient carries a BRAF-mutation? A comparison of predictive models. Oncotarget. 2016;7:36130–7. doi: 10.18632/oncotarget.9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verlinden I, van den Hurk K, Clarijs R, Willig AP, a Stallinga CMH, Roemen GMJM, van den Oord JJ, A zur Hausen, Speel E-JM, Winnepenninckx VJL. BRAFV600E immunopositive Melanomas Show Low Frequency of Heterogeneity and Association With Epithelioid Tumor Cells. Medicine (Baltimore) 2014;93:e285. doi: 10.1097/MD.0000000000000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. Europe PMC Funders; 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menzies AM, Lum T, Wilmott JS, Hyman J, Kefford RF, Thompson JF, Long G V, Scolyer RA. Intrapatient Homogeneity of BRAF. Am J Surg Pathol. 2014;38:377–82. doi: 10.1097/PAS.0000000000000136. [DOI] [PubMed] [Google Scholar]
- 18.Heinzerling L, Baiter M, Kühnapfel S, Schuler G, Keikavoussi P, Agaimy A, Kiesewetter F, Hartmann A, Schneider-Stock R. Mutation landscape in melanoma patients clinical implications of heterogeneity of BRAF mutations. Br J Cancer. 2013;109:2833–41. doi: 10.1038/bjc.2013.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manfredi L, Meyer N, Tournier E, Grand D, Uro-Coste E, Rochaix P, Brousset P, Lamant L. Highly Concordant Results Between Immunohistochemistry and Molecular Testing of Mutated V600E BRAF in Primary and Metastatic Melanoma. Acta Derm Venereol. 2015;96:630–4. doi: 10.2340/00015555-2326. [DOI] [PubMed] [Google Scholar]
- 20.Guadarrama-Orozco J, Ortega-Gómez A, Ruiz-García E, H Astudillo-de la Vega, Meneses-García A, Lopez-Camarillo C. Braf V600E mutation in melanoma: translational current scenario. Clin Transl Oncol. 2016. [DOI] [PubMed]
- 21.Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, Pavlick AC, Darvishian F, Christos P, Mazumdar M, Osman I, Polsky D. Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PLoS One. 2012;7:e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De Giorgi V, Massi D, Fonsatti E, Staibano S, Nappi O, Pagani E, Casula M, Manca A, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30:2522–9. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 23.Boursault L, Haddad V, Vergier B, Cappellen D, Verdon S, Bellocq J-P, Jouary T, Merlio J-P. Tumor Homogeneity between Primary and Metastatic Sites for BRAF Status in Metastatic Melanoma Determined by Immunohistochemical and Molecular Testing. Chammas R, editor. PLoS One. 201(8):e70826. doi: 10.1371/journal.pone.0070826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riveiro-Falkenbach E, Villanueva C a, Garrido MC, Ruano Y, García-Martín RM, Godoy E, Ortiz-Romero PL, Ríos-Martín JJ, Santos-Briz A, Rodríguez-Peralto JL. Intra- and Inter-tumoral Homogeneity of BRAFV600E Mutations in Melanoma Tumors. J Invest Dermatol. 2015. [DOI] [PubMed]
- 25.Ritterhouse LL, Barletta JA. BRAF V600E mutation-specific antibody: A review. Semin Diagn Pathol. 2015;32:400–8. doi: 10.1053/j.semdp.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Bruno W, Ghiorzo P, Battistuzzi L, Ascierto PA, Barile M, Gargiulo S, Gensini F, Gliori S, Guida M, Lombardo M, Manoukian S, Menin C, Nasti S, et al. Clinical genetic testing for familial melanoma in Italy: A cooperative study. J Am Acad Dermatol. 2009;61:775–82. doi: 10.1016/j.jaad.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 27.Ghiorzo P, Pastorino L, Queirolo P, Bruno W, Tibiletti MG, Nasti S, Andreotti V, Paillerets BB, Bianchi Scarrà G. Prevalence of the E318K MITF germline mutation in Italian melanoma patients: associations with histological subtypes and family cancer history. Pigment Cell Melanoma Res. 2013;26:259–62. doi: 10.1111/pcmr.12047. [DOI] [PubMed] [Google Scholar]
- 28.Bruno W, Pastorino L, Ghiorzo P, Andreotti V, Martinuzzi C, Menin C, Elefanti L, Stagni C, Vecchiato A, Rodolfo M, Maurichi A, Manoukian S, De Giorgi V, et al. Multiple primary melanomas (MPMs) and criteria for genetic assessment: MultiMEL, a multicenter study of the Italian Melanoma Intergroup. J Am Acad Dermatol. 2016;74:325–32. doi: 10.1016/j.jaad.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 29.Ghiorzo P, Gargiulo S, Pastorino L, Nasti S, Cusano R, Bruno W, Gliori S, Sertoli MR, Burroni A, Savarino V, Gensini F, Sestini R, Queirolo P, et al. Impact of E27X, a novel CDKN2A germ line mutation, on p16 and p14ARF expression in Italian melanoma families displaying pancreatic cancer and neuroblastoma. Hum Mol Genet. 2006;15:2682–9. doi: 10.1093/hmg/ddl199. [DOI] [PubMed] [Google Scholar]
- 30.Origone P, Gargiulo S, Mastracci L, Ballestrero A, Battistuzzi L, Casella C, Comandini D, Cusano R, AP Dei Tos, Fiocca R, Garuti A, Ghiorzo P, Martinuzzi C, et al. Molecular characterization of an Italian series of sporadic GISTs. Gastric cancer. 2013;16:596–601. doi: 10.1007/s10120-012-0213-y. [DOI] [PubMed] [Google Scholar]
- 31.Martinuzzi C, Pastorino L, Andreotti V, Garuti A, Minuto M, Fiocca R, Bianchi-scarrà G, Ghiorzo P, Grillo F, Mastracci L. A combination of immunohistochemistry and molecular approaches improves highly sensitive detection of BRAF mutations in papillary thyroid cancer. Endocrine. 2015. [DOI] [PubMed]
- 32.Grillo F, Pigozzi S, Ceriolo P, Calamaro P, Fiocca R, Mastracci L. Factors affecting immunoreactivity in long-term storage of formalin-fixed paraffin-embedded tissue sections. Histochem Cell Biol. 2015. [DOI] [PubMed]
- 33.Tetzlaff MT, Pattanaprichakul P, Wargo J, Fox PS, Patel KP, Estrella JS, Broaddus RR, Williams MD, Davies M, Routbort MJ, Lazar AJ, Woodman SE, Hwu W-J, et al. Utility of BRAF V600E Immunohistochemistry Expression Pattern as a Surrogate of BRAF Mutation Status in 154 Patients with Advanced Melanoma. Hum Pathol. 2015;46:1101–10. doi: 10.1016/j.humpath.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Light RJ. Measures of response agreement for qualitative data: Some generalizations and alternatives. Psychol Bull. 1971;76:365–77. [Google Scholar]
- 35.Matthias G, Lemon J, Fellows I, Puspendra S. IRR: Various Coefficients of Interrater Reliability and Agreement. R package version 0.84. 2012. https://cran.r-project.org/package=irr.
- 36.Core Team RR. R Foundation for Statistical Computing. Vienna, Austria: 2016. A language and environment for statistical computing. https://www.r-project.org/ [Google Scholar]
- 37.Chen T-L, Chang JW-C, Hsieh J-J, Cheng H-Y, Chiou C-C. A Sensitive Peptide Nucleic Acid Probe Assay for Detection of BRAF V600 Mutations in Melanoma. Cancer Genomics Proteomics. 2016;13:381–6. http://www.ncbi.nlm.nih.gov/pubmed/27566656. [PMC free article] [PubMed] [Google Scholar]
- 38.Kang S-H, Pyo JY, Yang S-W, Hong SW. Detection of BRAF V600E mutation with thyroid tissue using pyrosequencing: comparison with PNA-clamping and real-time PCR. Am J Clin Pathol. 2013;139:759–64. doi: 10.1309/AJCPN3ULH6YWBHPH. [DOI] [PubMed] [Google Scholar]
- 39.Ahn TS, Jeong D, Won M, · Haeil S, · Soyoung J, · Hyungjoo P, · Sang K, Bae B, Kim HJ, Jeon Y, · Moon, Lee S, Baek M. The BRAF mutation is associated with the prognosis in colorectal cancer. J Cancer Res Clin Oncol. 2014;140:1863–71. doi: 10.1007/s00432-014-1735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeo M-K, Jung M-K, Lee S-Y, Lee Y-M, Hur GM, Kim J-M. The usefulness of a novel fully automated PCR-based Idylla test for detection of the BRAF V600E mutation in thyroid tissue: comparison with PNA-clamping PCR, real-time PCR and pyrosequencing. J Clin Pathol. 2016. jclinpath-2016-204025. [DOI] [PubMed]
- 41.Sullivan RJ, Fisher DE. Understanding the biology of melanoma and therapeutic implications. Hematol Oncol Clin North Am. 2014;28:437–53. doi: 10.1016/j.hoc.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.FA San Lucas, Allenson K, Bernard V, Castillo J, Kim D, Ellis K, Ehli EA, Davies GE, Petersen JL, Li D, Wolff R, Katz M, Varadhachary G, et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol. 2015;26:2361–2. doi: 10.1093/annonc/mdv604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J, Goto Y, Murata H, Sakaizawa K, Uchiyama A, Saida T, Takata M. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br J Cancer. Nature Publishing Group; 2011;104:464–8. doi: 10.1038/sj.bjc.6606072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiappetta C, Proietti I, Soccodato V, Puggioni C, Zaralli R, Pacini L, Porta N, Skroza N, Petrozza V, Potenza C, C Della Rocca, Di Cristofano C. BRAF and NRAS mutations are heterogeneous and not mutually exclusive in nodular melanoma. Appl Immunohistochem Mol Morphol. 2015;23:172–7. doi: 10.1097/PAI.0000000000000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satzger I, Marks L, Kerick M, Klages S, Berking C, Herbst R, Völker B, Schacht V, Timmermann B, Gutzmer R. Allele frequencies of BRAFV600 mutations in primary melanomas and matched metastases and their relevance for BRAF inhibitor therapy in metastatic melanoma. Oncotarget. 2015;6:37895–905. doi: 10.18632/oncotarget.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casula M, Colombino M, Manca A, Caracò C, Botti G, Ascierto PA, Lissia A, Cossu A, Palmieri G. Low Levels of Genetic Heterogeneity in Matched Lymph Node Metastases from Patients with Melanoma. J Invest Dermatol. 2016;136:1917–20. doi: 10.1016/j.jid.2016.05.103. [DOI] [PubMed] [Google Scholar]
- 47.Long G V., Wilmott JS, Capper D, Preusser M, Zhang YE, Thompson JF, Kefford RF, von Deimling A, Scolyer RA. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am J Surg Pathol. 2013;37:61–5. doi: 10.1097/PAS.0b013e31826485c0. [DOI] [PubMed] [Google Scholar]
- 48.Kakavand H, Walker E, Lum T, Wilmott JS, Selinger CI, Smith E, Saw RPM, Yu B, Cooper WA, Long G V., O’Toole SA, Scolyer RA. BRAF V600E and NRAS Q61L/Q61R mutation analysis in metastatic melanoma using immunohistochemistry: a study of 754 cases highlighting potential pitfalls and guidelines for interpretation and reporting. Histopathology. 2016. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


