Abstract
BACKGROUND
Osteosarcoma is the most common primary bone malignancy. We meta-analyzed the prognostic value of altered miRNAs in patients with osteosarcoma.
METHODS
Sources from MEDLINE (from inception to August 2016) and EMBASE (from inception to August 2016) were searched. Studies of osteosarcoma with results of miRNA and studies that reported survival data were included and two authors performed the data extraction independently. Any discrepancies were resolved by a consensus. The outcome was overall survival and event-free survival assessed using hazard ratios (HRs).
RESULTS
After reviewing the full text of 65 articles, 25 studies including 2,278 patients were eligible in this study. The pooled HR for deaths was 1.40 (95% confidence interval [CI] 1.01-1.94, p=0.04) with random-effects model (χ2=113.08, p<0.00001, I2=79%) for patients of osteosarcoma with lower expression of miRNA. However, the pooled HR for events was not significant (HR 0.97, 0.63-1.48, p=0.87, χ2=72.65, p<0.00001, I2=79%). In pathway analysis of miRNAs, miRNA449a, 199-5p, 542-5p have common target genes.
CONCLUSIONS
Expression level of miRNA in patients of osteosarcoma is important as a prognostic factor.
Keywords: microRNA, osteosarcoma, prognosis, meta-analysis
INTRODUCTION
Osteosarcoma is the most common primary bony malignancies and first leading cause of sarcoma-related deaths in children and young adults. It mainly occurs in metaphyseal area of distal femur and proximal tibia. Osteosarcoma is highly aggressive tumor and most likely metastasizes to the lung [1]. Even though the survival rate has modestly increased over the last 2 decades by implicating radiotherapy and neo-adjuvant chemotherapy, five-year survival rate of metastatic osteosarcoma still remains in the range of 15% to 30%. For localized osteosarcoma which can be totally resectable, the five-year survival rate increases up to 70% [2]. Hence, excavating novel prognostic biomarkers of osteosarcoma is strongly needed to detect tumor at early stage. They will contribute to select better treatment options in earlier stages and predict the outcome more accurately.
MicroRNAs (miRNAs) are a class of small noncoding RNAs, which could interfere translation of many proteins after gene transcription [3]. Previous studies revealed that they are implicated on a variety of physiological processes including cellular differentiation, apoptosis, angiogenesis and cell proliferation [4]. Additionally, miRNAs play a pivotal role in tumorigenesis that can act either as tumor suppressor genes or oncogenes [3, 5]. Level of miRNAs expressed in several malignancies, including lung, kidney, liver, and cervical cancer were significantly different from that of normal tissue [6, 7]. Recent researches have demonstrated that various miRNAs are also altered in osteosarcoma tissue or blood sample [8]. These discoveries may indicate that miRNA could be potential prognostic biomarkers of osteosarcoma. Therefore, we meta-analyzed the prognostic value of altered miRNAs in patients with osteosarcoma.
RESULTS
Study characteristics
The electronic search identified 289 articles. Non-English language articles (n=3), non-human studies (n=13), conference abstracts (n=34), and 174 studies that did not meet the inclusion criteria based on their title and abstract were excluded. After reviewing the full text of 65 articles, 25 studies including 2,278 patients were eligible for inclusion in the study. The detailed procedure is shown in Figure 1. The studies included in this meta-analysis reported the prognostic value of 26 miRNAs in patients with osteosarcoma. Twenty-five studies assessing 26 miRNA expressions were included in the meta-analysis [9–33]. The prognostic value of miRNA expressions was assessed by analyzing overall survival (OS) in 2,278 patients, event free survival (EFS) in 1,569 patients. Visual inspection of the funnel plot suggested no evidence of publication bias (Figure 2B and 3B). The study characteristics are summarized in Table 1.
Figure 1. Flow chart.
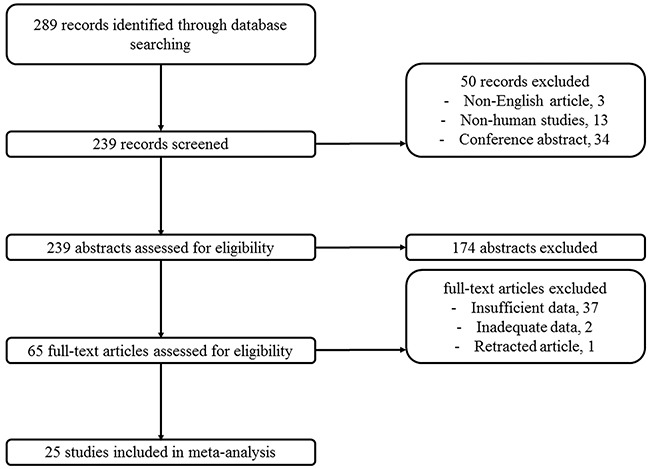
Figure 2. Forest plot A. and funnel plot B. for deaths of low expression of miRNA in osteosarcoma.
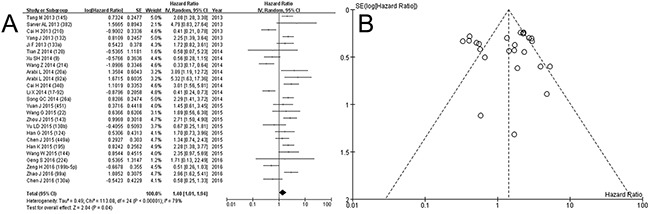
Figure 3. Forest plot A. and funnel plot B. for events of low expression of miRNA in osteosarcoma.
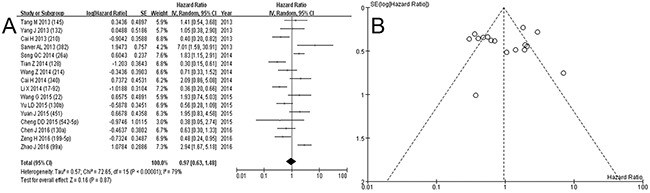
Table 1. Studies included in this meta-analysis.
| Author | Year of publication | Country | miRNA | No. of patients | Stage | Follow-up (months) | Endpoints | Expression associates with poor prognosis | Assay method | Sample |
|---|---|---|---|---|---|---|---|---|---|---|
| Cai H [10] | 2013 | China | 210 | 92 | - | 105a | OS, EFS | High | RT-PCR | Tissue |
| Li X [19] | 2014 | China | 17-92 cluster | 117 | I-III | 55a | OS, EFS | High | RT-PCR | Tissue |
| Tian Z [23] | 2014 | China | 128 | 100 | - | 38.89a | OS, EFS | High | RT-PCR | Tissue |
| Xu SH [27] | 2014 | China | 9 | 79 | I-III | 60a | OS | High | RT-PCR | Tissue |
| Wang Z [26] | 2014 | China | 214 | 92 | - | 107a | OS, EFS | High | RT-PCR | Tissue |
| Cheng DD [14] | 2015 | China | 542-5p | 40 | - | 40 | EFS | High | RT-PCR | Tissue |
| Yu LD [29] | 2015 | China | 130b | 68 | I-III | 36 | OS, EFS | High | RT-PCR | Tissue |
| Chen J [12] | 2016 | China | 130a | 86 | I-IV | 60a | OS, EFS | High | RT-PCR | Tissue |
| Zeng H [31] | 2016 | China | 199b-5p | 98 | I-IV | 60b | OS, EFS | High | RT-PCR | Tissue |
| Yang J [28] | 2013 | China | 132 | 166 | II-III | 105a | OS, EFS | Low | RT-PCR | Tissue |
| Tang M [22] | 2013 | China | 145 | 166 | II-III | 100a | OS, EFS | Low | RT-PCR | Tissue |
| Sarver AL [20] | 2013 | USA | 382 | 8 | - | 194a | OS, EFS | Low | RT-PCR | Tissue |
| Ji F [18] | 2013 | China | 133a | 92 | I-III | 60a | OS | Low | RT-PCR | Tissue |
| Song QC [21] | 2014 | China | 26a | 144 | II-III | 140a | OS, EFS | Low | RT-PCR | Tissue |
| Cai H [11] | 2014 | China | 340 | 92 | - | 100a | OS, EFS | Low | RT-PCR | Tissue |
| Arabi L [9] | 2014 | Switzerland | 20a | 57 | - | 112 | OS | Low | RT-PCR | Tissue |
| Arabi L [9] | 2014 | Switzerland | 92a | 57 | - | 112 | OS | Low | RT-PCR | Tissue |
| Zhou J [33] | 2015 | China | 143 | 45 | - | 36c | OS | Low | RT-PCR | Tissue |
| Yuan J [30] | 2015 | China | 451 | 118 | II-III | 80a | OS, EFS | Low | RT-PCR | Tissue |
| Wang W [25] | 2015 | China | 144 | 67 | I-IV | 80a | OS | Low | RT-PCR | Tissue |
| Wang G [24] | 2015 | China | 22 | 52 | I-IV | 60c | OS, EFS | Low | RT-PCR | Tissue |
| Han K [17] | 2015 | China | 195 | 107 | II-III | 90a | OS | Low | RT-PCR | Tissue |
| Han G [16] | 2015 | China | 124 | 105 | II-III | 80a | OS | Low | RT-PCR | Tissue |
| Chen J [13] | 2015 | China | 449a | 60 | I-III | 60b | OS | Low | RT-PCR | Tissue |
| Zhao J [32] | 2016 | China | 99a | 130 | II-III | 60a | OS, EFS | Low | RT-PCR | Tissue |
| Geng S [15] | 2016 | China | 224 | 40 | - | 60 | OS | Low | RT-PCR | Tissue |
- Follow-up; amedian, bheterogeneity of logrank test, cmean
- OS, overall survival; EFS, event free survival; RT-PCR, reverse transcription polymerase chain reaction
miRNA expression and prognosis of osteosarcoma
To analyze the prognostic value of low expression of miRNA in osteosarcoma, forest plots with OS and EFS are depicted in Figure 2 and 3. The pooled hazard ratio (HR) for deaths was 1.40 (95% confidence interval [CI] 1.01-1.94, p=0.04) with random-effects model (χ2=113.08, p<0.00001, I2=79%), which means more deaths with lower expression of miRNA in osteosarcoma. However, the pooled HR for events did not show the significant value (HR 0.97, 0.63-1.48, p=0.87, χ2=72.65, p<0.00001, I2=79%). Due to heterogeneity in pooled analysis of miRNA, we performed a subgroup analysis according to the expression level of miRNA. The HRs were calculated on the basis of low expression of miRNA, which means HR > 1 and < 1 implied poor and good prognosis for patients with low miRNA expression.
A. Worse prognosis with high miRNA expression
High expressions of miRNA210, 17-92 cluster, 128, 9, 214, 542-5p, 130b, 130a, 199b-5p were associated with poor prognosis of osteosarcoma patients, [10, 12, 14, 19, 23, 26, 27, 29, 31]. Eight studies were included to analyze OS with miRNA210, 17-92 cluster, 128, 9, 214, 130b, 130a, 199-5p in 732 patients [10, 12, 19, 23, 26, 27, 29, 31]. The HR ranged between 0.33 and 0.67 with a pooled HR for deaths of 0.45 (0.35-0.60, p<0.00001) with low expression of miRNA, and the test for heterogeneity gave no significant results (χ2=2.47, p=0.93, I2=0%) (Table 2). The EFS was analyzed with miRNA210, 17-92 cluster, 128, 214, 542-5p, 130b, 130a, 199b-5p based on 8 studies including 693 patients [10, 12, 14, 19, 23, 26, 29, 31]. The pooled HR of high miRNA for adverse events was 0.46 with a range from 0.30 to 0.71 (0.35-0.60, p<0.00001), and the test for heterogeneity gave no significant results (χ2=4.37, p=0.74, I2=0%) (Table 2). The forest plots for OS and EFS of high miRNA with worse prognosis are shown in Figure 4.
Table 2. Subgroup analysis according to miRNA expression with prognosis.
| No. of studies | miRNA | HR | 95% CI of HR | Heterogeneity I2 (%) | Model used | |
|---|---|---|---|---|---|---|
| Worse prognosis with high miRNA expression | ||||||
| OS | 8 | 210, 17-92, 128, 9, 214, 130b, 130a, 199b-5p | 0.46 | 0.35-0.60 | 0 | Fixed effect |
| EFS | 8 | 210, 17-92, 128, 214, 542-5p, 130b, 130a, 199b-5p | 0.46 | 0.35-0.60 | 0 | Fixed effect |
| Worse prognosis with low miRNA expression | ||||||
| OS | 16 | 132, 145, 382, 133a, 26a, 340, 20a, 92a, 143, 451, 144, 22, 195, 124, 449a, 99a, 224 | 2.25 | 1.90-2.66 | 0 | Fixed effect |
| EFS | 8 | 132, 145, 382, 26a, 340, 451, 22, 99a | 2.05 | 1.67-5.18 | 0 | Fixed effect |
- HR, hazard ratio; OS, overall survival; EFS, event free survival
Figure 4. Subgroup analysis: worse prognosis with high expression of miRNA; forest plots for deaths A. and events B. of low expression of miRNA in osteosarcoma.
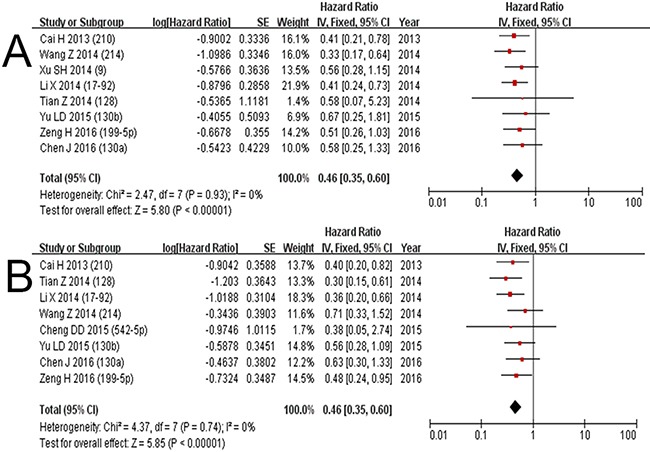
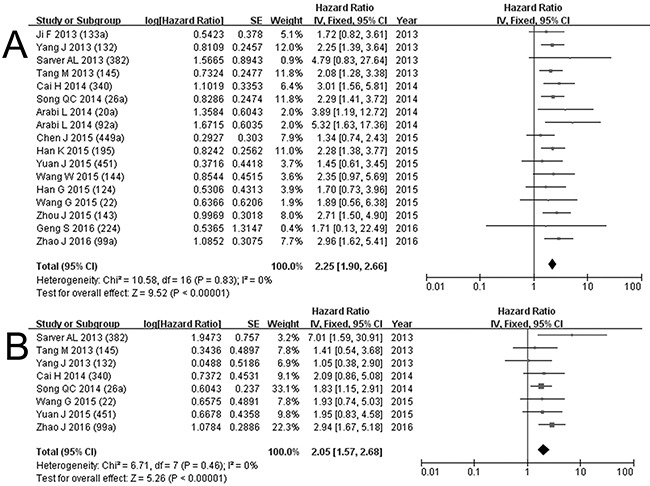
B. Worse prognosis with low miRNA expression
Low expression of miRNA132, 145, 382, 133a, 26a, 340, 20a, 92a, 143, 451, 144, 22, 195, 124, 449a, 99a, 224 were found to be associated with a poor prognosis [9, 11, 13, 15–18, 20–22, 24, 25, 28, 30, 32, 33]. Sixteen studies analyzed OS with miRNA132, 145, 382, 133a, 26a, 340, 20a, 92a, 143, 451, 144, 22, 195, 124, 449a, 99a, 224 in 1,506 patients [9, 11, 13, 15–18, 20–22, 24, 25, 28, 30, 32, 33], and eight studies (miRNA132, 145, 382, 26a, 340, 451, 22, 99a) containing 876 patients that reported EFS were included to calculate the combined HRs [11, 20–22, 24, 28, 30, 32]. The HR for deaths ranged widely between 1.34 and 5.32 with a combined HR of 2.25 (1.90-2.66, p<0.00001), and the pooled HR for events was 2.05 with a range from 1.05 to 7.01 (1.57-2.68, p<0.00001). There was no significant heterogeneity (OS: χ2=10.58, p=0.83, I2=0%, EFS: χ2=6.71, p=0.46, I2=0%). The forest plots for OS and EFS of low miRNA with worse prognosis are shown in Figure 5.
Figure 5. Subgroup analysis: worse prognosis with low expression of miRNA; forest plots for deaths A. and events B. of low expression of miRNA in osteosarcoma.
C. Pathway analysis of miRNAs
In a pathway analysis of miRNAs using mirPath v3.0 based on Tarbase, KEGG and GO, miRNA449a, 199-5p, 542-5p have target genes and signaling pathways, but others do not have known targets (Figure 6 and 7, Table 3) [34].
Figure 6. The number of target genes of miRNA449a, 199-5p and 542-5p.
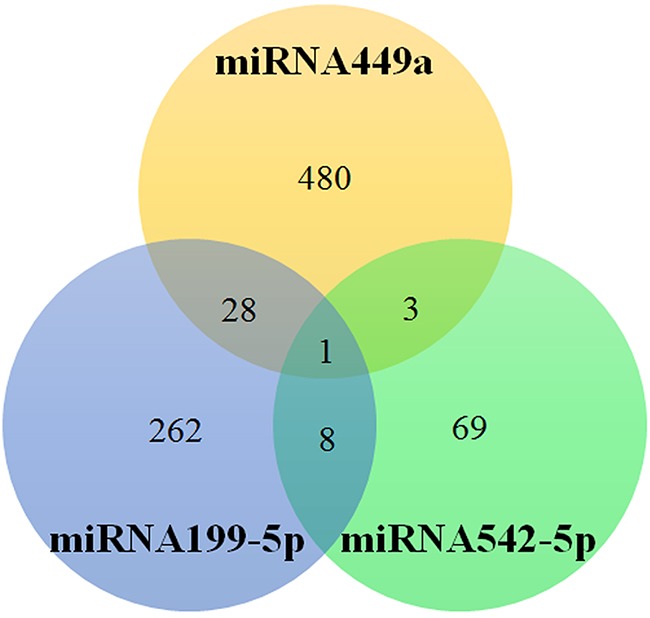
Figure 7. Heatmap of pathway analysis of miRNA449a, 199-5p and 542-5p by KEGG pathway A. and GO categories B.
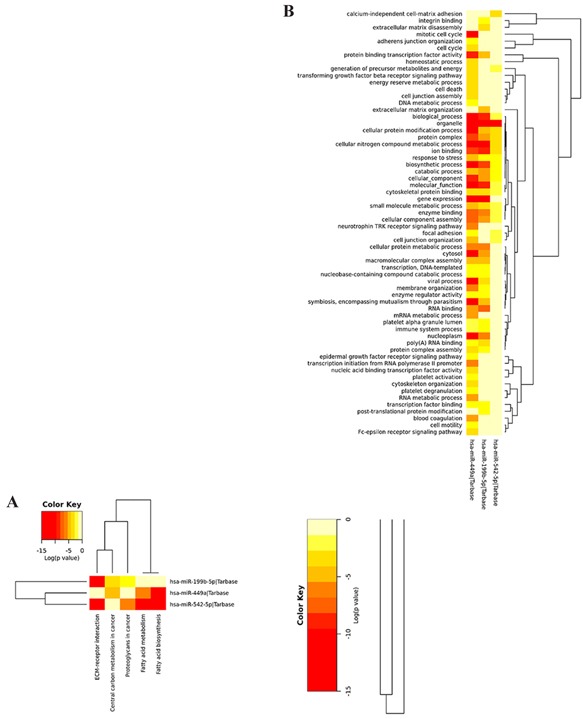
Table 3. pathway analysis of miRNA449a, 199b-5p, 542-5p.
| pathway analysis | p | no. of target genes | miRNA -449a | miRNA- 199b-5p | miRNA- 542-5p |
|---|---|---|---|---|---|
| KEGG pathway | |||||
| Fatty acid biosynthesis | 0 | 3 | Y | N | Y |
| Fatty acid metabolism | 0 | 3 | Y | N | Y |
| ECM-receptor interaction | 0 | 10 | N | Y | Y |
| Proteoglycans in cancer | 2.47E-07 | 15 | Y | Y | N |
| Central carbon metabolism in cancer | 1.58E-06 | 14 | Y | Y | N |
| GO Category | |||||
| molecular function | 0 | 789 | Y | Y | Y |
| cellular protein modification process | 0 | 155 | Y | Y | Y |
| biological process | 0 | 780 | Y | Y | Y |
| biosynthetic process | 0 | 255 | Y | Y | Y |
| cellular nitrogen compound metabolic process | 0 | 299 | Y | Y | Y |
| ion binding | 0 | 317 | Y | Y | Y |
| organelle | 0 | 596 | Y | Y | Y |
| protein complex | 1.33E-15 | 232 | Y | Y | Y |
| cellular component | 5.00E-15 | 784 | Y | Y | Y |
| enzyme binding | 2.80E-13 | 103 | Y | Y | Y |
| cellular component assembly | 5.09E-12 | 102 | Y | Y | Y |
| catabolic process | 2.86E-10 | 126 | Y | Y | Y |
| response to stress | 4.90E-08 | 136 | Y | Y | Y |
| cytoskeletal protein binding | 1.68E-07 | 61 | Y | Y | Y |
| small molecule metabolic process | 2.90E-07 | 130 | Y | Y | Y |
| gene expression | 0 | 70 | Y | Y | N |
| symbiosis, encompassing mutualism through parasitism | 0 | 58 | Y | Y | N |
| nucleoplasm | 2.22E-16 | 97 | Y | Y | N |
| cytosol | 1.89E-15 | 174 | Y | Y | N |
| viral process | 1.43E-14 | 50 | Y | Y | N |
| RNA binding | 2.72E-12 | 122 | Y | Y | N |
| cellular protein metabolic process | 4.33E-12 | 43 | Y | Y | N |
| protein binding transcription factor activity | 6.72E-12 | 49 | Y | Y | N |
| macromolecular complex assembly | 1.36E-08 | 64 | Y | Y | N |
| membrane organization | 9.90E-08 | 45 | Y | Y | N |
| cell junction organization | 5.04E-06 | 18 | Y | N | Y |
| poly(A) RNA binding | 5.87E-06 | 103 | Y | Y | N |
| protein complex assembly | 2.17E-05 | 52 | Y | Y | N |
| transcription, DNA-templated | 9.55E-05 | 127 | Y | Y | N |
| nucleobase-containing compound catabolic process | 0.000168 | 53 | Y | Y | N |
| enzyme regulator activity | 0.000207 | 51 | Y | Y | N |
| transcription factor binding | 0.000255 | 43 | Y | Y | N |
| generation of precursor metabolites and energy | 0.0004 | 20 | Y | N | Y |
| focal adhesion | 0.000601 | 35 | Y | N | Y |
| immune system process | 0.00162 | 81 | Y | Y | N |
| activation of signaling protein activity involved in unfolded protein response | 0.002297 | 9 | Y | Y | N |
| platelet alpha granule lumen | 0.003495 | 7 | Y | Y | N |
DISCUSSION
This study evaluated the prognostic value of miRNA in patients with osteosarcoma. As we described above, the combined HR of OS from the included studies was 1.40 (1.01-1.94, p=0.04), which provided that low expression of miRNAs was related with shorter OS in patients with osteosarcoma. However, the combined HR of EFS was not significant. Due to heterogeneity in pooled analysis of miRNA, we performed a subgroup analysis according to the expression level of miRNA. For a more detailed analysis, we performed a subgroup analysis according to miRNA expressions (high or low) related with poor prognosis. High expression of miRNA 210, 17-92 cluster, 128, 9, 214, 542-5p, 130b, 130a, and 199b-5p and low expression of miRNA 132, 145, 382, 133a, 26a, 340, 20a, 92a, 143, 451, 144, 22, 195, 124, 449a, 99a, 224 were associated with a poor prognosis of either deaths or events.
The possible role of miRNAs during osteosarcomagenesis is also investigated. Altered or aberrant expression of miRNA, its function during tumorigenesis, and potential target genes are summarized in Table 4. Nine miRNAs (miRNA 210, 17-92 cluster, 128, 9, 214, 542-5p, 130b, 130a, and 199b-5p) whose expression level correlated with poor prognosis function as an oncogene in osteosarcoma. Of that, miRNA 210 contributes its action in tumor initiation. miRNA 17-92 cluster, 214, 130b have been reported as they progress early stage osteosarcoma into advanced stage. And miRNA 9, 130a are associated with metastasis. On the contrary, 17 miRNAs (132, 145, 382, 133a, 26a, 340, 20a, 92a, 143, 451, 144, 22, 195, 124, 449a, 99a, 224) function as a tumor suppressor gene, which expressed low level when they were related with worse outcome. Among these miRNAs, miRNA 382, 133a, 26a, 340, 143, 144, 195 have shown an anti-metastatic activity. Especially, miRNA 133a, 143 could be utilized as a future therapeutic target for lung metastasis. Moreover, miRNA 132, 145, 92a might act as a therapeutic standard for chemotherapy.
Table 4. miRNAs with relative expression level associated with poor prognosis, their function and target or related gene.
| miRNA | Expression level associate with poor prognosis | Function | Target or related gene |
|---|---|---|---|
| miR-210[10] | high | Oncogene (Tumor Initiation) | N/D |
| miR-17–92[19] | high | Oncogene (Tumor Progression) | N/D |
| miR-128[23] | high | Oncogene | Inhibits PTEN |
| miR-9[27] | high | Oncogene Promote metastasis | N/D |
| miR-214[26] | high | Oncogene (Tumor Progression) | N/D |
| miR-542-5p [14] | high | Oncogene | Inhibits HUWE1 |
| miR-130b [29] | high | Oncogene (Tumor Progression) | Inhibits PPARγ |
| miR-130a [12] | high | Oncogene Promote metastasis | Inhibits PTEN |
| miR-199b-5p [31] | high | Oncogene | HES1 |
| miR-132 [28] | low | Discriminate good responders from poor responders | N/D |
| miR-145 [22] | low | Tumor suppressor Discriminate good responders from poor responders | N/D |
| miR-382 [20] | low | Oncogene Suppress metastasis | Stabilize MYC |
| miR-133a [18] | low | Tumor suppressor Suppress pulmonary metastasis | Inhibits Bcl-xL, Mcl-1 |
| miR-26a [21] | low | Tumor suppressor Suppress metastasis and recurrence | Inhibits EZH2 |
| miR-340 [11] | low | Tumor suppressor Suppress metastasis | N/D |
| miR-20a [9] | low | Tumor suppressor | Inhibits FAS |
| miR-92a [9] | low | Discriminate ifosfamide responder | Inhibits FAS |
| miR-143 [33] | low | Tumor suppressor Suppress pulmonary metastasis | MMP-13 Bcl-2 |
| miR-451 [30] | low | Tumor suppressor | N/D |
| miR-144 [25] | low | Tumor suppressor Suppress metastasis | Inhibits ROCK1, ROCK2 |
| miR-22 [24] | low | Tumor suppressor | Inhibits HMGB1 |
| miR-195 [17] | low | Suppress metastasis | Inhibits CCND1 |
| miR-124 [16] | low | Tumor suppressor | N/D |
| miR-449a [13] | low | Tumor suppressor | Inhibits Bcl-2 |
| miR-99a [32] | low | Tumor suppressor | mTOR |
| miR-224 [15] | Low | Tumor suppressor | Rac1 |
- N/D, not determined; PTEN, phosphatase and tensin homolog; HUWE1; PPARγ, peroxisome proliferator-activated receptor gamma; HES1, hes family bHLH transcription factor 1; MYC, v-myc avian myelocytomatosis viral oncogene homolog ; Bcl-xL, BCL2-like 1 isoform 1; Mcl-1, BCL2 family apoptosis regulator; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; FAS, tumor necrosis factor receptor superfamily member 6; MMP-13, matrix metallopeptidase 13; Bcl-2, B-cell lymphoma 2; ROCK, Rho-associated protein kinase; HMGB1, high mobility group box 1; CCND1, Cyclin D1; MTOR, mechanistic target of rapamycin; Rac1, Ras-related C3 botulinum toxin substrate 1
This is the first meta-analysis that evaluate the prognostic value of miRNAs in osteosarcoma. We demonstrated the association of the outcomes (OS, EFS) of osteosarcoma and level of miRNA expression. In addition, we measured HRs as the effectiveness endpoint in the current study. We did not measured the odds ratios or risk ratios whether these values represent only cross-sectional cumulative estimate and do not including the time factor [35]. Hence, HRs are most appropriate for our report which analyze time-to-event outcomes. The type of samples was homogenous. All of the samples which were experimented in included studies were obtained from tumor tissue of osteosarcoma patients. This might be increased reliability and decreased heterogeneity of our study. We excluded studies using serum or plasma because there were a few studies about serum or circulating miRNAs in patients with osteosarcoma [36, 37]. However, there are increasing interest in circulating miRNAs due to their comfortable accessibility by drawing patient's blood compared to miRNAs of tissue from operation and they could be easily repeated and monitored through.
From the pathway analysis, we found that several common targets and pathways of miRNA199b-5p, 449a and 542-5p, for example, fatty acid biosynthesis (target gene: Fatty Acid Synthase (FASN)) (Table 3). FASN is known as a therapeutic target in the treatment of osteosarcoma metastasis [38]. Pathways related to other malignancies were found in this analysis, such as breast cancer, and prostate cancer [38–41]. The target genes of miRNAs in various cancers may overlap, which suggest that miRNAs could be promising therapeutic targets in cancer treatment.
In this study, we investigated the prognostic value of miRNA in patients with osteosarcoma using a meta-analysis approach. Finally, we concluded that decreased miRNA expression in tumor tissue is associated with worse outcome of patients with osteosarcoma. However, we should also pay attention to increased expression of several miRNAs in tumor tissue of osteosarcoma. Furthermore, prospective studies with a large sample size are warranted to clarify the prognostic role of miRNAs in osteosarcoma.
MATERIALS AND METHODS
Data search and study election
We performed a systematic search if MEDLINE (from inception to August 2016) and EMBASE (from inception to August 2016) for English-language publications using the keywords “osteosarcoma”, “miRNA”, “prognosis”. All searches were limited to human studies. The inclusion criteria were studies of osteosarcoma that reported the results of miRNA expressions and survival data. Reviews, abstracts, and editorial materials were excluded. Two authors performed the searches and screening independently, and discrepancies were resolved by consensus.
Data extraction and statistical analysis
Data were extracted from the publications independently by two reviewers, and the following information was recorded: first author, year of publication, country, miRNA expression analyzed, number of patients, staging, and end points. The primary outcome was OS, which was defined as the time from the initiation of therapy until death from any cause. The secondary end point was EFS. Data regarding disease-free survival (DFS), progression-free survival (PFS), recurrence-free survival (RFS) were obtained from the included studies, and were redefined as EFS, which was measured from the date of initiation of therapy to the date of recurrence or metastasis [42, 43].
The effects of miRNA expressions on survival were assessed using hazard ratios. Survival data were extracted following a methodology suggested previously [44]. A univariate HR estimate and 95% CIs were extracted directly from each study, if provided by the authors. Otherwise, p values of the log-rank tests, 95% confidence intervals, number of events, and numbers of patients at risk were extracted to estimate the HR indirectly. Survival rates calculated from Kaplan-Meier curves were read using Engauge Digitizer version 3.0 (http://digitizer.sourceforge.net) to reconstruct the HR estimate and its variance, assuming that patients were censored at a constant rate during follow-up. The HRs were calculated on the basis of low expression of miRNA, which means HR > 1 and < 1 implied poor and good prognosis for patients with low miRNA expression. Heterogeneity among studies was assessed using χ2 tests and I2 statistics, as described previously [45]. Funnel plots were used to assess publication bias [46]. p values < 0.05 were considered to be statistically significant. The data from each study were analyzed using Review Manager (RevMan, Version 5.3 Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014). miRNA target genes and related pathways were predicted by DIANA-mirPath v3.0 (http://www.microrna.gr/miRPathv3) based on Tarbase, KEGG and GO [34] to interpret functional value of miRNAs.
Footnotes
CONFLICTS OF INTEREST
None
FUNDING
This study was supported by 2016 Post-Doc. Development Program of Pusan National University, Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1A6A3A11931738) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2015R1D1A1A01059035).
Author contributions
Yun Hak Kim: write the manuscript
Tae Sik Goh, Chi-Seung Lee: study design
Sae Ock Oh, Jeung Il Kim, Seung Hyeon Jeung: statistical analysis
Kyoungjune Pak; write the manuscript
REFERENCES
- 1.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–718. [PubMed] [Google Scholar]
- 2.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452:1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Keys DN, Au-Young JK, Chen C. MicroRNAs in embryonic stem cells. J Cell Physiol. 2009;218:251–255. doi: 10.1002/jcp.21607. [DOI] [PubMed] [Google Scholar]
- 5.Zhao G, Cai C, Yang T, Qiu X, Liao B, Li W, Ji Z, Zhao J, Zhao H, Guo M, Ma Q, Xiao C, Fan Q, Ma B. MicroRNA-221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One. 2013;8:e53906. doi: 10.1371/journal.pone.0053906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottai G, Pasculli B, Calin GA, Santarpia L. Targeting the microRNA-regulating DNA damage/repair pathways in cancer. Expert Opin Biol Ther. 2014;14:1667–1683. doi: 10.1517/14712598.2014.950650. [DOI] [PubMed] [Google Scholar]
- 8.Nugent M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer Manag Res. 2014;6:15–25. doi: 10.2147/CMAR.S53928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arabi L, Gsponer JR, Smida J, Nathrath M, Perrina V, Jundt G, Ruiz C, Quagliata L, Baumhoer D. Upregulation of the miR-17-92 cluster and its two paraloga in osteosarcoma - reasons and consequences. Genes Cancer. 2014;5:56–63. doi: 10.18632/genesandcancer.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai H, Lin L, Cai H, Tang M, Wang Z. Prognostic evaluation of microRNA-210 expression in pediatric osteosarcoma. Med Oncol. 2013;30:499. doi: 10.1007/s12032-013-0499-6. [DOI] [PubMed] [Google Scholar]
- 11.Cai H, Lin L, Cai H, Tang M, Wang Z. Combined microRNA-340 and ROCK1 mRNA profiling predicts tumor progression and prognosis in pediatric osteosarcoma. Int J Mol Sci. 2014;15:560–573. doi: 10.3390/ijms15010560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Yan D, Wu W, Zhu J, Ye W, Shu Q. MicroRNA-130a promotes the metastasis and epithelial-mesenchymal transition of osteosarcoma by targeting PTEN. Oncol Rep. 2016;35:3285–3292. doi: 10.3892/or.2016.4719. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Zhou J, Chen X, Yang B, Wang D, Yang P, He X, Li H. miRNA-449a is downregulated in osteosarcoma and promotes cell apoptosis by targeting BCL2. Tumour Biol. 2015;36:8221–8229. doi: 10.1007/s13277-015-3568-y. [DOI] [PubMed] [Google Scholar]
- 14.Cheng DD, Yu T, Hu T, Yao M, Fan CY, Yang QC. MiR-542-5p is a negative prognostic factor and promotes osteosarcoma tumorigenesis by targeting HUWE1. Oncotarget. 2015;6:42761–42772. doi: 10.18632/oncotarget.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng S, Gu L, Ju F, Zhang H, Wang Y, Tang H, Bi Z, Yang C. MicroRNA-224 promotes the sensitivity of osteosarcoma cells to cisplatin by targeting Rac1. J Cell Mol Med. 2016;20:1611–1619. doi: 10.1111/jcmm.12852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Kinahan PE, Mankoff DA, Linden HM. The Value of Establishing the Quantitative Accuracy of PET/CT Imaging. Journal of nuclear medicine. 2015;56:1133–1134. doi: 10.2967/jnumed.115.159178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han K, Chen X, Bian N, Ma B, Yang T, Cai C, Fan Q, Zhou Y, Zhao TB. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget. 2015;6:8875–8889. doi: 10.18632/oncotarget.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y, Wang Z, Wang Z, Cheng P, Tong D, Li C, Tang H. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56:220–226. doi: 10.1016/j.bone.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Yang H, Tian Q, Liu Y, Weng Y. Upregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcoma. Neoplasma. 2014;61:453–460. doi: 10.4149/neo_2014_056. [DOI] [PubMed] [Google Scholar]
- 20.Sarver AL, Thayanithy V, Scott MC, Cleton-Jansen AM, Hogendoorn PC, Modiano JF, Subramanian S. MicroRNAs at the human 14q32 locus have prognostic significance in osteosarcoma. Orphanet J Rare Dis. 2013;8:7. doi: 10.1186/1750-1172-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song QC, Shi ZB, Zhang YT, Ji L, Wang KZ, Duan DP, Dang XQ. Downregulation of microRNA-26a is associated with metastatic potential and the poor prognosis of osteosarcoma patients. Oncol Rep. 2014;31:1263–1270. doi: 10.3892/or.2014.2989. [DOI] [PubMed] [Google Scholar]
- 22.Tang M, Lin L, Cai H, Tang J, Zhou Z. MicroRNA-145 downregulation associates with advanced tumor progression and poor prognosis in patients suffering osteosarcoma. Onco Targets Ther. 2013;6:833–838. doi: 10.2147/OTT.S40080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian Z, Guo B, Yu M, Wang C, Zhang H, Liang Q, Jiang K, Cao L. Upregulation of micro-ribonucleic acid-128 cooperating with downregulation of PTEN confers metastatic potential and unfavorable prognosis in patients with primary osteosarcoma. Onco Targets Ther. 2014;7:1601–1608. doi: 10.2147/OTT.S67217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Shen N, Cheng L, Lin J, Li K. Downregulation of miR-22 acts as an unfavorable prognostic biomarker in osteosarcoma. Tumour Biol. 2015;36:7891–7895. doi: 10.1007/s13277-015-3379-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Zhou X, Wei M. MicroRNA-144 suppresses osteosarcoma growth and metastasis by targeting ROCK1 and ROCK2. Oncotarget. 2015;6:10297–10308. doi: 10.18632/oncotarget.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Cai H, Lin L, Tang M, Cai H. Upregulated expression of microRNA-214 is linked to tumor progression and adverse prognosis in pediatric osteosarcoma. Pediatr Blood Cancer. 2014;61:206–210. doi: 10.1002/pbc.24763. [DOI] [PubMed] [Google Scholar]
- 27.Xu SH, Yang YL, Han SM, Wu ZH. MicroRNA-9 expression is a prognostic biomarker in patients with osteosarcoma. World J Surg Oncol. 2014;12:195. doi: 10.1186/1477-7819-12-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Gao T, Tang J, Cai H, Lin L, Fu S. Loss of microRNA-132 predicts poor prognosis in patients with primary osteosarcoma. Mol Cell Biochem. 2013;381:9–15. doi: 10.1007/s11010-013-1677-8. [DOI] [PubMed] [Google Scholar]
- 29.Yu LD, Jin RL, Gu PC, Ling ZH, Lin XJ, Du JY. Clinical significance of microRNA-130b in osteosarcoma and in cell growth and invasion. Asian Pac J Trop Med. 2015;8:752–756. doi: 10.1016/j.apjtm.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 30.Yuan J, Lang J, Liu C, Zhou K, Chen L, Liu Y. The expression and function of miRNA-451 in osteosarcoma. Med Oncol. 2015;32:324. doi: 10.1007/s12032-014-0324-x. [DOI] [PubMed] [Google Scholar]
- 31.Zeng H, Zhang Z, Dai X, Chen Y, Ye J, Jin Z. Increased Expression of microRNA-199b-5p Associates with Poor Prognosis Through Promoting Cell Proliferation, Invasion and Migration Abilities of Human Osteosarcoma. Pathol Oncol Res. 2016;22:253–260. doi: 10.1007/s12253-015-9901-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, Chen F, Zhou Q, Pan W, Wang X, Xu J, Ni L, Yang H. Aberrant expression of microRNA-99a and its target gene mTOR associated with malignant progression and poor prognosis in patients with osteosarcoma. Onco Targets Ther. 2016;9:1589–1597. doi: 10.2147/OTT.S102421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Wu S, Chen Y, Zhao J, Zhang K, Wang J, Chen S. microRNA-143 is associated with the survival of ALDH1+CD133+ osteosarcoma cells and the chemoresistance of osteosarcoma. Exp Biol Med (Maywood) 2015;240:867–875. doi: 10.1177/1535370214563893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL, Maniou S, Karathanou K, Kalfakakou D, Fevgas A, Dalamagas T, Hatzigeorgiou AG. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA: mRNA interactions. Nucleic Acids Res. 2015;43:D153–159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michiels S, Piedbois P, Burdett S, Syz N, Stewart L, Pignon JP. Meta-analysis when only the median survival times are known: a comparison with individual patient data results. Int J Technol Assess Health Care. 2005;21:119–125. doi: 10.1017/s0266462305050154. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Zhang Y, Zhang X, Zhang M, Liu H, Zhang S, Qi B, Sun X. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed Pharmacother. 2015;75:153–158. doi: 10.1016/j.biopha.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J Bone Oncol. 2016;5:74–79. doi: 10.1016/j.jbo.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu SJ, Zhi H, Chen PZ, Chen W, Lu F, Ma GS, Dai JC, Shen C, Liu NF, Hu ZB, Wang H, Shen HB. Fatty acid desaturase 1 polymorphisms are associated with coronary heart disease in a Chinese population. Chin Med J (Engl) 2012;125:801–806. [PubMed] [Google Scholar]
- 39.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikitovic D, Berdiaki K, Chalkiadaki G, Karamanos N, Tzanakakis G. The role of SLRP-proteoglycans in osteosarcoma pathogenesis. Connect Tissue Res. 2008;49:235–238. doi: 10.1080/03008200802147589. [DOI] [PubMed] [Google Scholar]
- 41.Richardson AD, Yang C, Osterman A, Smith JW. Central carbon metabolism in the progression of mammary carcinoma. Breast Cancer Res Treat. 2008;110:297–307. doi: 10.1007/s10549-007-9732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pak K, Suh S, Kim SJ, Kim IJ. Prognostic value of genetic mutations in thyroid cancer: a meta-analysis. Thyroid. 2015;25:63–70. doi: 10.1089/thy.2014.0241. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Q, Feng Y, Mao X, Qie M. Prognostic value of fluorine-18-fluorodeoxyglucose positron emission tomography or PET-computed tomography in cervical cancer: a meta-analysis. Int J Gynecol Cancer. 2013;23:1184–1190. doi: 10.1097/IGC.0b013e31829ee012. [DOI] [PubMed] [Google Scholar]
- 44.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 45.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egger M, G Davey Smith, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]


