Abstract
We assessed the performance of a rapid, single-well, real-time PCR assay for the detection of rifampin-resistant Mycobacterium tuberculosis by using clinical isolates from north India and Mexico, regions with a high incidence of tuberculosis. The assay uses five differently colored molecular beacons to determine if a short region of the M. tuberculosis rpoB gene contains mutations that predict rifampin resistance in most isolates. Until now, the assay had not been sufficiently tested on samples from countries with a high incidence of tuberculosis. In the present study, the assay detected mutations in 16 out of 16 rifampin-resistant isolates from north India (100%) and in 55 of 64 rifampin-resistant isolates from Mexico (86%) compared to results with standard susceptibility testing. The assay did not detect mutations (a finding predictive of rifampin susceptibility) in 37 out of 37 rifampin-susceptible isolates from India (100%) and 125 out of 126 rifampin-susceptible isolates from Mexico (99%). DNA sequencing revealed that none of the nine rifampin-resistant isolates from Mexico, which were misidentified as rifampin susceptible by the molecular beacon assay, contained a mutation in the region targeted by the molecular beacons. The one rifampin-susceptible isolate from Mexico that appeared to be rifampin resistant by the molecular beacon assay contained an S531W mutation, which is usually associated with rifampin resistance. Of the rifampin-resistant isolates that were correctly identified in the molecular beacon assay, one contained a novel L530A mutation and another contained a novel deletion between codons 511 and 514. Overall, the molecular beacon assay appears to have sufficient sensitivity (89%) and specificity (99%) for use in countries with a high prevalence of tuberculosis.
Multidrug-resistant tuberculosis (MDR-TB) is an increasing problem worldwide (13). MDR-TB is associated with significant mortality (12, 23) and has resulted in serious institutional outbreaks (5). Rapid diagnostic assays for MDR-TB should address these problems by enabling early isolation and treatment of patients with this disease (9, 17). Rifampin resistance is an excellent marker for multidrug-resistant Mycobacterium tuberculosis, as 90% of rifampin-resistant M. tuberculosis strains are also isoniazid resistant and, hence, are classified as multidrug resistant (20). Rifampin resistance is also amenable to detection by rapid genotypic assays, because approximately 95% of all rifampin-resistant strains contain mutations localized in an 81-bp core region of the bacterial RNA polymerase gene, rpoB (11, 17). Moreover, virtually all mutations that occur in this region result in rifampin resistance. By contrast, nearly all rifampin-susceptible M. tuberculosis isolates have the same wild-type nucleotide sequence in this region (11, 17, 19).
Various molecular methods have been developed to rapidly detect mutations in the M. tuberculosis rpoB core region, including the line probe assay (3), single-strand conformational polymorphism (SSCP) PCR (2, 20), and real-time PCR (6, 21, 22). Researchers developed a molecular beacon-based real-time PCR assay for this purpose (14, 15) and later converted this method into a multicolor, single-tube assay format (4). The single-well molecular beacon assay used five molecular beacons, each hybridizing to a different target segment within the rpoB core region and each labeled with a differently colored fluorophore. Each molecular beacon was designed to be so specific that it could not bind to its target if the target sequence differed from the wild-type M. tuberculosis rpoB sequence by even a single nucleotide substitution (10). Because molecular beacons fluoresce only when they are bound to their targets (24), the absence of fluorescence from any fluorophore in the assay indicates the presence of a mutation and thus predicts rifampin resistance (4). The assay thus has the advantage that it can detect unknown mutations in the rpoB region. The assay was simple, rapid, specific, and highly sensitive in tests on isolates of M. tuberculosis from New York City and Madrid (15). It also correctly predicted that 11 clinical sputum samples collected in Rio de Janeiro (Brazil) were rifampin susceptible (4). However, the ability of the assay to detect rpoB mutations in countries with a high incidence of tuberculosis, where different mutations could cause rifampin resistance, had not been tested. Here we assess the suitability of the single-tube molecular beacon assay to detect mutations in the rpoB gene of clinical M. tuberculosis isolates from the high-incidence countries India and Mexico.
MATERIALS AND METHODS
M. tuberculosis isolates.
A total of 243 isolates of M. tuberculosis from patients from north India and Mexico were tested for mutations associated with resistance to rifampin by the molecular beacon assay. Thirty-seven rifampin-susceptible and 16 rifampin-resistant isolates were obtained from 53 patients with tuberculosis in the outpatient department of Respiratory Medicine at the Vallabhbhai Patel Chest Institute in Delhi, India, between January 2001 and January 2002. The Vallabhbhai Patel Chest Institute serves as a referral center for patients with respiratory diseases in north India. A large number (33%) of these patients had histories of previous treatment at the time of collection of their sputa. The isolates were biochemically characterized with nitrate reduction, niacin production, catalase (7), and BACTEC NAP tests (Becton Dickinson Microbiology Systems, Sparks, Md.). All 53 isolates were also characterized by IS6110 fingerprinting (1). Another 190 isolates of M. tuberculosis were obtained from the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, in Mexico City, Mexico, which serves as a reference center for patients with tuberculosis. The Mexican isolates were obtained from three different geographical regions (Mexico City, Huauchinango, and Orizaba) between 1995 and 2002 and were identified by BACTEC (Becton Dickinson) and the NAP test (Accu-Probe). Fifty-nine of the isolates had been characterized by IS6110 typing (25). Of the 190 Mexican isolates, 64 were rifampin resistant. No isolate from either country was rifampin monoresistant.
Susceptibility testing.
The susceptibility of the Delhi M. tuberculosis isolates to isoniazid, rifampin, ethambutol, and streptomycin was tested by the standard proportion method (7). Resistance was defined as greater than 1% growth in the presence of 0.2 μg of isoniazid/ml, 1 μg of rifampin/ml, 5 μg of ethambutol/ml, and 2 μg of streptomycin/ml (7). The susceptibility of the Mexican isolates to the primary antituberculosis drugs was determined by the 460 TB BACTEC system (Becton Dickinson) at the Instituto Nacional Ciencias Médicas y Nutrición, as described previously (2).
Sample preparation for PCR.
The M. tuberculosis reference strain H37Rv was used as a positive control in the molecular beacon assays. Genomic DNA was extracted from H37Rv and clinical M. tuberculosis isolates by treatment with cetyltrimethylammonium bromide (CTAB) (Sigma, St. Louis, Mo.) in the presence of 0.7 M sodium chloride, as described previously (25).
Oligonucleotide sequences.
The nucleotide sequences of the molecular beacons and primers used in this study have been described previously (4). Five molecular beacons were designed so that they hybridized to different segments of the wild-type sequence of the M. tuberculosis rpoB core region. Each 15- to 20-bp-long probe sequence was selected so that the probe-target hybrid was just strong enough to overcome the strength of the hairpin stem under assay conditions. The five molecular beacons were labeled with five differently colored fluorophores so that they could be well distinguished from each other in a single reaction as described previously (4).
Assay conditions.
PCR was performed in 96-well microtiter plates (Applied Biosystems, Foster City, Calif.) as described previously (4). Fluorescence was measured in every well during the annealing step throughout the course of each reaction. The spectral data were automatically analyzed by the computer program controlling the spectrofluorometric thermal cycler to determine the fluorescence intensity contributed by each of the differently colored molecular beacons. The presence of a mutation within the 81-bp rpoB core region was detected by the absence of a characteristic rising fluorescence signal from one of the five molecular beacons in the assay. Conversely, M. tuberculosis isolates were predicted to be rifampin susceptible when all five of the molecular beacons in the assay generated a characteristic rising signal.
DNA sequencing and SSCP analysis.
The sequences of rpoB core region amplicons were analyzed in a subset of the study isolates by automated DNA sequencing with an Applied Biosystems 3100 capillary sequencer using the primers described above. Fourteen isolates from Mexico had been previously analyzed by SSCP PCR, as described previously (2).
RESULTS
Molecular beacon assay results.
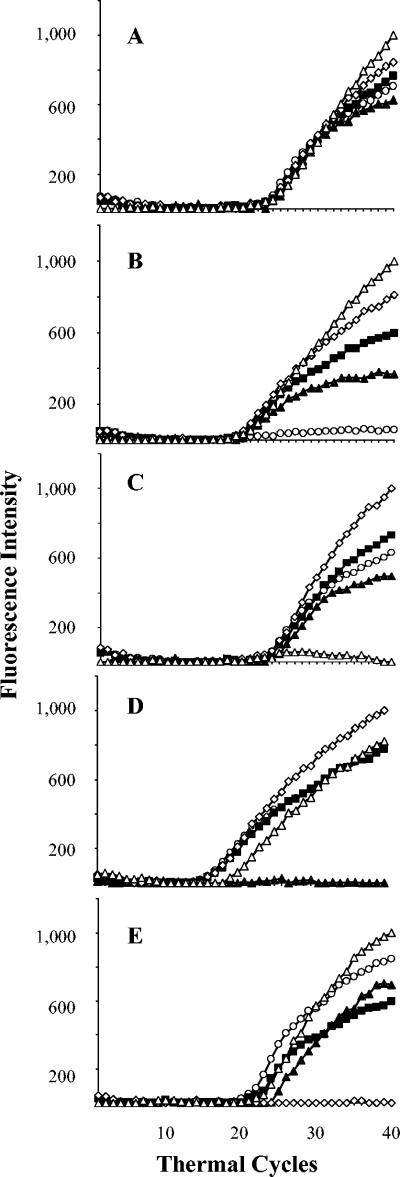
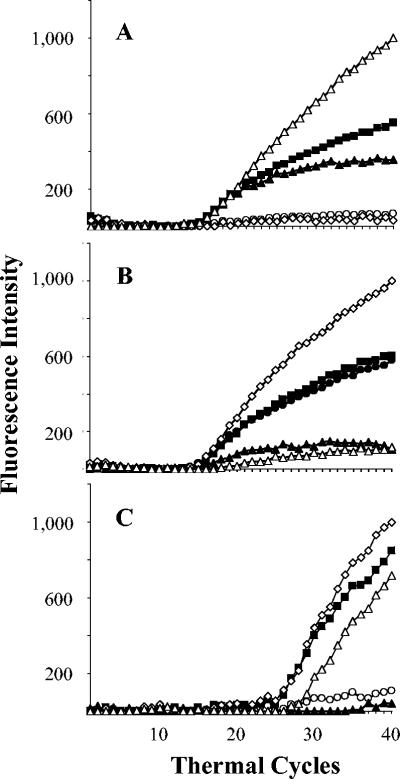
The results of the molecular beacon assays are summarized in Table 1. Typical assay results are shown in Fig. 1 and 2. Overall, the sensitivity of the assay in both populations was 89%, specificity was 99%, positive predictive value was 99%, and negative predictive value was 95%. Fluorescence signals in all five molecular beacons developed in all 37 of the rifampin-susceptible isolates from Delhi, India (specificity, 100%), including 5 rifampin-susceptible isolates that had previously been misidentified as being rifampin resistant by the standard proportion method. The rifampin susceptibility of these five isolates was confirmed by repeat susceptibility testing after the results of the molecular beacon assays were known. All 16 of the rifampin-resistant isolates from Delhi produced a flat signal for at least one of the molecular beacon probes (sensitivity, 100%) (Fig. 1). Probe E most commonly detected a mutation, followed by probes B, D, and A (Table 2). None of the isolates from Delhi appeared to contain a mutation in the region of probe C. Two molecular beacons failed to fluoresce in each of three isolates from Delhi (Fig. 2). In one isolate, both probe A and probe D failed to fluoresce; in a second isolate, probe B and probe E failed to fluoresce; and in a third isolate, probe A and probe B failed to fluoresce. These results suggested that the three isolates contained more than one mutation in the rpoB core region.
TABLE 1.
Comparison of molecular beacon assay results with those of phenotypic susceptibility testing
| Origin | Total no. of isolates | Sensitivitya | Specificityb | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|---|
| India | 53 | 100 (16/16) | 100 (37/37) | 100 | 100 |
| Mexico | 190 | 86 (55/64) | 99 (125/126) | 98 | 93 |
| Total | 243 | 89 (71/80) | 99 (162/163) | 99 | 95 |
Ability of the molecular beacon assay to detect resistance, expressed as a percentage (number of isolates resistant by both methods/number resistant by phenotypic susceptibility testing).
Ability of the molecular beacon assay to detect susceptibility, expressed as a percentage (number of isolates susceptible by both methods/number susceptible by phenotypic susceptibility testing).
FIG. 1.
Typical real-time PCR results for selected rifampin-susceptible and rifampin-resistant M. tuberculosis isolates. (A) A rifampin-susceptible isolate in which all five differently colored molecular beacons hybridized to the rpoB core region. (B to E) Rifampin-resistant isolates in which either (B) probe A, (C) probe B, (D) probe D, or (E) probe E failed to fluoresce. None of the isolates had a flat signal for probe C. The fluorescence of each molecular beacon is indicated as follows: ○, probe A; ▵, probe B; ▪, probe C; ▴, probe D; and ⋄, probe E.
FIG. 2.
Detection of double mutations or deletions. No fluorescence increase was observed for two differently colored probes in three of the isolates, suggesting the presence of multiple mutations. (A) Probes A and D failed to fluoresce; (B) probes B and E failed to fluoresce; and (C) probes and A and B failed to fluoresce. The fluorescence of each molecular beacon is indicated as follows: ○, probe A; ▴, probe B; ▪, probe C; ⋄, probe D; and ▵, probe E.
TABLE 2.
Distribution of assay results obtained with M. tuberculosis clinical isolates in which at least one flat fluorescence signal was observed
| Molecular beacon | No. of positive detections of fluorescence signal
|
||
|---|---|---|---|
| India | Mexico | Total | |
| A | 4a | 0 | 4 |
| B | 5b | 9 | 14 |
| C | 0 | 0 | 0 |
| D | 1 | 11 | 12 |
| E | 9 | 36 | 45 |
Double mutations were observed in two isolates (flat signals for probes A-D and A-B).
Double mutations were observed in two isolates (flat signals for probes A-B and B-E).
Fluorescence signals in all five molecular beacons developed in 124 of the 125 rifampin-susceptible Mexican isolates (specificity, 99%). A fluorescence signal failed to develop with probe E in one of the Mexican isolates identified as susceptible to rifampin by BACTEC analysis. Fifty-five of the 64 rifampin-resistant isolates from Mexico presented a flat signal for at least one of the molecular beacon probes, while fluorescence developed in all five molecular beacons in nine of the rifampin-resistant isolates (sensitivity, 86%). Probe E was again the most common molecular beacon with a flat signal, followed by probe D (Table 2). None of the isolates from Mexico gave a negative signal with probes A and C or a negative signal with more than one molecular beacon simultaneously.
DNA sequencing.
The rpoB core region was sequenced in the three Indian isolates that gave a single negative signal with probe A. All three isolates were found to have the L511P (CTG→CCG) mutation (Table 3). The Delhi isolates with two negative signals each were also sequenced. One contained a novel deletion between codons 511 and 514 (wild-type sequence TGAGCCAAT was deleted). The second isolate contained both the L511P (CTG→CCG) and the H526Q (CAC→CAG) mutations, and the third isolate contained both the N516A (GAC→GCC) and the L533P (CTG→CCG) mutations. The mutations in each isolate were consistent with the probes that failed to give a positive signal. The isolates from Delhi and Mexico that gave a flat signal in probe E were also sequenced. The most common mutations in this group were concentrated in codon 531 (Table 3). The most common mutation at this codon was S531L (TCG→TTG), which occurred in 7 out of 9 (78%) and 23 out of 36 (64%) of the Delhi and Mexican isolates, respectively. The second most common mutation was S531W (TCG→TGG), which occurred in 1 out of 9 (11%) and 8 out of 36 (22%) of the Delhi and Mexican isolates, respectively. A mutation that has not previously been identified, L530A, was found in a Mexican isolate that gave a flat signal in probe E.
TABLE 3.
Mutations detected in the rpoB core region from isolates with flat fluorescence signals for probe A or E
| Probe | Mutant codon | Nucleic acid substitution | Amino acid substitution | No. of mutant isolates
|
||
|---|---|---|---|---|---|---|
| India | Mexico | Total | ||||
| A | 511 | CTG→CCG | L→P | 3a | 0 | 3 |
| E | 530 | CTG→GCT | L→A | 0 | 1 | 1 |
| 531 | TCG→TTG | S→L | 7 | 23 | 30 | |
| TCG→TGG | S→W | 1 | 8 | 9 | ||
| TCG→TTC | S→F | 0 | 2 | 2 | ||
| 533 | CTG→CCG | L→P | 1 | 1 | 2 | |
| CTG→GCG | L→A | 0 | 1 | 1 | ||
A fourth isolate with a negative signal for probe A had a deletion between codons 511 and 514.
Nine of the rifampin-resistant isolates from Mexico gave positive fluorescence signals in all five probes, suggesting that their rpoB core region was a wild-type DNA sequence. No mutations were seen by DNA sequencing in the rpoB core region in eight of these isolates; the ninth isolate had an I572F (ATC→TTC) mutation located outside of the core region targeted by the five molecular beacons. One Mexican isolate that was susceptible to rifampin showed a negative fluorescence signal for probe E. DNA sequencing of this isolate showed that an S531W mutation (normally strongly associated with rifampin resistance) was present. Unfortunately, this isolate had lost viability during freezing storage and rifampin susceptibility could not be retested.
Comparison of the molecular beacon assay to single-strand conformational polymorphism analysis.
Twelve rifampin-resistant and two rifampin-susceptible Mexican isolates that were investigated in this study had previously been characterized by single-strand conformational polymorphism PCR (2). The SSCP PCR assay identified 4 of the 12 rifampin-resistant isolates as rifampin-resistant rpoB mutants, while the molecular beacon assay correctly identified 11 of these isolates as rifampin resistant. One rifampin-resistant isolate was identified as susceptible by both SSCP PCR and by the molecular beacon assay. DNA sequencing of this isolate revealed the existence of an I572F mutation outside the rpoB core region. Of the two rifampin-susceptible isolates, both were identified as susceptible by SSCP PCR. In contrast, one of the susceptible isolates was identified as rifampin resistant by the molecular beacon assay. This isolate, already described above, contained an S531W mutation in the rpoB core region. Thus, it is likely that this isolate was actually rifampin resistant and was correctly identified by the molecular beacon assay.
Correlation between IS6110 type and molecular beacon assay results.
IS6110 typing was carried out to confirm that the isolates of M. tuberculosis tested represented a diverse population. Many of the isolates studied (79% of the Indian strains and 38% of the Mexican strains) had unique banding patterns. Eleven of the Indian isolates and 28 of the Mexican isolates were identified as belonging to 5 and 14 clusters, respectively. However, the clusters were not associated with any specific mutation in the rpoB gene. These results suggest that the isolates studied were genetically unrelated and most likely developed rifampin resistance independently.
DISCUSSION
This study demonstrates that the molecular beacon assay effectively detects rifampin resistance in clinical M. tuberculosis isolates from countries with a high incidence of tuberculosis. Other advantages of this assay include the single-well format, the ability to combine PCR and post-PCR analysis into a single step, and the virtual elimination of cross-contamination conferred by the ability to perform assays in closed tubes. The assay detected mutations in the rpoB core region targeted by the molecular beacons every time a mutation was present. Conversely, the assay identified the core region as containing the wild-type sequence every time that mutations were absent from this region. The main limitation of the assay is that it is unable to detect rifampin resistance caused by mutations outside of the rpoB core region. Indeed, the assay had a sensitivity of 100% for the isolates from Delhi, but it had a sensitivity of 86% for isolates from Mexico, where it mistakenly identified 9 out of 64 rifampin-resistant isolates as being rifampin susceptible because these isolates did not have mutations in the rpoB core region. Screening assays should be highly specific, and the molecular beacon assay had an overall specificity of 99%. The assay identified 37 out of 37 rifampin-susceptible Indian isolates as being rifampin susceptible, and it identified 125 of 126 rifampin-susceptible Mexican isolates as being rifampin susceptible. Furthermore, the sole rifampin-susceptible isolate that was misidentified as being rifampin resistant contained an S531W mutation that has previously been shown to be associated with rifampin resistance (11, 16). Thus, it is likely that this susceptible isolate was, in fact, rifampin resistant. Unfortunately, this sample was not viable for repeat susceptibility testing. A potential problem of the assay is that it would be expected to detect silent rpoB mutations when they were present and would falsely identify such mutants as rifampin resistant. However, silent mutations are exceedingly rare in M. tuberculosis (19), thus, this problem does not have an important effect on specificity. In fact, it is worth noting that in the present study, the assay correctly identified five rifampin-susceptible isolates from India that had been initially classified as rifampin resistant by conventional susceptibility testing (but later confirmed to be susceptible by repeat testing). This observation suggests that the specificity of the molecular beacon assay may sometimes be higher than that of conventional susceptibility testing.
We found that four isolates from Delhi gave negative signals with probe A. Three of these isolates had mutations in codon 511, and the fourth had a novel deletion spanning codons 511 to 514. Mutations at codon 511 have been found by other workers in India (8). No rifampin-resistant isolate from Mexico contained a mutation in this region. This difference could reflect regional strain variations or differences in host factors. None of the 243 isolates showed a negative fluorescence signal with probe C, which targets rpoB codons 518 to 522. Earlier studies from India have reported mutations in codon 518, but only when they were accompanied by mutations in codon 531 (8). Other studies have reported mutations at codons 518, 521, and 522 at frequencies of only 0.8, 1.5, and 3%, respectively (18). Thus, it is possible that probe C could be omitted from future assays without a major effect on assay sensitivity. We also compared the results from the molecular beacon assays to results obtained with SSCP PCR in 14 isolates from Mexico City. Our results show that the molecular beacon assays were much more sensitive in detecting rifampin resistance in this group of isolates.
In summary, the molecular beacon assay was as effective at detecting mutations associated with rifampin resistance in M. tuberculosis isolates from northern India and Mexico as has previously been reported for isolates from the United States and Spain (15). The assay also identified rifampin-susceptible isolates that had previously been misidentified as resistant, further supporting the utility of a genetic approach to susceptibility testing. The assay was also more effective than SSCP PCR at detecting rifampin-resistant isolates. The assay is not dependent on probes hybridizing to specific mutant codons; hence, new and unknown mutations arising in a population, such as those identified in this study, can be easily detected with the same set of probes. With real-time instruments becoming more affordable, we anticipate that the assay can become economically feasible for developing countries in the near future. Ultimately, this assay will enable more rapid diagnosis, earlier treatment, and prompt implementation of infection control procedures to reduce the morbidity, mortality, and the spread of drug-resistant tuberculosis.
Acknowledgments
This work was supported by National Institutes of Health grants AI-46669 and EB-00277. M.V.-B. received an Overseas Associateship from the Department of Biotechnology of the Indian government. D.A. and F.K. are among a group of coinvestigators who hold patents in molecular beacons technology, and they receive income from licensees.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Boswoth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City, an analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 2.Bobadilla, M., A. Ponce-de-Leon, C. Arenas-Huertero, G. Vargas-Alarcon, M. Kato-Maeda, P. M. Small, P. Couary, G. M. Ruiz-Palacios, and J. Sifuentes-Osornio. 2001. rpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis identified by polymerase chain reaction single-strand conformational polymorphism. Emerg. Infect. Dis. 7:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Beenhouwer, H., Z. Lhiang, G. Jannes, W. Mijs, L. Machtelinckx, R. Rossau, H. Traore, and F. Portaels. 1995. Rapid detection of rifampicin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuber. Lung Dis. 76:425-430. [DOI] [PubMed] [Google Scholar]
- 4.El-Hajj, H., S. A. E. Marras, S. Tyagi, F. R. Kramer, and D. Alland. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 39:4131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frieden, T. R., L. F. Sherman, K. L. Maw, P. I. Fujiwara, J. T. Crawford, B. Nivin, V. Sharp, D. Hewlett, K. Brudney, D. Alland, and B. N. Kreiswirth. 1996. A multi-institutional outbreak of highly drug-resistant tuberculosis: epidemiology and clinical outcomes. JAMA 276:1229-1235. [PubMed] [Google Scholar]
- 6.García de Viedma, D., M. del Sol Diaz Infantes, F. Lasala, F. Chaves, L. Alcalá, and E. Bouza. 2002. New real-time PCR able to detect in a single tube multiple rifampin-resistance mutations and high-level isoniazid-resistance mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 40:988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology—a guide for the level III laboratory. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, Ga.
- 8.Mani, C., N. Selvakumar, S. Narayanan, and P. R. Narayanan. 2001. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J. Clin. Microbiol. 39:2987-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani, C., N. Selvakumar, N. Gajendiran, B. Panigrahi, P. Venkatesan, and P. R. Narayanan. 2003. Standardisation and evaluation of DNA-lanthanide fluorescence spectroscopy for determining rifampicin resistance in Mycobacterium tuberculosis clinical isolates. Int. J. Tuber. Lung Dis. 7:873-878. [PubMed] [Google Scholar]
- 10.Marras, S. A. E., F. R. Kramer, and S. Tyagi. 1999. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. 14:151-156. [DOI] [PubMed] [Google Scholar]
- 11.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pablos-Mendez, A., T. R. Sterling, and T. R. Frieden. 1996. The relationship between delayed or incomplete treatment and all-cause mortality in patients with tuberculosis. JAMA 276:1223-1228. [DOI] [PubMed] [Google Scholar]
- 13.Pablos-Mendez, A., M. C. Raviglione, A. Laszlo, N. Binkin, H. L. Rieder, F. Bustreo, D. L. Cohn, C. S. Lambregts-van Weezenbeek, S. J. Kim, P. Chaulet, P. Nunn, et al. 1998. Global surveillance for antituberculosis-drug resistance, 1994-1997. N. Engl. J. Med. 338:1641-1649. [DOI] [PubMed] [Google Scholar]
- 14.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 15.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 17.Riska, P. F., W. R. Jacobs, Jr., and D. Alland. 2000. Molecular determinants of drug resistance in tuberculosis. Int. J. Tuber. Lung Dis. 4:S4-S10. [PubMed] [Google Scholar]
- 18.Siddiqi, N., M. Shamim, S. Hussain, R. K. Choudhary, N. Ahmed, Prachee, S. Banerjee, G. R. Savithri, M. Alam, N. Pathak, et al. 2002. Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob Agents Chemother. 46:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 21.Torres, M. J., A. Criado, J. C. Palomares, and J. Aznar. 2000. Use of real-time PCR and fluorimetry for rapid detection of rifampin and isoniazid resistance-associated mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3194-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres, M. J., A. Criado, M. Ruiz, A. C. Llanos, J. C. Palomares, and J. Aznar. 2003. Improved real-time PCR for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. Diagn. Microbiol. Infect. Dis. 45:207-212. [DOI] [PubMed] [Google Scholar]
- 23.Turett, G. S., E. E. Telzak, L. V. Torian, S. Blum, D. Alland, I. Weisfuse, and B. A. Fazal. 1995. Improved outcomes for patients with multidrug-resistant tuberculosis. Clin. Infect. Dis. 21:1238-1244. [DOI] [PubMed] [Google Scholar]
- 24.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 25.van Embden, J., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406. [DOI] [PMC free article] [PubMed] [Google Scholar]