Abstract
A nonphotochromogenic, rapidly growing Mycobacterium strain was isolated in pure culture from the sputum and the bronchoalveolar fluid of a patient with hemoptoic pneumonia by using axenic media and an amoebal coculture system. Both isolates grew in less than 7 days at 24 to 37°C with an optimal growth temperature of 30°C. The isolates exhibited biochemical and antimicrobial susceptibility profiles overlapping those of Mycobacterium abscessus, Mycobacterium chelonae, and Mycobacterium immunogenum, indicating that they belonged to M. chelonae-M. abscessus group. They differed from M. abscessus in β-galactosidase, β-N-acetyl-β-glucosaminidase, and β-glucuronidase activities and by the lack of nitrate reductase and indole production activities, as well as in their in vitro susceptibilities to minocycline and doxycycline. These isolates and M. abscessus differed from M. chelonae and M. immunogenum by exhibiting gelatinase and tryptophane desaminase activities. Their 16S rRNA genes had complete sequence identity with that of M. abscessus and >99.6% similarity with those of M. chelonae and M. immunogenum. Further molecular investigations showed that partial hsp65 and sodA gene sequences differed from that of M. abscessus by five and three positions over 441 bp, respectively. Partial rpoB and recA gene sequence analyses showed 96 and 98% similarities with M. abscessus, respectively. Similarly, 16S-23S rRNA internal transcribed spacer sequence of the isolates differed from that of M. abscessus by a A→G substitution at position 60 and a C insertion at position 102. Phenotypic and genotypic features of these two isolates indicated that they were representative of a new mycobacterial species within the M. chelonae-M. abscessus group. Phylogenetic analysis suggested that these isolates were perhaps recently derived from M. abscessus. We propose the name of “Mycobacterium massiliense” for this new species. The type strain has been deposited in the Collection Institut Pasteur as CIP 108297T and in Culture Collection of the University of Göteborg, Göteborg, Sweden, as CCUG 48898T.
During the last few years, the number of nontuberculous mycobacteria (NTM) reported in various clinical situations has greatly increased because of opportunistic infections in immunocompromised patients and improved culture and identification techniques (18). The 16S rRNA gene sequence analysis of NTM led to the description of 40 new species since 1992 and contributed to the description of new clinical forms (19, 35, 43, 44). Particularly, mycobacteria of the Mycobacterium chelonae-Mycobacterium abscessus group (M. chelonae, M. abscessus, and Mycobacterium immunogenum) emerged as opportunistic pathogens (45, 47-49, 51), causing hypersensitivity pneumonitis in automobile production workers and chronic lung disease in elderly women with bronchiectasis and in young adults with cystic fibrosis (4, 9, 16, 36, 49). Precise species identification in this group of mycobacteria remains difficult. Only two biochemical tests, i.e., sodium chloride tolerance and utilization of citrate, allow species identification of these organisms (37, 49), but these tests require 4 weeks to complete (8). Identification of species of the M. chelonae-M. abscessus group by using high-performance liquid chromatography of mycolic acids has been limited by profile similarities (49). The 16S rRNA gene sequence of M. abscessus differs from the respective sequences of M. chelonae and M. immunogenum by only four and eight positions, respectively, over 1,483 bp (25, 49). We therefore developed partial PCR sequencing of rpoB gene for improved identification of NTM (1), and we also demonstrated its usefulness for taxonomy (2). When applying this tool to a collection of 23 M. chelonae-M. abscessus group clinical isolates, we found two isolates from the same patient that exhibited 96.0% similarity only with the sequence of M. abscessus type strain, although they shared 100% similarity of the 16S rRNA sequence and similar standard phenotypic profile. We further characterized these isolates by extensive phenotypic and molecular methods and found that they were representative of a hitherto-undescribed rapidly growing Mycobacterium (RGM) species. The name “Mycobacterium massiliense” sp. nov. is proposed for this new species.
CASE REPORT
A 50-year-old woman presented with an 8-year history of bronchiectasis and hemoptysis. She had no history of past tuberculosis but received substitutive corticosteroid treatment for an Addison's disease for 2 years. No bacteriological investigations had been performed during the course of her illness. The patient had had close contacts with horses, dogs, and cats and presented with a fever of 38°C, subcutaneous nodules of both forearms, impaired respiratory function, and micronodular lesions in the upper lobe of the right lung. Blood examination was unremarkable apart from a biological inflammatory syndrome. An NTM was isolated from the sputum sample and from a bronchoalveolar lavage obtained during bronchoscopy 2 months later by using amoebal coculture according to our laboratory protocol (26) and conventional medium inoculation. Direct Ziehl-Neelsen staining showed acid-fast bacilli. Histological examination of the forearm nodules disclosed swelling and nongranulomatous, nonspecific inflammatory infiltrate with lymphocytic cells predominantly present around the blood vessels. Ziehl-Neelsen and periodic acid-Schiff (PAS) stainings were negative, and the culture remained sterile. After isolation and identification of a M. chelonae-M. abscessus group strain in respiratory samples, the patient was treated with oral clarithromycin (1 g/day) for 1 month and oral minocycline (100 mg/day) for an additional 6 months. Clinical improvement was noted at the 6-month follow-up examination with disappearance of the cutaneous nodules and marked improvement of the respiratory function.
MATERIALS AND METHODS
Identification and characterization of the isolates.
Sputum and bronchoalveolar samples were decontaminated by using the N-acetyl-l-cysteine sodium hydroxide (NALC-NaOH) protocol (21, 23). Briefly, an equal volume of digestant (3% NaOH, 1.45% sodium citrate, 0.5% NALC [Merck, Darmstadt, Germany]) was added to up to 10 ml of specimen. After being vortexed, the mixture was shaken for 30 min and neutralized by adding sterile phosphate buffer (bioMérieux, La Balme les Grottes, France) to a final volume of 50 ml and then centrifuged at 3,000 rpm for 10 min. The supernatant was discarded, and the sediment was resuspended in 2 ml of phosphate buffer. Half of the sediment was frozen at −80°C, and the other part was used for acid-fast staining and inoculation into BACTEC 9000MB broth according to the manufacturer's instructions (BD Biosciences, Sparks, Md.). Mycobacterial isolates were subcultured on Middlebrook 7H10 agar, Lowenstein-Jensen (LJ) (egg-based) slants (bioMérieux), and 5% sheep-blood agar (bioMérieux). Solid and liquid cultures were inspected twice weekly for colonies and turbidity, respectively.
Amoebal coculture.
Respiratory specimens were cocultivated with amoebae according to our laboratory protocol for respiratory specimen processing (26). An Acanthamoeba polyphaga strain, Linc-AP1, kindly provided by T. J. Rowbotham (Public Health Laboratory, Leeds, United Kingdom), was grown at 28°C for 4 days in 150-cm3 culture flasks (Corning) with 30 ml of peptone-yeast extract-glucose broth to reach an average of 5 × 105 amoebae/ml (14, 15, 26). Amoebae were then harvested by centrifugation at 2,000 rpm for 10 min, and the pellet was resuspended twice in 30 ml of Page's modified Neff's amoeba saline (PAS) (14, 26). Then, 1 ml of a suspension of 5 × 105 A. polyphaga/ml was distributed into each well of a 12-well Costar microplate (Corning), and the decontaminated sputum sample was inoculated onto amoebal microplates. The microplates were then centrifuged at 4,000 rpm for 30 min, followed by incubation at 33°C under a humidified 5% CO2 atmosphere. The amoebal cocultures were subcultured onto fresh amoebae on day 4 and then examined daily for the occurrence of amoebal lysis. The presence of intra-amoebal bacteria was detected, after gentle shaking and cytocentrifugation at 800 rpm for 10 min as described by Gimenez (12), by Ziehl-Neelsen and Gram stainings.
Electron microscopy.
Inoculated A. polyphaga were washed overnight in monophosphate buffer (pH 12) and fixed for 1 h at 4°C at room temperature with 1% osmium tetroxide. Dehydration was performed by successive washes in increasing acetone concentrations (25 to 100%). After the preparations were incubated for 1 h in a suspension of acetone-Epon and overnight in Epon, they were embedded in araldite (Fluka, St. Quentin Fallavier, France). Thin sections were cut from blocks by using an Ultracut microtome (Reichert-Leica, Marseille, France), deposited on copper grids coated with Formvar (Sigma-Aldrich, Taufkirchen, Germany), and stained for 10 min with a solution of methanol-uranyl acetate and lead nitrate with sodium citrate in water (14). Grids were examined with a Morgani 268D electron microscope (Philips, Eindhoven, The Netherlands).
Phenotypic analysis.
We observed the colony morphology, pigmentation, and the ability of the isolates to grow at various temperatures (24, 30, 37, 42, and 45°C) on 5% sheep blood agar, Middlebrook 7H10 agar, and LJ slants. The following biochemical and physiological values were determined: the activities of arylsulfatase and catalase, the extent of iron uptake, the degradation of p-aminosalicylic acid, and growth on MacConkey agar without crystal violet and on LJ slants in the presence of 5% sodium chloride (21, 30). Additional biochemical tests were performed by inoculation of API Coryne and API 20E (bioMérieux) strips (42) according to the instructions of the manufacturer, with the exception that incubation time was increased to 3 days at 30°C under a highly humidified atmosphere. Every test was done three times in order to ensure the reproducibility of results.
Susceptibility testing.
Suspensions were prepared by emulsifying colonies into 5 ml of sterile water to achieve a density equal to a 1.0 McFarland turbidity standard by visual examination. Suspensions were mixed vigorously on a vortex mixer for 15 to 20 s and then used to inoculate the entire surface of a 5% sheep blood agar plate. The MICs of rifampin, ciprofloxacin, sparfloxacin, ofloxacin, minocycline, clarithromycin, doxycycline, amikacin, penicillin, imipenem, amoxicillin, ceftriaxone, cefotaxime, metronidazole, teicoplanin, and vancomycin were determined by incubation with the respective E-test (AB Biodisk, Solna, Sweden) at 30°C on 5% sheep blood agar for 3 days. For each drug, the MIC was recorded as the point of intersection between the zone edge and the E-test strip. The strains were classified as either susceptible, intermediate, or resistant to a particular antibiotic according to breakpoints recommended by the National Committee for Clinical Laboratory Standards (29) Additional disk diffusion method on 5% sheep blood agar for 3 days at 30°C was used to determine the susceptibility to trimethoprim-sulfamethoxazole (1.25 μg/23.75 μg), tobramycin (10 μg), pipemidic acid (30 μg), and cefoxitin (30 μg). Using a disk diffusion method for certain antimicrobials, the zone sizes for resistance used were those of Grange and Stanford (13). Growth on LJ slants in the presence of isoniazid (0.1, 1, and 10 μg/ml), streptomycin (4 μg/ml), ethambutol (2 μg/ml), and para-aminosalicylic acid (0.5 μg/ml) was also evaluated. Each susceptibility test was done in triplicate in three separate days in order to ensure the reproducibility of results.
Measurement of G+C content of DNA.
After culture on 5% sheep blood agar, DNA extraction, purification, degradation, and G+C content determinations by HPLC were done as described by Mesbah et al. (27), except that a Waters 625 LC system with a Waters 486 Tenable Absorbance Detector and a Waters 746 Data Module (Millipore, Saint Quentin en Yvelines, France) were used. Triplicate determinations were made.
Sequence and phylogenetic analyses.
DNA was extracted from colonies grown on 5% sheep blood agar by using the Fast-Prep device and the FastDNA kit according to the recommendations of the manufacturer (Bio 101, Inc., Carlsbad, Calif.). We performed the amplification and sequencing of the 16S rRNA (50), hsp65 (40), rpoB (1), sodA (2), and recA (3) genes. The 16S-23S rRNA internal transcribed spacer (ITS) was amplified and sequenced by using the primer pair Sp1 and Sp2 (33). Products of sequencing reactions were recorded with an ABI Prism 3100 DNA sequencer according to the standard protocol of the supplier (Perkin-Elmer Applied Biosystems). The percentages of similarity between the sequences were determined by using the CLUSTAL W program supported by the PBIL website (http://npsa-pbil.ibcp.fr/cgi-bin/npsa). For phylogenetic analyses, sequences were trimmed in order to start and finish at the same nucleotide position for all of the strains under study. Multisequence alignment was performed by using the CLUSTAL X program (version 1.81) in the PHYLIP software package (41). Phylogenetic trees were obtained from DNA sequences by using the neighbor-joining method with Kimura's two-parameter distance correction model with 1,000 bootstrap replications in the MEGA version 2.1 software package (24). The sequences determined in the present study for “M. massiliense” were deposited in GenBank under the following access numbers: 16S rRNA, AY593980; hsp65, AY596465; sodA, AY593975; recA, AY593979; rpoB, AY593981; and 16S-23S ITS, AY593978.
RESULTS
Phenotypic characteristics and susceptibility testing.
The two isolates were strictly aerobic, nonmotile, non-spore-forming, acid-fast, gram-positive rods. The colonies on 5% sheep blood agar were nonphotochromogenic and intermediate between smooth and rough. Growth occurred by between 2 and 4 days on 5% sheep blood agar, Middlebrook 7H10 agar, and LJ slants at between at 24 and 37°C; no growth was observed at 42 and 45°C. Both isolates grew at 30°C on MacConkey agar without crystal violet and LJ slants containing 5% NaCl. They exhibited pyrazinamidase, pyrrolidonyl arylamidase, phosphatase alkaline, β-galactosidase, α-glucosidase, esculine (β-glucosidase), N-acetyl-β-glucosaminidase, gelatinase, catalase, and 3-day arylsulfatase activities and produced acetoin. The isolates lacked nitrate reductase, β-glucuronidase, urease, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, sodium citrate utilization, and tryptophane desaminase activities and H2S production. They did not show iron uptake and degradation of para-aminosalicylic acid. Also, they did not utilize glucose, ribose, xylose, mannitol, maltose, lactose, saccharose, glycogene, inositol, sorbitol, rhamnose, melobiose, amygdaline, or arabinose as the sole carbon source. Antibiotic susceptibility profiles for the two isolates included susceptibility to amikacin (MIC, 32 μg/ml), clarithromycin (MIC, 0.064 μg/ml), minocycline (MIC, 4 μg/ml),and doxycycline (MIC, 0.75 μg/ml) and resistance to cefoxitin (MIC, >2 μg/ml), rifampin (MIC, >32 μg/ml), ciprofloxacin (MIC, >32 μg/ml), sparfloxacin (MIC > 32 μg/ml), ofloxacin (MIC, >32 μg/ml), streptomycin (MIC, >4 μg/ml), tobramycin (MIC, >8 μg/ml), penicillin (MIC, >32 μg/ml), amoxicillin (MIC, >32 μg/ml), imipenem (MIC, >32 μg/ml), ceftriaxone (MIC, >256 μg/ml), cefotaxime (MIC, >256 μg/ml), vancomycin (MIC, >256 μg/ml), teicoplanin (MIC, >256 μg/ml), metronidazole (MIC, >256 μg/ml), trimethoprim-sulfamethoxazole (MIC, >8 μg/ml), isoniazid (MIC, >10 μg/ml), and ethambutol (MIC, >2 μg/ml). All of the phenotypic and antibiotic susceptibility tests gave identical results over three replicates. Biochemical and susceptibility characteristics that differentiated the isolates from the species of the M. chelonae-M. abscessus group are summarized in Tables 1 and 2.
TABLE 1.
Comparison of biochemical characteristics of “M. massiliense” and closely related mycobacterial speciesa
| Characteristic | Presence (+) or absence (−) of various characteristics in:
|
|||
|---|---|---|---|---|
| “M. massiliense” | M. abscessus | M. chelonae | M. immunogenum | |
| Growth at 24°C | + | + | + | + |
| Growth at 30°C | + | + | + | + |
| Growth at 37°C | + | + | + | + |
| Growth at 42°C | − | − | − | − |
| Growth at 45°C | − | − | − | − |
| Pigmentation | − | − | − | − |
| Arylsulfatase (3 days) | + | + | + | + |
| Catalase | + | + | + | + |
| Semiquantitative catalase | + | + | + | + |
| 5% NaCl tolerance (30°C) | + | + | − | − |
| MacConkey (without crystal violet) | + | + | + | + |
| Degradation of p-aminosalicylate | − | − | − | − |
| Iron uptake | − | − | − | − |
| Nitrate reductase | − | + | + | − |
| Pyrazinamidase | + | + | + | ± |
| Pyrrolidonyl arylamidase | + | + | + | + |
| Phosphatase alkaline | + | + | + | + |
| β-Glucuronidase | − | + | − | − |
| β-Galactosidase | + | − | − | + |
| α-Glucosidase | + | + | + | + |
| N-Acetyl-β-glucosaminidase | + | − | − | − |
| Esculin | + | + | + | + |
| Urease | − | − | − | − |
| Gelatinase | + | + | − | − |
| Arginine dihydrolase | − | − | − | − |
| Lysine decarboxylase | − | − | − | − |
| Ornithine decarboxylase | − | − | − | − |
| Citrate | − | − | + | − |
| H2O production | − | − | − | − |
| Tryptophane desaminase | + | + | − | − |
| Indol production | − | + | − | − |
| Acetoin production | + | + | + | + |
| Glucose | − | − | − | − |
| Ribose | − | − | − | − |
| Xylose | − | − | − | − |
| Mannitol | − | − | − | − |
| Maltose | − | − | − | − |
| Lactose | − | − | − | − |
| Saccharose | − | − | − | − |
| Glycogen | − | − | − | − |
| Inositol | − | − | − | − |
| Sorbitol | − | − | − | − |
| Rhamnose | − | − | − | − |
| Meliobiose | − | − | − | − |
| Amygdaline | − | − | − | − |
| Arabinose | − | − | − | − |
Results were reproducible over triplicate testing. Colony morphology was rough/smooth for all four species.
TABLE 2.
Antimicrobial susceptibility test results for “M. massiliense” and closely related mycobacterial speciesa
| Antimicrobial | MIC (μg/ml) for:
|
|||
|---|---|---|---|---|
| “M. massiliense” | M. abscessus | M. chelonae | M. immunogenum | |
| Clarithromycin | 0.064 | 1.5 | 0.125 | 0.75 |
| Minocycline | 4 | >256 | 16 | >256 |
| Doxycycline | 0.75 | >256 | >256 | >256 |
| Cefoxitin (disc, 30 μg) | >32 | >32 | >32 | >32 |
| Colistin (disc, 50 μg) | >2 | >2 | >2 | >2 |
| Rifamplin | >32 | >32 | >32 | >32 |
| Ciprofloxacin | >32 | >32 | 0.5 | >32 |
| Sparfloxacin | >32 | >32 | 8 | >32 |
| Ofloxacin | >32 | >32 | 2 | >32 |
| Amikacin | 48 | 32 | 48 | >256 |
| Imipenem | >32 | >32 | >32 | >32 |
| Penicillin | >32 | >32 | >32 | >32 |
| Amoxicillin | >256 | >256 | >256 | >256 |
| Ceftriaxone | >256 | >256 | >256 | >256 |
| Cefotaxime | >256 | >256 | >256 | >256 |
| Vancomycin | >256 | >256 | >256 | >256 |
| Teicoplanin | >256 | >256 | >256 | >256 |
| Metronidazole | >256 | >256 | >256 | >256 |
| Tobramycin (disc, 10 μg) | >8 | >8 | >8 | >8 |
| Trimethoprim-sulfamethoxazole (disc, 1.25 μg + 23.75 μg) | >8 and >152 | >8 and >152 | >8 and >152 | >8 and >152 |
| Acid pipemidic (disc, 20 μg) | >16 | >16 | >16 | >16 |
| Isoniazid | >10 | >10 | NTb | NT |
| Streptomycin | >4 | >4 | NT | NT |
| Ethambutol | >2 | >2 | NT | NT |
Results were reproducible over triplicate testing.
NT, not tested.
Intra-amoebal survival and growth.
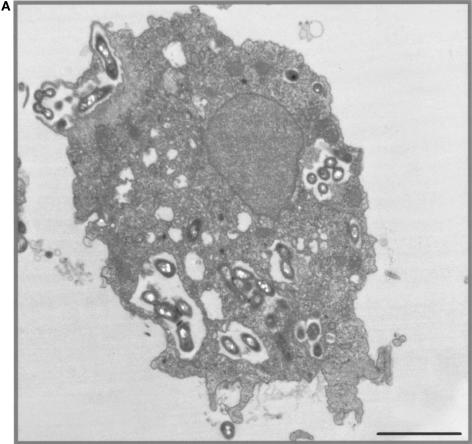
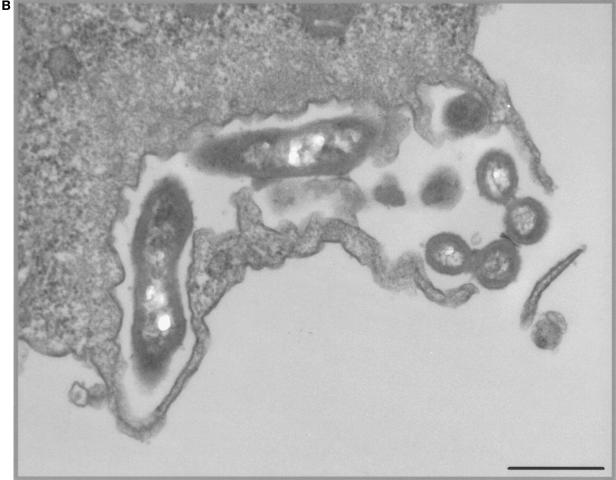
No amoebal lysis was observed during the time of coculture. Acid-fast and Gimenez stainings, and transmission electron microscopy demonstrated mycobacterial internalization by A. polyphaga during coculture. After 3 days of coculture, electron microscopy observation disclosed the intravacuolar location with one to five mycobacteria per vacuole (Fig. 1A). They appeared as slightly curved or straight bacilli, 0.4 μm in width and 2 μm in length. Cell walls exhibited a trilamellar structure, including waxes with characteristic mycolic acid with long-branched chains (Fig. 1B).
FIG. 1.
(A) Intravacuolar location of “M. massiliense” within A. polyphaga, as shown by electron microscopy. Magnification, ×39,200. Bar, 2 μm. (B) Trilamellar structure of the cell wall of “M. massiliense,” as seen by electron microscopy. The electron micrograph shows the rod element. Magnification, ×30,800. Bar, 0.2 μm.
G+C content of DNA.
The G+C content of the DNA was 65 ± 3 mol%.
Genotypic analyses of the 16S rRNA, hsp65, sodA, recA, and rpoB genes and of the 16S-23S rRNA ITS.
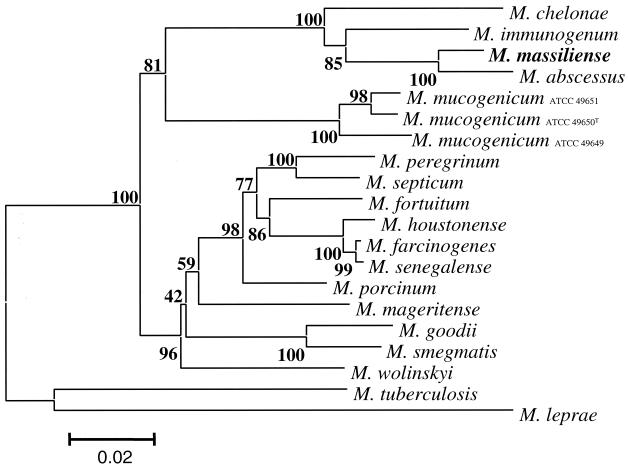
The 16S rRNA gene sequence of both isolates had complete identity with that of M. abscessus CIP 104536T over 1,483 bp. The 441-bp hsp65 gene region described by Telenti et al. (40) differed by 5, 31, and 35 nucleotides from that of M. abscessus, M. chelonae, and M. immunogenum, respectively. The partial 441-bp sodA gene region (2) differed by 3, 26, and 31 nucleotides from that of M. abscessus, M. chelonae, and M. immunogenum, respectively. The partial 915-bp recA gene sequence analysis (3) showed 98.0, 89.8, and 91.8% similarity with M. abscessus, M. chelonae, and M. immunogenum, respectively. The partial 723-bp rpoB gene sequence analysis (1) showed 96.0, 95.4, and 97.0% sequence similarity with M. abscessus, M. chelonae, and M. immunogenum, respectively. Based on the 16S-23S rRNA ITS (33), this new species differed from M. abscessus by the substitution of A→G at position 60 and the insertion of a C at position 102 of the spacer sequence (Fig. 2). Further 16S-23S rRNA ITS sequence analysis showed 19 and 32 position differences from M. chelonae and M. immunogenum, respectively, over 224-bp. A phylogenetic tree based on combined rpoB and recA gene partial sequences by incorporating 17 RGM species and the new taxon is shown in Fig. 3. A bootstrap value of 100% in the neighbor-joining tree supported the fork separating “M. massiliense” from M. abscessus. Parsimony and maximum-likelihood methods confirmed that “M. massiliense” clustered with species of the M. chelonae-M. abscessus group. Its lineage was clearly different from that of M. abscessus and quite distant from other recognized species (Fig. 3).
FIG. 2.
Alignment of the of partial 16S-23S rRNA ITS region (120 nucleotides) of “M. massiliense” (GenBank accession number AY593978) and M. abscessus (GenBank accession number AY593976).
FIG. 3.
Phylogenetic tree of the combined rpoB+recA gene sequences of “M. massiliense” and 17 RGM prepared by using the neighbor-joining method and Kimura's two-parameter distance correction model. The support of each branch, as determined from 1,000 bootstrap samples, is indicated by the value at each node (as a percentage). M. tuberculosis and M. leprae were used as the outgroups. The scale bar represents a 2% difference in nucleotide sequences.
DISCUSSION
When we applied rpoB to the identification of clinical isolates belonging to the M. chelonae-M. abscessus group (1), we found two isolates from the same patient sharing 96.0% sequence similarity only with that of M. abscessus type strain, suggesting that they were significantly distant from known taxons of this group. Indeed, in a previous study, we showed that an RGM belonged to the same species if it had <2% sequence divergence with one of the 20 studied species and that it belonged to a new species if it exhibited >3% sequence divergence from the corresponding type strain with the partial 723-bp rpoB sequence (2). We therefore further characterized these two isolates by a polyphasic approach comprising biochemical and antimicrobial susceptibility tests and genotypic analysis. One carbohydrate source and three enzymatic activities clearly separated both isolates from M. abscessus CIP 104536T, since “M. massiliense” exhibited β-galactosidase and N-acetyl-β-glucosaminidase activities, whereas M. abscessus did not. “M. massiliense” had no β-glucuronidase activity and did not produce indole, in contrast to M. abscessus. Interestingly, “M. massiliense” and M. abscessus both differed from M. chelonae and M. immunogenum type strains by exhibiting gelatinase and tryptophane desaminase activities. “M. massiliense” and M. immunogenum were negative for the nitrate reduction, whereas M. abscessus and M. chelonae were positive. Discrepancies in the result of nitrate reduction test with that previously reported (37) may due to the fact that we used miniaturized API strip instead of conventional method (30). “M. massiliense” did not exhibit both iron uptake and urease activities, like the three species of the M. chelonae-M. abscessus group (37, 49). However, it exhibited some reactions typical of M. abscessus, such as the inability to utilize citrate, sorbitol, inositol, and mannitol as sole carbon sources (37) (Table 1).
Previous antibiotic susceptibility testing of a large collection of 45 M. abscessus isolates found them to be resistant to doxycycline (52), in contrast to the two “M. massiliense” isolates that were susceptible. This could be a useful marker for differentiating “M. massiliense” from M. abscessus. However, the clinical relevance of in vitro antibiotic susceptibility testing of “M. massiliense” is unknown; interestingly, the patient was cured after treatment with clarithromycin and minocycline, and “M. massiliense” was susceptible in vitro to these antibiotics. However, “M. massiliense” shared resistance to tobramycin (MIC, >8 μg/ml) and ciprofloxacin (MIC, >32 μg/ml) in common with M. abscessus (5, 39, 52).
We previously showed that 16S rRNA gene sequence did not discriminate all NTM taxa (1, 2). We proposed that rpoB gene sequence difference of >3% and a recA gene sequence difference of >2% discriminated NTM species (1-3).The 16S rRNA sequence of both isolates showed complete identity with that of the M. abscessus type strain. However, the 16S rRNA gene sequence poorly discriminates mycobacterial species such as Mycobacterium gastri and Mycobacterium kansasii (11); Mycobacterium senegalense, Mycobacterium farcinogenes, and “Mycobacterium houstonense” (formerly M. fortuitum third biovariant sorbitol positive) (2); and species of the Mycobacterium tuberculosis complex (11, 44). The two isolates exhibited 96.0% partial rpoB sequence similarity with that of the M. abscessus. Also, their partial recA gene sequence (3) showed 98.0% similarity with M. abscessus. Blackwood et al. found that in mycobacteria, the intraspecies sequence similarity of this region ranged from 98.7 to 100% (3). Furthermore, “M. massiliense” and M. abscessus were clearly differentiated by hsp65 and sodA gene sequence analyses despite the low number of informative sites in those genes (five and three nucleotides, respectively) (2). On the other hand, M. fortuitum and M. senegalense, as well as M. peregrinum and M. septicum, which have the highest degree of 16S rRNA similarity (>99.6%) are separated from one another by only three nucleotides in their partial hsp65 gene sequences (2, 31). Likewise, M. senegalense and “M. houstonense” have complete 16S rRNA gene sequence identity over 1,483 bp but differed from each other by two nucleotides only within the sodA partial sequence. Furthermore, based on the 16S-23S rRNA ITS, the new taxon differed from M. abscessus by one substitution and one insertion. One substitution in 16S-23S rRNA ITS discriminated sequevars among M. farcinogenes and Mycobacterium avium isolates (17, 28). Roth et al. found that insertion or deletion in the 16S-23S rRNA ITS discriminated different species (32), with the exception of members of the M. tuberculosis complex (10). Recently, we showed that the bootstrap values at the nodes of a few clusters with regard to the partial sodA and hsp65-based trees were too low to induce much confidence compared to the recA and rpoB gene-derived phylogenies (2), probably due to inadequate samples sizes (6). Furthermore, good congruence was obtained between the 16S rRNA and the combined rpoB and recA phylogenetic trees (2). We therefore combined the latter two genes to determine the evolutionary relationships of “M. massiliense” within the M. chelonae-M. abcessus group. This analysis demonstrated that “M. massiliense” was more closely related to M. abscessus than to M. chelonae and M. immunogenum.
One of the two isolates reported here was cultured by using an amoeba coculture system (14, 26) initially described for the isolation of Legionella-like amoebal pathogen in the sputum of a woman with persistent pneumonia (34). Several other mycobacterial species were observed in amoebae. Mycobacterium leprae was shown to survive in free-living amoebae, but no mycobacterial multiplication occurred (20). Further experiments showed that Mycobacterium avium, Mycobacterium marinum, Mycobacterium ulcerans, Mycobacterium simiae, and “Mycobacterium habane” could enter free-living amoebae; Mycobacterium smegmatis, Mycobacterium fortuitum, and Mycobacterium phlei were in large numbers in the amoebae, eventually inducing amoebal lysis (7, 22, 38).
In the present study, we describe a new RGM species, “M. massiliense,” isolated in amoebic coculture from the lower respiratory tract of a 50-year-old woman with hemoptoic pneumonia. The same strain was recovered two times over a 2-month period. An NTM is generally considered to be pathogenic if it isolated (i) repeatedly from different samples during several weeks or in large amounts, (ii) in the presence of a coexisting bronchopulmonary disease, or (iii) when the clinical presentation and X-ray pattern is suggestive (pseudotuberculosis) (16, 46). The new taxon could therefore be considered pathogenic. Additional arguments in favor of a pathogenic role are the facts that the closely related species M. abscessus has been shown to cause pulmonary disease and that the underlying diseases included bronchiectasis, cystic fibrosis, gastroesophageal disorders, and prior granulomatous disease such as sarcoidosis or tuberculosis (9, 16, 36). A study of M. abscessus pulmonary disease emphasized striking similarities to M. avium complex lung disease of the type known as nodular bronchiectasis (16). These similarities suggested a common pathogenicity or host susceptibility (16). Patients infected with M. avium complex and most patients with M. abscessus pulmonary disease had underlying nodular bronchiectasis (46). Approximately 20% of patients with M. abscessus infection eventually developed M. avium complex infection (16). In a later study, M. abscessus has been identified by using the microbiologic and radiographic criteria of the American Thoracic Society (16) due to the lack of molecular identification methods at that time. The use of the partial rpoB gene for further genetic analysis would be helpful to better assess the clinical relevance of the species or subspecies that are associated with disease since the M. abscessus type strain was first isolated from a patient with abscess (25).
Since these mycobacteria were isolated in Marseille, we propose that the isolates be included in the genus Mycobacterium as “Mycobacterium massiliense,” a name that refers to Massilia, the old Greek and Roman name for Marseille.
Acknowledgments
We thank Christian de Fontaine, Lina Barrassi, and Nicolas Aldrovandi for technical assistance and Babu V. A. K. Gundi for expert review of the manuscript.
REFERENCES
- 1.Adékambi, T., P. Colson, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adékambi, T., and M. Drancourt. Dissection of phylogenic relationships among nineteen rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA, and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 3.Blackwood, K. S., C. He, J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J. Clin. Microbiol. 38:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microb. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown-Elliott B. A., R. J. Wallace, Jr., C. J. Crist, L. Mann, and R. W. Wilson. 2002. Comparison of in vitro activities of gatifloxacin and ciprofloxacin against four taxa of rapidly growing mycobacteria. Antimicrob. Agents Chemother. 46:3283-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull, J. J., J. Huelsenbeck, C. W. Cunningham, D. L. Swofford, and P. J. Wadell. 1993. Partitioning and combining data in phylogenetic analysis. Syst. Biol. 42:384-397. [Google Scholar]
- 7.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conville, P. S., and F. G. Witebsky. 1998. Variables affecting results of sodium chloride tolerance test for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 36:1555-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen, R. A., C. L. Cannon, E. J. Mark, and A. A. Colin. 2000. Mycobacterium abscessus infection in cystic fibrosis: colonization or infection? Am. J. Respir. Crit. Care Med. 161:641-645. [DOI] [PubMed] [Google Scholar]
- 10.Frothingham, R., H. G. Hills, and K. H. Wilson. 1994. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32:1639-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillman, L. M., J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2001. Identification of Mycobacterium species by multiple-fluorescence PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 39:3085-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimenez, D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain. Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 13.Grange, J. M., and J. L. Stanford. 1974. Reevaluation of Mycobacterium fortuitum (synonym: Mycobacterium ranae). Int. J. Syst. Bacteriol. 24:320-329. [Google Scholar]
- 14.Greub, G., and D. Raoult. 2002. Crescent bodies of Parachlamydia acanthamoeba and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl. Environ. Microbiol. 68:3076-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greub, G., B. La Scola, and D. Raoult. 2004. Amoebae-resisting bacteria isolated from human nasal swabs by amoebal coculture. Emerg. Infect. Dis. 10:470-477. [DOI] [PubMed] [Google Scholar]
- 16.Griffith, D. E., W. M. Girard, and R. J. Wallace, Jr. 1993. Clinical features of pulmonary disease caused by rapidly growing mycobacteria: an analysis of 154 patients. Am. Rev. Respir. Dis. 147:1271-1278. [DOI] [PubMed] [Google Scholar]
- 17.Hamid, M. E., A. Roth, O. Landt, R. M. Kroppenstedt, M. Goodfellow, and H. Mauch. 2002. Differentiation between Mycobacterium farcinogenes and Mycobacterium senegalense strains based on 16S-23S ribosomal DNA internal transcribed spacer sequences. J. Clin. Microbiol. 40:707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanna, B. A., A. Ebrahimzadeh, L. B. Elliott, M. A. Morgan, S. M. Novak, S. Rusch-Gerdes, M. Acio, D. F. Dunbar, T. M. Holmes, C. H. Rexer, C. Savthyakumar, and A. M. Vannier. 1999. Multicenter evaluation of the BACTEC MGIT 960 system for recovery of mycobacteria. J. Clin. Microbiol. 37:748-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, T., W. R. Butler, F. Hollis, M. M. Floyd, S. R. Toney, Y. W. Tang, C. Steele, and R. J. Leggiadro. 2003. Characterization of a novel rapidly growing Mycobacterium species associated with sepsis. J. Clin. Microbiol. 41:5650-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jadin, J. B. 1975. Amibes limax: vecteurs possibles de mycobactéries et de Mycobacterium leprae. Acta Leprol. 59:57-67. [Google Scholar]
- 21.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. CDC publication 86-8230. U.S. Department of Health and Human Services, Centers for Disease Control, Atlanta, Ga.
- 22.Krishna-Prasad, B. N., and S. K. Gupta. 1978. Preliminary report on engulfment and retention of mycobacteria by trophozoites of axenically grown Acanthamoeba castellanii Douglas. Curr. Sci. 47:245-247. [Google Scholar]
- 23.Kubica, G. P., W. E. Dye, M. L. Cohn, and G. Middlebrook. 1963. Sputum digestion and decontamination with N-acetyl-l-cysteine-sodium hydroxide for culture of mycobacteria. Am. Rev. Respir. Dis. 87:775-779. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. 2001. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 25.Kusunoki, S., and T. Ezaki. 1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp abscessus (Kubica et al) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 42:240-245. [DOI] [PubMed] [Google Scholar]
- 26.La Scola, B., L. Mezi, P. J. Weiller, and D. Raoult. 2001. Isolation of Legionella anisa using an amoebic coculture procedure. J. Clin. Microbiol. 39:365-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 28.Mijs, W., P. de Haas, R. Rossau, T. Van der Laan, L. Rigouts, F. Portaels, and D. van Soolingen. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and “M. avium subsp. hominissuis” for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505-1518. [DOI] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 2003. Susceptibility testing of Mycobacteria, Nocardia, and other aerobic actinomycetes. Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, Pa. [PubMed]
- 30.Pfyffer, G. E., B. A. Brown-Elliot, and R. J. Wallace, Jr. 2003. Mycobacterium: general characteristics, isolation, and staining procedures, p. 532-559. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 31.Ringet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth, A., M. Fischer, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 38:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowbotham, T. J. 1983. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 36:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selvarangan, R., W. K. Wu, T. T. Nguyen, L. D. Carlson, C. K. Wallis, S. K. Stiglich, Y. C. Chen, K. C. Jost, Jr., J. L. Prentice, R. J. Wallace, Jr., S. L. Barrett, B. T. Cookson, and M. B. Coyle. 2004. Characterization of a novel group of mycobacteria and proposal of Mycobacterium sherrisii sp. nov. J. Clin. Microbiol. 42:52-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sermet-Gaudelus, I., M. Le Bourgeois, C. Pierre-Audigier, C. Offredo, D. Guillemot, S. Halley, C. Akoua-Koffi, V. Vincent, V. Sivadon-Tardy, A. Ferroni, P. Berche, P. Scheinmann, G. Lenoir, and J. L. Gaillard. 2003. Mycobacterium abscessus and children with cystic fibrosis. Emerg. Infect. Dis. 9:1587-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silcox, V. A., R. C. Good, and M. M. Floyd. 1981. Identification of clinically significant Mycobacterium fortuitum complex isolates. J. Clin. Microbiol. 14:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swenson, J. M., R. J. Wallace, Jr., V. A. Silcox, and C. Thornsberry. 1985. Antimicrobial susceptibility of five subgroups of Mycobacterium fortuitum and Mycobacterium chelonae. Antimicrob. Agents Chemother. 28:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. Böttger, and T. Bodmer. 1993. Rapidly growing mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torkko, P., S. Suomalainen, E. Iivanainen, E. Tortoli, M. Suutari, J. Seppanen, L. Paulin, and M. L. Katila. 2002. Mycobacterium palustre sp. nov., a potentially pathogenic, slowly growing mycobacterium isolated from clinical and veterinary specimens and from Finnish stream waters. Int. J. Syst. Evol. Microbiol. 52:1519-1525. [DOI] [PubMed] [Google Scholar]
- 43.Tortoli, E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turenne, C. Y., A. A. Suchak, J. N. Wolfe, A. Kabani, and L. E. Nicolle. 2003. Soft tissue infection caused by a novel pigmented, rapidly growing Mycobacterium species. J. Clin. Microbiol. 41:2779-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, R. C. Good, J. A. Tschen, and M. Seabury Stone. 1983. Spectrum of disease due to rapidly growing mycobacteria. Rev. Infect. Dis. 5:657-679. [DOI] [PubMed] [Google Scholar]
- 46.Wallace, R. J., Jr., J. Glassroth, D. E. Griffith, K. N. Olivier, J. L. Cook, and F. Gordin. 1997. American Thoracic Society: diagnosis and treatment of disease caused by nonnontuberculous mycobacteria. Am. J. Med. Crit. Care Med. 156:S1-S15. [DOI] [PubMed] [Google Scholar]
- 47.Wallace, R. J., Jr., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52:453-490. [DOI] [PubMed] [Google Scholar]
- 48.Wallace, R. J., Jr., Y. Zhang, R. W. Wilson, L. Mann, and H. Rossmore. 2002. Presence of a single genotype of the newly described species Mycobacterium immunogenum in industrial metalworking fluids associated with hypersensitivity pneumonitis. Appl. Environ. Microbiol. 68:5580-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson, R. W., V. A. Steingrube, E. C. Böttger, B. Springer, B. A. Brown-Elliot, V. Vincent, K. C. Jost, Jr., Y. Zhang, M. J. Garcia, S. H. Chiu, G. O. Onyi, H. Rossmoore, D. R. Nash, and R. J. Wallace, Jr. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 51:1751-1764. [DOI] [PubMed] [Google Scholar]
- 50.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolinskyi, E. 1992. Mycobacterial diseases other than tuberculosis. Clin. Infect. Dis. 15:1-12. [DOI] [PubMed] [Google Scholar]
- 52.Yakrus, M. A., M. S. Hernandez, M. M. Floyd, D. Sikes, W. R. Butler, and B. Metchock. 2001. Comparison of methods for identification of Mycobacterium abscessus and Mycobacterium chelonae isolates. J. Clin. Microbiol. 39:4103-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]