Abstract
The genomic sequences of Salmonella enterica subsp. enterica strains CT18, Ty2 (serovar Typhi), and LT2 (serovar Typhimurium) were analyzed for potential variable number tandem repeats (VNTRs). A multiple-locus VNTR analysis (MLVA) of 99 strains of S. enterica supsp. enterica based on 10 VNTRs distinguished 52 genotypes and placed them into four groups. All strains tested were independent human isolates from France and did not reflect isolates from outbreak episodes. Of these 10 VNTRs, 7 showed variability within serovar Typhi, whereas 1 showed variability within serovar Typhimurium. Four VNTRs showed high Nei's diversity indices (DIs) of 0.81 to 0.87 within serovar Typhi (n = 27). Additionally, three of these more variable VNTRs showed DIs of 0.18 to 0.58 within serovar Paratyphi A (n = 10). The VNTR polymorphic site within multidrug-resistant (MDR) serovar Typhimurium isolates (n = 39; resistance to ampicillin, chloramphenicol, spectinomycin, sulfonamides, and tetracycline) showed a DI of 0.81. Cluster analysis not only identified three genetically distinct groups consistent with the present serovar classification of salmonellae (serovars Typhi, Paratyphi A, and Typhimurium) but also discriminated 25 subtypes (93%) within serovar Typhi isolates. The analysis discriminated only eight subtypes within serovar Typhimurium isolates resistant to ampicillin, chloramphenicol, spectinomycin, sulfonamides, and tetracycline, possibly reflecting the emergence in the mid-1990s of the DT104 phage type, which often displays such an MDR spectrum. Coupled with the ongoing improvements in automated procedures offered by capillary electrophoresis, use of these markers is proposed in further investigations of the potential of MLVA in outbreaks of salmonellosis, especially outbreaks of typhoid fever.
Repetitive DNA is abundant in eucaryotic cells and has been increasingly described in procaryotes. Tandem repeats that represent a single locus and that show interindividual length polymorphisms are called “variable number tandem repeats” (VNTRs). VNTRs and other short-sequence DNA tandem repeats in procaryotic genomes appear to provide useful information on both the functional and the evolutionary aspects of bacterial genetic diversity (34). All bacterial genome sequences released so far contain tandem repeats at various densities, depending on the species (17). The increasing availability of whole-genome sequences is an invaluable source of VNTRs, which has opened the way to multiple-locus VNTR analysis (MLVA) for the typing of bacteria. MLVAs have been proposed so far for Bacillus anthracis (17), Yersinia pestis (27), Francisella tularensis (10), Mycobacterium tuberculosis (16), Legionella pneumophila (28), Pseudomonas aeruginosa (24), Escherichia coli O157:H7 (19), and Salmonella enterica subsp. enterica serovars Typhimurium (18) and Typhi (20).
Serovars of S. enterica subsp. enterica include pathogens that differ widely in their host range spectra (for a review, see reference 3). Among these are serovar Typhimurium, a common cause of salmonellosis that affects humans and animals worldwide, and serovar Typhi, a food- or waterborne life-threatening illness that affects 17 million people each year, with approximately 600,000 deaths (5). The internationally standardized Vi phage typing system described by Craigie and Yen (6) defines more than 100 Vi phage types of serovar Typhi, and similar systems have been developed for serovar Typhimurium (2). The stability of phage type provides an opportunity to monitor the spread of a clone over decades. Adversely, most isolates are distributed among only a few phage types. For example, it has been demonstrated that the DT104 phage type of multidrug-resistant (MDR) serovar Typhimurium is a single clone that has spread pandemically in Europe (30). Similarly, 42 of 48 isolates of MDR serovar Typhi isolates were assigned to Vi phage type E1 (14). The introduction of molecular typing methods, such as ribotyping (1), randomly amplified polymorphic DNA analysis (9), amplified fragment length polymorphism analysis (12), and pulsed-field gel electrophoresis (PFGE) (33), has greatly improved the ability to discriminate between epidemiologically related and unrelated isolates in outbreaks of typhoid fever (29) and other salmonelloses (26). PFGE remains a valuable investigational tool because its high discriminatory power allows investigators to make decisions of epidemiological importance.
However, all of these methods suffer from one or more significant drawbacks, including insufficient discriminatory power, poor reproducibility between laboratories, and difficulties with the comparison and accumulation of results by different laboratories. Multilocus sequence typing (MLST) (21) and its recent introduction for the characterization of Salmonella strains (15) has opened the way to standardization of data handling and the development of web-based resources for querying databases. When applicable, MLVA typing is very low cost, is accessible to any laboratory equipped with minimum molecular biology equipment, and is open to large-scale standardization. Moreover, MLVA remains fully compatible with automated fragment size determination by capillary electrophoresis and unambiguous nucleotide sequence determination to those who have these resources. Recently, the nonfastidious VNTR-based DNA fingerprinting of Salmonella has demonstrated the potential of MLVA for the typing of S. enterica subsp. enterica serotypes Typhimurium (18) and Typhi (20). In the context of bioterrorism countermeasure deployment or, more importantly, for epidemiological follow-up in less developed countries, the present report contributes to the development of MLVA through the evaluation of multiple VNTR markers for the discrimination of about a hundred S. enterica subsp. enterica isolates, with an emphasis on both serovars Typhi and Typhimurium.
MATERIALS AND METHODS
Strains and DNA preparation.
Ninety-nine strains of S. enterica subsp. enterica, including serovars Typhi (27 isolates) and Typhimurium (39 isolates) as well as other serovars (33 isolates), were analyzed (Fig. 1). Twenty-six serovar Typhimurium isolates were originally isolated from human stool samples in the context of the control of disease in symptomatic and nonsymptomatic food handlers in restaurants. Twenty-six serovar Typhi isolates and nine serovar Paratyphi A isolates were originally isolated from blood or stool samples from symptomatic patients. All were single patient isolates. The travel histories of the individuals were unknown because the original clinical records were not available. Isolation was done locally by French regional military health laboratories (Aquitaine, Bretagne, Ile de France, Lorraine, Midi-Pyrénées, Basse Normandie, Provence-Alpes-Côte d'Azur, Rhône-Alpes, and, to a lesser extent, laboratories overseas), as were identification by standard techniques (with the API 20E system and by serotyping) and antibiotic susceptibility testing with disks. All serovar Typhimurium isolates were ampicillin, chloramphenicol, spectinomycin, sulfonamide, and tetracycline resistant (commonly abbreviated ACSSuT). The phage types of the ACSSuT Typhimurium isolates were not determined. Original isolates cultured on nutrient agar slopes were sent to the clinical microbiology laboratory of the Bégin Military Hospital (Saint Mandé, France) and stored frozen at −80°C in 10% glycerol until they were needed.
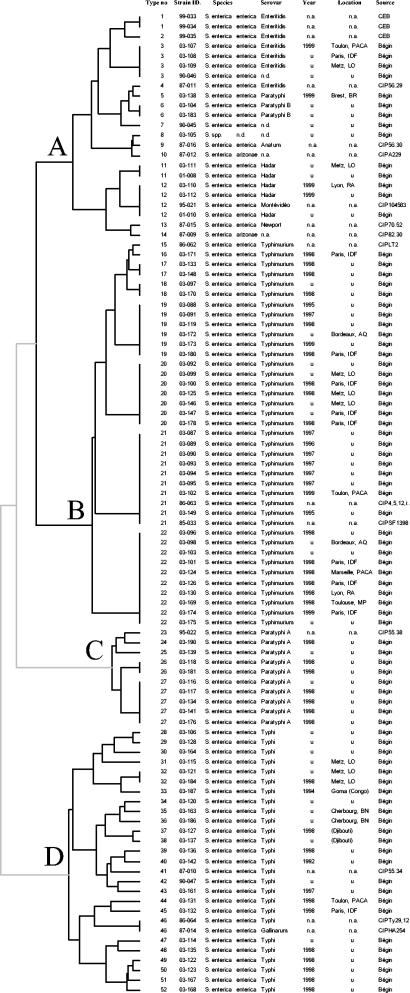
FIG. 1.
Dendrogram deduced from cluster analysis of the 99 isolates obtained from Bégin Military Hospital, Saint Mandé, France (Bégin); CEB; and the Collection de l'Institut Pasteur (CIP). n.d., not determined; n.a., not applicable; u, unknown (see also Materials and Methods); AQ, Aquitaine; BR, Bretagne; IDF, Ile de France; LO, Lorraine; MP, Midi-Pyrénées; BN, Basse Normandie; PACA, Provence-Alpes-Côte d'Azur; RA, Rhône-Alpes.
Three strains of Salmonella serovar Enteritidis were from the collection of bacterial strains maintained by the Centre d'Études du Bouchet (CEB), Vert le Petit, France. They consisted of three distinct preservations of isolates that have been recovered after the 2nd, 44th, and 99th inoculations of guinea pigs, respectively, in the 1960s. Other serovars were purchased at different times from the Collection de l'Institut Pasteur (Paris, France).
DNA was isolated with the MagNa Pure system and the MagNa Pure LC DNA isolation kit III (Roche Diagnostics, Meylan, France), according to the instructions of the manufacturer. The DNA was extracted from the collection strains in larger amounts by a method described elsewhere (36).
Identification of tandem repeats.
The complete genome sequences of Salmonella serovar Typhi CT18 (25) and Ty2 (7) and of Salmonella serovar Typhimurium LT2 (23) were analyzed by using the tandem repeats database described elsewhere (8, 17) and accessible at http://minisatellites.u-psud.fr to identify tandem repeats with different sizes in the two genomes. Tandem repeats were named according to their chromosomal locations and by use of the Salmonella serovar Typhimurium LT2 genome. When applicable, previously described VNTRs were named according to their original nomenclature (18, 20).
VNTR amplification and genotyping.
Primer sets specific for potential VNTR loci present in the CT18, Ty2, or LT2 sequence were designed with Primer3 software (31). The theoretical sizes of the amplicons were determined with the BLAST program by comparison with the CT18, Ty2, or LT2 sequence at http://minisatellites.u-psud.fr. Amplifications were performed in mixtures of 25 μl containing 1 ng of DNA, 1 U of Taq DNA polymerase (Qbiogen, Illkirch, France), 0.5 μM each flanking primer, 200 μM each deoxynucleoside triphosphate, and 1× incubation mixture with 2 mM MgCl2 (Qbiogen). PCRs were run on a GeneAmp 9600 PCR system (Roche Diagnostics). An initial denaturation at 96°C for 5 min was followed by 30 cycles of a three-step cycling protocol (96°C for 30 s, 62°C for 1 min, and 72°C for 1 min) and a final elongation step at 72°C for 10 min. PCR products (2 to 5 μl) were run on 2% standard agarose gel (ICN Biomedicals, Orsay, France) in 0.5× TBE buffer (10× TBE is 890 mM Tris base, 890 mM boric acid, and 20 mM EDTA [pH 8.3]) at 10 V/cm. Samples were manipulated and dispensed with multichannel pipettes (Biohit, Bonnelles, France), which were also used for gel loading, in order to reduce the risk of errors. Gel lengths of 20 cm were used. Gels were stained with ethidium bromide, visualized under UV light, and photographed. Allele sizes were estimated by using a 20-bp ladder (Bio-Rad, Marnes la Coquette, France) as a size marker. Each 50 wells of gel contained eight regularly spaced size marker lanes. In addition, strain LT2 was included as a control for size assignments (one LT2 control for each set of five DNA samples), as described previously (16).
Data analysis.
Tagged image file format files of the gels and the resulting data were managed with the BioNumerics software package (version 3.5; Applied-Maths, Sint-Martens-Latem, Belgium) to estimate the sizes of the alleles. Allele sizes were converted into motif copy numbers in the tandem array, imported into BioNumerics software, and then subjected to cluster analysis by using the categorical coefficient and Ward clustering parameters. This implies that an equivalent weight is given to any multistate character at any locus, whatever the repeat number is. The polymorphism information index or Nei's diversity index (DI) was calculated for each marker as 1 − Σ(allele frequency)2.
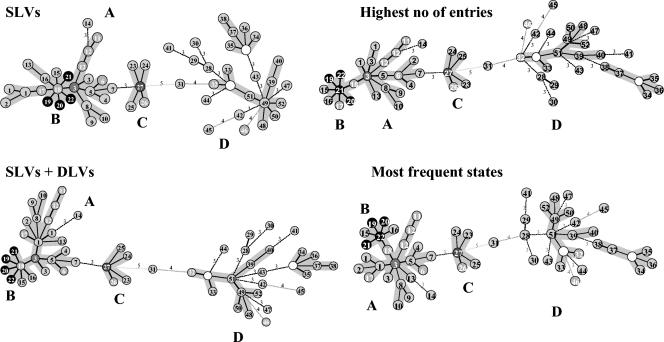
The categorical coefficient was used to calculate the minimum-spanning tree (MST) with BioNumerics software. The creation of hypothetical genotypes was allowed. When solutions with identical calculated distances were obtained, BioNumerics software applies a priority rule based on criteria other than distance. Four priority rules are available and were tested: (i) the highest number of single-locus variants (SLVs; when two types have an equal distance to a linkage position in the tree, the type that has the highest number of SLVs is linked first), (ii) the highest number of SLVs and double-locus variants (DLVs; when two types have an equal distance to a linkage position in the tree, the types that differ in two states are considered equally), (iii) the highest number of entries (the program counts how many entries that each unique type contains; when equivalent linkage possibilities exist, the type that has the highest number of entries is linked first), and (iv) the most frequent state (the program calculates a frequency table for each state of characters, and types are ranked on the basis of the frequencies of their characters; when equivalent possibilities exist, the types that have the highest rank are linked first).
The absence of allele at a given locus (i.e., no amplification, despite repeated attempts) is neutral for cluster analysis (i.e., types 1, 3, and 12) (Table 3 and Fig. 1). When the MST is constructed, the absence of an allele was considered as a result (zero) by the BioNumerics software; thus, a distance of 1 is calculated between two strains that differ only by the absence of one allele in one strain (i.e., types 1, 3, and 12; see Fig. 3).
TABLE 3.
Numbers of repetitions at 10 loci for 99 S. enterica subsp. enterica strains
| Group | Serovar | Type | Strain | No. of repetitions at the following locus:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sal02 | Sal04 | Sal06 | Sal10 | TR1c | Sal15 | STTR5b | Sal20 | Sal22 | Sal23 | ||||
| A | Enteritidis | 1 | 99-033 | 3 | 1 | —a | 2 | 2 | 3 | 11 | 10 | 3 | 4 |
| A | Enteritidis | 1 | 99-034 | 3 | 1 | 3 | 2 | 2 | 3 | 11 | 10 | 3 | 4 |
| A | Enteritidis | 2 | 99-035 | 3 | 1 | 5 | 2 | 2 | 3 | 11 | 10 | 3 | 5 |
| A | Enteritidis | 3 | 03-107 | 3 | 1 | — | 2 | 2 | 3 | 12 | 10 | 3 | 4 |
| A | Enteritidis | 3 | 03-108 | 3 | 1 | 3 | 2 | 2 | 3 | 12 | 10 | 3 | 4 |
| A | Enteritidis | 3 | 03-109 | 3 | 1 | 3 | 2 | 2 | 3 | 12 | 10 | 3 | 4 |
| A | NDd | 3 | 90-046 | 3 | 1 | 3 | 2 | 2 | 3 | 12 | 10 | 3 | 4 |
| A | Enteritidis | 4 | 87-011 | 3 | 1 | 3 | 2 | 2 | 3 | 8 | 10 | 3 | 5 |
| A | Paratyphi B | 5 | 03-138 | 3 | 1 | 3 | 2 | 2 | 3 | 12 | 10 | 3 | 5 |
| A | Paratyphi B | 6 | 03-104 | 3 | 1 | 3 | 2 | 2 | 3 | 13 | 10 | 3 | 5 |
| A | Paratyphi B | 6 | 03-183 | 3 | 1 | 3 | 2 | 2 | 3 | 13 | 10 | 3 | 5 |
| A | ND | 7 | 90-045 | 3 | 1 | 3 | 2 | 2 | 3 | 10 | 9 | 3 | 5 |
| A | ND | 8 | 03-105 | 3 | 1 | 3 | 2 | 2 | 2 | 24 | 10 | 3 | 4 |
| A | Anatum | 9 | 87-016 | 3 | 1 | 3 | 2 | 2 | 2 | 19 | 10 | 3 | 4 |
| A | NAe | 10 | 87-012 | — | 1 | 3 | 2 | 2 | 2 | 7 | 10 | 3 | 4 |
| A | Hadar | 11 | 03-111 | 3 | 1 | — | 2 | 2 | 3 | 23 | 11 | 3 | 4 |
| A | Hadar | 11 | 01-008 | 3 | 1 | — | 2 | 2 | 3 | 23 | 11 | 3 | 4 |
| A | Hadar | 12 | 03-110 | 3 | 1 | 3 | 2 | 2 | 3 | 22 | 11 | 3 | 4 |
| A | Hadar | 12 | 03-112 | 3 | 1 | — | 2 | 2 | 3 | 22 | 11 | 3 | 4 |
| A | Montevideo | 12 | 95-021 | 3 | 1 | 3 | 2 | 2 | 3 | 22 | 11 | 3 | 4 |
| A | Hadar | 12 | 01-010 | 3 | 1 | — | 2 | 2 | 3 | 22 | 11 | 3 | 4 |
| A | Newport | 13 | 87-015 | 3 | 1 | 3 | 2 | 2 | 3 | 9 | 15 | 3 | 4 |
| A | NA | 14 | 87-009 | — | 1 | — | 2 | 2 | 3 | 9 | 12 | 3 | 4 |
| B | Typhimurium | 15 | 86-062 | 3 | 1 | 3 | 3 | 2 | 3 | 13 | 10 | 3 | 4 |
| B | Typhimurium | 16 | 03-171 | 3 | 1 | 3 | 3 | 2 | 3 | 9 | 10 | 3 | 4 |
| B | Typhimurium | 17 | 03-133 | 3 | 1 | 3 | 3 | 2 | 3 | 11 | 10 | 3 | 4 |
| B | Typhimurium | 17 | 03-148 | 3 | 1 | 3 | 3 | 2 | 3 | 11 | 10 | 3 | 4 |
| B | Typhimurium | 18 | 03-097 | 3 | 1 | 3 | 3 | 2 | 3 | 12 | 10 | 3 | 4 |
| B | Typhimurium | 18 | 03-170 | 3 | 1 | 3 | 3 | 2 | 3 | 12 | 10 | 3 | 4 |
| B | Typhimurium | 19 | 03-088 | 3 | 1 | 3 | 3 | 2 | 3 | 17 | 10 | 3 | 4 |
| B | Typhimurium | 19 | 03-091 | 3 | 1 | 3 | 3 | 2 | 3 | 17 | 10 | 3 | 4 |
| B | Typhimurium | 19 | 03-119 | 3 | 1 | 3 | 3 | 2 | 3 | 17 | 10 | 3 | 4 |
| B | Typhimurium | 19 | 03-172 | 3 | 1 | 3 | 3 | 2 | 3 | 17 | 10 | 3 | 4 |
| B | Typhimurium | 19 | 03-173 | 3 | 1 | 3 | 3 | 2 | 3 | 17 | 10 | 3 | 4 |
| B | Typhimurium | 19 | 03-180 | 3 | 1 | 3 | 3 | 2 | 3 | 17 | 10 | 3 | 4 |
| B | Typhimurium | 20 | 03-092 | 3 | 1 | 3 | 3 | 2 | 3 | 14 | 10 | 3 | 4 |
| B | Typhimurium | 20 | 03-099 | 3 | 1 | 3 | 3 | 2 | 3 | 14 | 10 | 3 | 4 |
| B | Typhimurium | 20 | 03-100 | 3 | 1 | 3 | 3 | 2 | 3 | 14 | 10 | 3 | 4 |
| B | Typhimurium | 20 | 03-125 | 3 | 1 | 3 | 3 | 2 | 3 | 14 | 10 | 3 | 4 |
| B | Typhimurium | 20 | 03-146 | 3 | 1 | 3 | 3 | 2 | 3 | 14 | 10 | 3 | 4 |
| B | Typhimurium | 20 | 03-147 | 3 | 1 | 3 | 3 | 2 | 3 | 14 | 10 | 3 | 4 |
| B | Typhimurium | 20 | 03-178 | 3 | 1 | 3 | 3 | 2 | 3 | 14 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 03-087 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 03-089 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 03-090 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 03-093 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 03-094 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 03-095 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 03-102 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 86-063 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 03-149 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 21 | 85-033 | 3 | 1 | 3 | 3 | 2 | 3 | 15 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-096 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-098 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-103 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-101 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-124 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-126 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-130 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-169 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-174 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| B | Typhimurium | 22 | 03-175 | 3 | 1 | 3 | 3 | 2 | 3 | 16 | 10 | 3 | 4 |
| C | Paratyphi A | 23 | 95-022 | 15 | 1 | 3 | 1 | 2 | 3 | 8 | 9 | 4 | 5 |
| C | Paratyphi A | 24 | 03-190 | 12 | 1 | 3 | 1 | 2 | 3 | 9 | 9 | 4 | 5 |
| C | Paratyphi A | 25 | 03-139 | 8 | 1 | 3 | 1 | 2 | 3 | 10 | 10 | 4 | 5 |
| C | Paratyphi A | 26 | 03-118 | 9 | 1 | 3 | 1 | 2 | 3 | 11 | 9 | 4 | 5 |
| C | Paratyphi A | 26 | 03-181 | 9 | 1 | 3 | 1 | 2 | 3 | 11 | 9 | 4 | 5 |
| C | Paratyphi A | 27 | 03-116 | 9 | 1 | 3 | 1 | 2 | 3 | 10 | 9 | 4 | 5 |
| C | Paratyphi A | 27 | 03-117 | 9 | 1 | 3 | 1 | 2 | 3 | 10 | 9 | 4 | 5 |
| C | Paratyphi A | 27 | 03-134 | 9 | 1 | 3 | 1 | 2 | 3 | 10 | 9 | 4 | 5 |
| C | Paratyphi A | 27 | 03-141 | 9 | 1 | 3 | 1 | 2 | 3 | 10 | 9 | 4 | 5 |
| C | Paratyphi A | 27 | 03-176 | 9 | 1 | 3 | 1 | 2 | 3 | 10 | 9 | 4 | 5 |
| D | Typhi | 28 | 03-106 | 9 | 2 | 5 | 2 | 9 | 2 | 15 | 15 | 4 | 5 |
| D | Typhi | 29 | 03-128 | 9 | 2 | 5 | 2 | 9 | 2 | 13 | 15 | 4 | 5 |
| D | Typhi | 30 | 03-164 | 10 | 2 | 6 | 2 | 9 | 2 | 12 | 15 | 4 | 5 |
| D | Typhi | 31 | 03-115 | 9 | 2 | 3 | 4 | 13 | 2 | 10 | 15 | 4 | 5 |
| D | Typhi | 32 | 03-121 | 11 | 2 | 5 | 2 | 13 | 2 | 16 | 15 | 4 | 5 |
| D | Typhi | 32 | 03-184 | 11 | 2 | 5 | 2 | 13 | 2 | 16 | 15 | 4 | 5 |
| D | Typhi | 33 | 03-187 | 11 | 2 | 5 | 1 | 17 | 2 | 16 | 15 | 4 | 5 |
| D | Typhi | 34 | 03-120 | 14 | 2 | 5 | 2 | 10 | 2 | 10 | 20 | 4 | 5 |
| D | Typhi | 35 | 03-163 | 13 | 2 | 5 | 2 | 10 | 2 | 10 | 18 | 4 | 5 |
| D | Typhi | 36 | 03-186 | 14 | 2 | 5 | 2 | 10 | 2 | 11 | 21 | 4 | 5 |
| D | Typhi | 37 | 03-127 | 14 | 2 | 5 | 2 | 8 | 2 | 12 | 18 | 4 | 5 |
| D | Typhi | 38 | 03-137 | 14 | 2 | 5 | 2 | 8 | 2 | 12 | 17 | 4 | 5 |
| D | Typhi | 39 | 03-136 | 16 | 2 | 5 | 2 | 17 | 2 | 14 | 17 | 4 | 5 |
| D | Typhi | 40 | 03-142 | 16 | 2 | 5 | 2 | 12 | 2 | 13 | 17 | 4 | 5 |
| D | Typhi | 41 | 87-010 | 12 | 2 | 5 | 2 | 7 | 2 | 13 | 19 | 4 | 5 |
| D | Typhi | 42 | 90-047 | 11 | 2 | 5 | 2 | 11 | 2 | 14 | 14 | 4 | 5 |
| D | Typhi | 43 | 03-161 | 14 | 2 | 5 | 2 | 15 | 2 | 14 | 13 | 4 | 5 |
| D | Typhi | 44 | 03-131 | 12 | 2 | 6 | 2 | 17 | 2 | 16 | 21 | 4 | 5 |
| D | Typhi | 45 | 03-132 | 18 | 2 | 6 | 2 | 18 | 2 | 17 | 14 | 4 | 5 |
| D | Typhi | 46 | 86-064 | 15 | 2 | 8 | 2 | 12 | 2 | 16 | 16 | 4 | 5 |
| D | Gallinarum | 46 | 87-014 | 15 | 2 | 8 | 2 | 12 | 2 | 16 | 16 | 4 | 5 |
| D | Typhi | 47 | 03-114 | 18 | 2 | 4 | 2 | 15 | 2 | 14 | 16 | 4 | 5 |
| D | Typhi | 48 | 03-135 | 17 | 2 | 4.5 | 2 | 17 | 2 | 15 | 16 | 4 | 5 |
| D | Typhi | 49 | 03-122 | 17 | 2 | 5 | 2 | 17 | 2 | 14 | 16 | 4 | 5 |
| D | Typhi | 50 | 03-123 | 17 | 2 | 5 | 2 | 16 | 2 | 14 | 16 | 4 | 5 |
| D | Typhi | 51 | 03-167 | 17 | 2 | 5 | 2 | 17 | 2 | 14 | 15 | 4 | 5 |
| D | Typhi | 52 | 03-168 | 17 | 2 | 5 | 2 | 17 | 3 | 14 | 16 | 4 | 5 |
FIG. 3.
Salmonella population modeling. The number in each circle indicates the genotype identified in Table 3. The number of isolates for a given genotype is arbitrarily visualized by different cell shadings or fonts: gray or black, one isolate; sequential increase in shading from gray to black circles with white numbers, two, three, four, and five or more isolates. The empty circle indicates a hypothetical genotype not present in the population analyzed. The distance between neighboring genotypes is expressed as the number of allelic changes and is outlined by different shapes of lines: short bold line, one change; long thin line, two changes; black dotted lines, three to five changes, as indicated.
RESULTS
The resource described previously (8) and accessible from http://minisatellites.u-psud.fr was used to explore the complete genome sequences of serovars Typhi (CT18 and Ty2) and Typhimurium (LT2). The tandem repeats detected predominantly had a repeat unit length with multiples of 3 over a range of 15 to 24 bp. For example, 7, 23, 104, and 7 tandem repeats with unit lengths of 6, 12, 21, and 30 bp, respectively, were detected in the CT18 genome. The Ty2 genome harbors 4, 30, 106, and 5 tandem repeats of the corresponding lengths; and the LT2 genome harbors 5, 25, 117, and 9 tandem repeats of the corresponding lengths.
All VNTR loci considered herein were located on the chromosome. Fourteen, 33, and 35 tandem repeats with repeat units longer than 6 bp were predicted to be polymorphic between CT18 and Ty2, CT18 and LT2, and Ty2 and LT2, respectively. Six tandem repeats were predicted to be of different lengths in all three strains. Altogether, 41 tandem repeats with two to three distinct alleles among the three genomes were identified (the list of repeats is available on request and from http://bacterial-genotyping.igmors.u-psud.fr/) (16). All these tandem repeats represented potential VNTR loci for the typing of serovars Typhi and Typhimurium and were evaluated here. Thirteen primer pairs either produced multiple amplification products, even under stringent PCR conditions (eight pairs), or failed to amplify their target loci (five pairs). These 13 loci were not investigated further. Among the 28 primer pairs that efficiently amplified a unique DNA fragment, 26 pairs yielded a product of the expected size. Two primer pairs (STTR5 and Sal23) yielded a product of an unexpected size, corresponding to the presence of 13 (instead of 15) and 4 (instead of 3) copies of the repeat unit, respectively. Lindstedt et al. (18) also identified this discrepancy when they used primer STTR5.
A collection of 99 clinical isolates collected from various geographical regions in France between 1993 and 1999, with an emphasis on serotypes Typhi and Typhimurium, was investigated. All 28 VNTR loci were amplified from all isolates of serovars Typhi, Typhimurium, and Paratyphi. The two methods used for DNA extraction had no effect on the amplification patterns (data not shown). The results presented herein are based on the ultimate selection of the most polymorphic markers observed: Sal02, Sal04, Sal06, Sal10, TR1, Sal15, STTR5, Sal20, TR5, and Sal23 (Table 1). Within this selection, however, one isolate of S. enterica subsp. arizonae was not amplified with either Sal02 or Sal06 and four isolates of S. enterica subsp. enterica serotype Hadar were not amplified with Sal06. As a quality control, the LT2 reference DNA tested at a frequency of one control per group of five strains processed generated identical VNTR genotypes, thus demonstrating the reproducibility of the typing system setup (data not shown).
TABLE 1.
VNTR markers attributes and primers selected for amplification of VNTRsa
| Locus (alias) and primer | Sequence (5′ to 3′) | Locus name in CT18 | Product size (bp) in CT18 (copy no.) | Alias in Ty2 | Product size (bp) in Ty2 (copy no.) | Alias in LT2 | Product size (bp) in LT2 (copy no.) |
|---|---|---|---|---|---|---|---|
| Sal02 L | GGAAAGACTGGCGAACAAAT | CT18_0666_6 bp | 149 (10) | Ty2_2315_6 bp | 179 (15) | LT2_0681_6 bp | 107 (3) |
| Sal02 R | TCGCCAATACCATGAGTACG | ||||||
| Sal04 L | TCGCACAGATGACCAATTTT | CT18_0740_20 bp | 194 (2) | Ty2_2240_20 bp | 194 (2) | LT2_0764_20 bp | 174 (1) |
| Sal04 R | GATCGACGCTCACTGCTTC | ||||||
| Sal06 L | TTGGTCGCGGAACTATAACTG | CT18_0764_6 bp | 174 (5) | Ty2_2216_6 bp | 180 (6) | LT2_0789_6 bp | 162 (3) |
| Sal06 R | CTTCGTCTGATTGCCACTCC | ||||||
| Sal10 L | AAGCGACGTTCTTCTGCAAC | CT18_2016_12 bp | 196 (2) | Ty2_1009_12 bp | 196 (2) | LT2_2053_12 bp | 208 (3) |
| Sal10 R | TGGAATATGATGGCATGACG | ||||||
| TR1c (Sal11) L | GCCAACGATCGCTACTTTTT | CT18_2017_7 bp | 239 (12) | Ty2_1009_7 bp | 232 (11) | LT2_2053_7 bp | 169 (2) |
| TR1c (Sal11) R | GCGCATACTACACCGATCAC | ||||||
| Sal15 L | GTGACCGGTTGAGTTTGCAT | CT18_2917_12 bp | 189 (2) | Ty2_2903_12 bp | 189 (2) | LT2_3067_12 bp | 201 (3) |
| Sal15 R | GGCAGGTTGTACCAGTTCGT | ||||||
| STTR5d (Sal16) L | CCATGGCTGCAGTTAATTTCT | CT18_3041_6 bp | 224 (14) | Ty2_3027_6 bp | 236 (16) | LT2_3184_6 bp | 230 (15b) |
| STTR5d (Sal16) R | TGATACGCTTTTGACGTTGC | ||||||
| Sal20 L | CAGCCGACACAACTTAACGA | CT18_3643_3 bp | 193 (16) | Ty2_3629_3 bp | 196 (17) | LT2_4301_3 bp | 175 (10) |
| Sal20 R | ACTGTACCGTGCGCGTTT | ||||||
| TR5c (Sal22) L | GCCAGAGGGTTCATTTTCAA | CT18_4624_7 bp | 184 (6) | Ty2_4607_7 bp | 170 (4) | LT2_4645_7 bp | 163 (3) |
| TR5c (Sal22) R | ATGCGACGCCGTTTTACTAC | ||||||
| Sal23 L | CCCGCACACTAAGGAGAGAC | CT18_4738_12 bp | 262 (3.3) | Ty2_4721_12 bp | 262 (3.3) | LT2_4774_12 bp | 250 (3b) |
| Sal23 R | ACCGCGTTAGTGGCTAACAT |
Description and nomenclature refer to genome sequence data for serovars Typhi (CT18 and Ty2) and Typhimurium (LT2) and tandem repeat finder analysis.
The LT2 strain used in the study as a reference control (CIP 86-62, ATCC 43971) harbors 13 and 4 repetitions at loci STTR5 and Sal23, respectively.
Previously characterized (18).
Previously characterized (20).
A comprehensive view of the allelic variability at selected loci and among the isolates in the collection is given in Table 2. Two to 15 alleles were observed. Tandem repeats that displayed the highest allelic size variations had the shortest repeat units (i.e., less than 7 bp) (Table 2). Genotype data for 10 loci and for all 99 isolates are presented in Table 3.
TABLE 2.
Features of selected VNTR loci-observed in 99 strains of S. enterica subsp. enterica
| Locus (alias) | Alias in LT2 | No. of alleles | Copy no. of each allele observed (size range [bp]) | DI
|
|||
|---|---|---|---|---|---|---|---|
| All strains (n = 99) | Serovar Typhi (n = 27) | Serovar Typhimurium (n = 39) | Serovar Paratyphi A (n = 10) | ||||
| Sal02 | LT2_0681_6 bp | 12 | 3, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 (107-197) | NAc | 0.87 | 0 | 0.48 |
| Sal04 | LT2_0764_20 bp | 2 | 1, 2 (174-194) | 0.40 | 0 | 0 | 0 |
| Sal06 | LT2_0789_6 bp | 5 | 3, 4, 5, 6, 8 (162-252) | NA | 0.43 | 0 | 0 |
| TR1a (Sal11) | LT2_2053_7 bp | 12 | 2, 7, 8, 9, 10, 11, 12, 13, 15, 16, 17, 18 (169-281) | 0.46 | 0.87 | 0 | 0 |
| Sal10 | LT2_2053_12 bp | 4 | 1, 2, 3, 4 (184-220) | 0.60 | 0.14 | 0 | 0 |
| Sal15 | LT2_3067_12 bp | 2 | 2, 3 (189-201) | 0.42 | 0.07 | 0 | 0 |
| STTR5b (Sal16) | LT2_3184_6 bp | 15 | 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 19, 22, 23, 24 (182-284) | 0.90 | 0.82 | 0.81 | 0.58 |
| Sal20 | LT2_4301_3 bp | 13 | 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 (172-208) | 0.67 | 0.81 | 0 | 0.18 |
| TR5a (Sal22) | LT2_4645_7 bp | 2 | 3, 4 (163-170) | 0.47 | 0 | 0 | 0 |
| Sal23 | LT2_ 4774_12 bp | 2 | 4, 5 (250-262) | 0.49 | 0 | 0 | 0 |
DIs, which were calculated from the allele frequencies observed in the collection, reflects to some extent the usefulness of a VNTR locus for typing purposes. DIs ranged from 0.40 to 0.90 (Table 2). When serovar Typhi (27 isolates) alone is considered, the DIs ranged from 0.14 to 0.87 (Table 2). The four most variable loci (Sal02, TR1, STTR5, and Sal20) exhibited DIs of 0.87, 0.87, 0.82, and 0.81, respectively. They displayed allelic distributions that ranged from 3 to 18, 2 to 18, 7 to 24, and 9 to 21 repetitions, respectively (Table 2). Moreover, a continuous range of repetitions was observed at loci Sal02 (8 to 18 repetitions), STTR5 (7 to 17 repetitions), and Sal20 (9 to 21 repetitions) (Table 2). This distribution may reflect a well-balanced diversity of the population investigated, in contrast to missing alleles that would have been observed within an unbalanced population due to sampling bias or, more likely, relatively high mutation rates for these loci.
The unique locus (STTR5) that was polymorphic in both serovar Typhi and serovar Typhimurium had similar DIs: 0.82 and 0.81, respectively (Table 2). All other markers displayed a unique allele within the serovar Typhimurium ACSSuT isolates investigated here. Additionally, three markers were found to be variable within serovar Paratyphi A, with DIs ranging from 0.18 to 0.58 (Table 2).
In order to determine the extent of genetic diversity among the strains tested, clustering analysis was performed as described in Materials and Methods. Fifty-two combinations of motifs with unique allele copy numbers were observed among the 99 isolates (Fig. 1 and Table 3). There were four main groups, denoted groups A to D, each of which comprised smaller groups or individual isolates (Fig. 1). Fixed allelic differences existed between major groups, as demonstrated by the presence of an allele with two repeat units at locus Sal04 which was characteristic of group D (Table 3). Similarly, a repeat of three units at Sal10 was characteristic of group B. Fixed but composite allelic differences also defined major groups (Table 3). For example, the pattern of one, four, and five repeats at loci Sal04, Sal22, and Sal23, respectively, identified group C (Table 3).
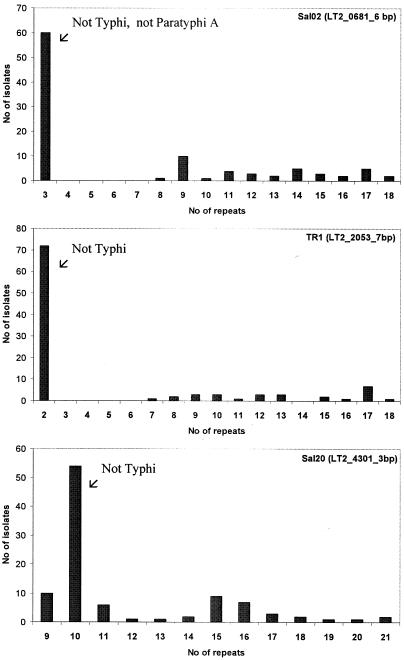
Variable alleles at Sal02 (Fig. 2) were encountered in serovars Typhi and Paratyphi A, in contrast to the predominant allele made up of three repeats, which was present in all remaining isolates except S. enterica subsp. arizonae. For the latter isolate, the absence of amplification is presumably due to some divergence at flanking regions of the tandem repeat and suggests that primers should be redesigned to consider this subspecies. Variables alleles at TR1 (Fig. 2) were encountered only in serovar Typhi, in contrast to the predominant allele made of two repeats which was present in all remaining isolates. Variable alleles at Sal20 (Fig. 2) were encountered in serovar Typhi and to a lower extent in serovar Paratyphi A, in contrast to the predominant allele made up of 10 repeats, which was present in all remaining isolates (Fig. 2). Conversely, STTR5 exhibited allele variability independently of the serovar status, thus implying that a given allele (i.e., 11 repeats) can be observed in all groups, groups A, B, C, and D (Table 3).
FIG. 2.
Restraint allele distribution among the 99 isolates at variable loci Sal02, TR1, and Sal20.
On the basis of the composite allelic distribution (Table 3), population modeling was deduced by construction of MSTs (Fig. 3) by using different priority rules, as detailed in Materials and Methods. Such MSTs offer a more detailed view of the diversity of the test strain panel and highlight closer subtypes that differ by very few allelic changes. For example, all serovar Typhimurium isolates differed by no more than one allelic change (Table 3). When both SLVs and DLVs or the most frequent state settings were used, genotype 17 (strains 03-133 and 03-148) was excluded from the serovar Typhimurium group due to the 11 repeats at STTR5 harbored by these two strains. This illustrates the splitting effects of such more variable markers. All four MSTs displayed very similar topologies, as suggested by the very similar grouping of strains (groups A, B, C, and D), identical linkages between groups (the groups were always branched together through the identical succession of genotypes 18, 3, 5, 7, 27, and 31), and identical calculated distances within this succession. In contrast to serovar Typhimurium, serovar Typhi isolates were much more heterogeneous and the subtree topology was more speculative. The serovar Typhi isolate of genotype 31 (strain 03-115) always linked the serovar Typhi subgroup to the other part of the MST. There was a discrete evolution of the genotype pattern of the serovar Enteritidis isolates between the 2nd, 44th, and 99th challenges in guinea pigs (Fig. 3). Isolate CEB99-35 (99th passage) differed from isolates CEB99-33 (2nd passage) and CEB99-34 (44th passage) at two loci, Sal06 and Sal23 (Table 3).
DISCUSSION
The capability to trace S. enterica serovar Typhi epidemiologically remains of great importance because of the incidence of typhoid fever (5) and the emergence of MDR strains (12, 14). Moreover, S. enterica serovar Typhimurium may be responsible for food-borne acute enterocolitis and the spread of MDR DT104 isolates worldwide. These two serotypes were among the very few human pathogens and/or potential bioterrorism threat agents not typed so far by MLVA, until the recent reports of Lindstedt et al. (18) (serovar Typhimurium) and Liu et al. (20) (serovar Typhi). In the present study, additional candidate VNTR loci were characterized from serovar Typhimurium strain LT2, and a selection of suitable markers for typing purpose was used to discriminate among S. enterica isolates, with an emphasis on serovars Typhi and Typhimurium. Some very polymorphic loci were not selected in the present work due to the presence of multiple bands for some strains, which made allele assignment uncertain. However, different PCR conditions or primer choices may solve this issue if additional markers are needed.
Two discrepancies between theoretical and observed alleles of STTR5 and Sal23 for strain LT2 were likely due to sequencing errors or strain divergence among strain collections. The repeat of 15 units at the STTR5 locus has already been observed (18). Additionally, all serovar Typhimurium isolates analyzed here harbored four, but not three, repeats at monomorphic locus Sal23.
Seven of the 28 VNTRs tested in this study (Sal02, Sal06, TR1, Sal10, Sal15, STTR5, and Sal20) proved to be polymorphic within serovar Typhi, thus discriminating 25 of the 27 isolates tested (93%). Similarly, three of the five VNTRs tested by Liu et al. (20) discriminated 59 isolates into 49 genotypes. It is reasonable to assume that inclusion of the highly variable marker TR2 (DI = 0.95 [20]) would have further increased the level of discrimination among the isolates in our collection. Indeed, one of the two pairs of isolates that remained identical (types 32 and 46 in Fig. 1) exhibited distinct amplification profiles (data not shown). STTR5 is similarly very highly variable; and markers such as these, although they are probably not appropriate for phylogenetic investigations, may be of great use when studying local outbreaks. STTR5 similarly separates the 39 serovar Typhimurium ACSSuT isolates into eight genotypes. The phage types of these isolates were not determined, and the proportion of DT104 isolates is unknown, although it is probably high. The DI of 0.81 obtained for this collection (n = 39) is consistent with the DI of 0.85 observed for a collection of 78 isolates of serovar Typhimurium, with an emphasis on phage type DT104, and with the DI of 0.73 observed when only the 37 DT104 isolates were considered (18; B. A. Lindstedt, personal communication). Genotypic variability within DT104 isolates has already been documented by amplified fragment length polymorphism analysis (12) and PFGE (22). The population of serovar Typhi investigated here appeared to be more diverse than the populations of the other serovars; and this is in accordance with the high degree of heterogeneity observed by others among isolates originating from Asia (13, 14), India (32), and Chile (11).
Multilocus enzyme electrophoresis studies based on 25 chromosomal loci demonstrated that identity according to serovar does not necessarily reflect a close genetic relationship (4). MLST based on three genes confirms this observation (15). The results by Kotetishvili et al. (15) suggested that the discriminatory ability of MLST for the typing of Salmonella is better than that of serotyping or PFGE typing. It will be of interest to compare MLVA with MLST for determination of the genetic relatedness of various Salmonella strains and serotypes. The tendency observed here by MLVA for genetically distinct groups to be consistent with the serovar classification will need to be assessed with strains from reference collections.
The alleles among the serovars tested in this study showed a restrained distribution, as suggested by the predominance of some repeat arrays made up of short motifs (Fig. 2). This was especially striking for Sal02, TR1, and Sal20. Short repeat motifs are possibly believed to allow bacterial adaptation to different environments (35), and the restrained distribution observed may support this idea. Although it is assumed that variation occurs randomly, unknown mechanisms result in the generation of longer repeats arrays in serovar Typhi (the Sal02, TR1, and Sal20 loci) and serovar Paratyphi A (the Sal02 locus). It is thus tempting to speculate from the present data that host adaptation has generated host-adapted variants with specific VNTR arrays.
The collection of serovar Typhimurium investigated previously (18) was geographically biased toward northern Europe, whereas the present study considers isolates originating from France. Some differences in the allele distributions at STTR5 may be due to these different geographical origins. For example, the alleles at this locus with 12 and 13 repeat units were rare in our collection (Table 3). Similarly, in the case of serovar Typhi, the allelic distribution at locus TR1 differed in the two populations investigated so far. The collection investigated here showed a deficit for the alleles with 12 repeat units (Table 3), whereas the collection from Asia (20) showed a strong deficit for the allele with 17 repeat units. Analysis of larger collections from various origins will address this question more accurately, once a reference set of tandem repeat loci has been more formally established. MSTs outlined didactically the population analyzed in terms of distance between isolates and numbers of isolates. Moreover, and perhaps most importantly, it allowed the creation of hypothetical types which correspond to missing links between subgroups within the MSTs, thus highlighting isolates that are not present in a collection.
In conclusion, we carried out MLVA in order to discern genetic similarities and differences among a random sample of serovar Typhi and Typhimurium isolates. A high level of subtype discrimination among the Typhi isolates was achieved. MLVA may be applied to the rapid typing of pathogens and to achieving a high level of discrimination of numerous pathogens and contributes to an improved responsiveness to outbreak investigations. The present work is intended to follow up this trend by developing easy-to-use DNA fingerprinting capabilities for Salmonella. Typing by MLVA can be achieved with basic equipment. The data obtained can easily be compared to published genotypes, either at a local level or by querying data via the website http://bacterial-genotyping.igmors.u-psud.fr/. The potential Internet-based querying tools will help make MLVA, like MLST, a good candidate for a robust and geographically widespread control program.
Acknowledgments
This work was supported by grant DGA/DSA/SPNum 02-3601 from the Délégation Générale pour l'Armement, Ministère de la Défense, as a part of the identification and typing of human pathogens related to bioterrorism.
We thank the referees for the significant improvements that they have suggested.
REFERENCES
- 1.Altwegg, M., F. W. Hickman-Brenner, and J. J. Farmer III. 1989. Ribosomal RNA gene restriction patterns provide increased sensitivity for typing Salmonella typhi strains. J. Infect. Dis. 160:145-149. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, E. S., L. R. Ward, and M. J. de Saxe. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (London) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäumler, A. J., R. M. Tsolis, T. A. Ficht, and L. G. Adams. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 5.Chin, J. 2000. Control of communicable diseases manual, p. 623. American Public Health Association, Washington, D.C.
- 6.Craigie, J., and C. H. Yen. 1938. The demonstration of types of B typhosus by means of preparations of type II Vi phage. II The stability and epidemiological significance of V form types of B typhosus. Can. Public Health J. 29:484-496. [Google Scholar]
- 7.Deng, W., S.-R. Liou, G. Plunkett, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denoeud, F., and G. Vergnaud. 2004. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a Web-based resource. BMC Bioinformatics 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fald, A. A., A. V. Nguyen, and M. I. Khan. 1995. Analysis of Salmonella enteritidis isolates by arbitrary primed PCR. J. Clin. Microbiol. 33:987-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fica, A. E., S. Prat-Miranda, A. Fernandez-Ricci, K. D'Ottone, and F. C. Cabello. 1996. Epidemic typhoid in Chile: analysis by molecular and conventional methods of Salmonella typhi strain diversity in epidemic (1977 and1981) and nonepidemic (1990) years. J. Clin. Microbiol. 34:1701-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebreyes, A. W., and C. Altier. 2002. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium. J. Clin. Microbiol. 40:2813-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampton, M. D., L. R. Ward, B. Rowe, and E. J. Threlfall. 1998. Molecular fingerprinting of multidrug-resistant Salmonella enterica serotype Typhi. Emerg. Infect. Dis. 4:317-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harnett, N., S. McLeod, Y. Au Yong, J. Wan, S. Alexander, R. Khakhria, and C. Krishnan. 1998. Molecular characterization of multiresistant strains of Salmonella typhi from South Asia isolated in Ontario, Canada. Can. J. Microbiol. 44:356-363. [PubMed] [Google Scholar]
- 15.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Flèche, P., M. Fabre, F. Denoeud, J.-L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Flèche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindstedt, B. A., E. Heir, E. Gjernes, and G. Kapperud. 2003. DNA fingerprinting of Salmonella enterica subsp. enterica serovar Typhimurium with emphasis on phage type DT104 based on variable number of tandem repeat loci. J. Clin. Microbiol. 41:1469-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindstedt, B. A., T. Vardun, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Escherichia coli O157 using PCR multiplexing and multi-colored capillary electrophoresis. J. Microbiol. Methods 58:213-222. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y., M.-A. Lee, E.-E. Ooi, Y. Mavis, A.-L. Tan, and H.-H. Quek. 2003. Molecular typing of Salmonella enterica serovar Typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats. J. Clin. Microbiol. 41:4388-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markogiannakis, A., P. T. Tassios, M. Lambiri, L. R. Ward, J. Kourea-Kremastinou, N. J. Legakis, The Greek Nontyphoidal Salmonella Study Group, and A. C. Vatopoulos. 2000. Multiple clones within multidrug-resistant Salmonella enterica serotype Typhimurium phage type DT104. J. Clin. Microbiol. 38:1269-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 24.Oteniente, L., S. Brisse, P. T. Tassios, and G. Vergnaud. 2003. Evaluation of the polymorphism associated with tandem repeats for Pseudomonas aeruginosa strain typing. J. Clin. Microbiol. 41:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 26.Peters, T. M., C. Maguire, E. J. Threlfall, I. S. T. Fisher, N. Gill, and A. J. Gatto. 2003. The Salm-gene project—a European collaboration for DNA fingerprinting for food-related salmonellosis. Eurosurveillance 8:46-50. [DOI] [PubMed] [Google Scholar]
- 27.Pourcel, C., F. André-Mazeaud, H. Neubauer, F. Ramisse, and G. Vergnaud. 2004. Tandem repeats analysis for the high resolution phylogenetic analysis of Yersinia pestis. BMC Microbiol. 4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pourcel, C., Y. Vidgop, F. Ramisse, G. Vergnaud, and C. Tram. 2003. Characterization of a tandem polymorphism in Legionella pneumophila and its use for genotyping. J. Clin. Microbiol. 41:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pradier, C., O. Keita-Perse, E. Bernard, C. Gisbert, M. J. Vezolles, A. Armengaud, D. Carles, F. Grimont, J. C. Desenclos, and P. Dellamonica. 2000. Outbreak of typhoid fever on the French Riviera. Eur. J. Clin. Microbiol. Infect. Dis. 19:464-467. [DOI] [PubMed] [Google Scholar]
- 30.Prager, R., A. Liesegang, W. Rabsch, B. Gericke, W. Thiel, W. Voigt, R. Helmut, L. Ward, and H. Tschape. 1999. Clonal relationship of Salmonella enterica serovar Typhimurium phage type DT104 in Germany and Austria. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 289:399-414. [DOI] [PubMed] [Google Scholar]
- 31.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365. In S. Misener and S. Krawetz (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 32.Shanahan, P. M. A., M. V. Jesudason, C. J. Thomson, and S. G. B. Amyes. 1998. Molecular analysis of and identification of antibiotic resistance genes in clinical isolates of Salmonella typhi from India. J. Clin. Microbiol. 36:1595-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thong, K. L., Y. M. Cheong, S. Puthucheary, C. L. Koh, and T. Pang. 1994. Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulse-field gel electrophoresis. J. Clin. Microbiol. 32:1135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Belkum, A. 1999. Short sequence repeats in microbial pathogenesis and evolution. Cell. Mol. Life Sci. 56:729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Belkum, A., S. Scherer, L. Van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yabuuchi, E., Y. Kosako, H. Oyaizu, I. Yano, H. Hotta, Y. Hashimoto, T. Ezaki, and M. Arakawa. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36:1251-1275. [DOI] [PubMed] [Google Scholar]