Abstract
The commercial BD ProbeTec ET (BDPT) system for the detection of Neisseria gonorrhoeae and Chlamydia trachomatis from clinical specimens was compared with our in-house LightCycler real-time PCR (LC-PCR) assays. Specimens initially positive by the BDPT system were retested by our LC-PCR assays. Our results for C. trachomatis testing indicate a 91.2% agreement when the results of 114 clinical specimens, initially positive by BDPT over a wide range of method-other-than-acceleration (MOTA) scores, were retested by our LC-PCR assay. The agreement between the two systems improved to 96% when only MOTA scores of >30,000 were retested by the LC-PCR assay. The overall agreement between the two systems for Neisseria gonorrhoeae detection from 155 clinical specimens was only 77.4%, with agreement particularly low (24.1%) for MOTA scores ranging from 2,000 to 19,999. Repeat testing of specimens with the BDPT only closely correlated with that seen by others demonstrating that reproducibility of the BDPT system for specimens initially within the MOTA score range from 2,000 to 9,999 is problematic, especially for Neisseria gonorrhoeae testing. With our study, we proposed an algorithm for C. trachomatis and N. gonorrhoeae testing which involves screening with the BDPT system followed by selective use of our in-house LC-PCR assays.
The arrival of nucleic acid amplification tests for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae DNA from clinical specimens provided an opportunity to improve testing for laboratories receiving specimens where organism viability was an issue and high throughput was important. However, nucleic acid amplification tests, because of their greater sensitivity compared to conventional culture techniques, have the disadvantage of producing a greater number of false positive results (2). This is especially problematic in patient populations which are at low prevalence for C. trachomatis or N. gonorrhoeae because the effect of false positives on the positive predictive value is more pronounced at lower prevalence rates.
Our laboratory began with the BD ProbeTec ET (BDPT) (Becton Dickinson and Company, Franklin Lakes, N.J.) more than 3 years ago because of its high sensitivity, lower cost, and high-throughput capability. The BDPT can amplify DNA from both C. trachomatis and N. gonorrhoeae in separate wells, and plates can optionally contain wells which monitor inhibition of amplification for each specimen. The BDPT assay functions by means of strand displacement amplification with amplified DNA being detected by means of fluorescent energy transfer probes (5). After switching to nucleic acid amplification testing, we soon realized the potential for reporting a higher number of false positives compared to prior conventional testing based on our analyses of the BDPT package insert. The BDPT assay produces a method-other-than-acceleration (MOTA) score for each specimen with lower MOTA scores (2,000 to 9,999) being more prone to contain false positives (supported with discrepant analysis), for both C. trachomatis and N. gonorrhoeae testing, compared to specimens producing higher MOTA scores.
We therefore incorporated a testing algorithm which involved repeat testing when initial results gave a MOTA score between 2,000 and 9,999. Those specimens negative on repeat testing were deemed indeterminate, and another specimen was requested. However, the recent results of others (2) and our own internal examination revealed that for lower MOTA score ranges, the reproducibility of the BDPT assay is problematic with the possibility that, even with repeat testing with only the BDPT assay, false positive results could still be reported. A recent publication stated that when testing low prevalence populations, all positive tests should be considered presumptive and additional testing should be considered (1). The same publication also suggested approaches to additional testing and included repeat testing of the original specimen “with a different test that uses a different target, antigen, or phenotype and a different format” as one of four possibilities.
Therefore, the purpose of our study was to see how we could best utilize an in-house LightCycler PCR (LC-PCR) assay for both C. trachomatis and N. gonorrhoeae testing as a means of improving our BDPT assay screening results. Our study compared the BDPT assay directly with our LC-PCR assay over a wide range of MOTA scores for both C. trachomatis and N. gonorrhoeae tests. With this study, we devised a new algorithm for both C. trachomatis and N. gonorrhoeae testing which incorporates the use of our LC-PCR assays.
MATERIALS AND METHODS
Becton Dickinson ProbeTec ET (BDPT).
Specimen processing and BDPT performance followed the manufacturer's guidelines (Becton Dickinson and Company). For a description of the BDPT assay, the reader should refer to earlier publications (5, 9).
Patient population.
Our specimens were collected from U.S. Army, U.S. Air Force, and U.S. Navy active-duty personnel and their dependents. During the period from January to September 2003, 21,981 specimens were tested, and, of these, 18,376 specimens were endocervical, 2,589 were urethral, and 523 were urine. The high number of endocervical specimens is the result of a required annual screen. Our positive prevalence rate during this period was 7% for C. trachomatis and 1.6% for N. gonorrhoeae, with 0.3% of specimens positive for both C. trachomatis and N. gonorrhoeae. The positive BDPT N. gonorrhoeae specimens used in this study were collected from April to September 2003 and the BDPT-positive C. trachomatis specimens were collected in August and September 2003. Only Becton Dickinson-approved specimen sources with Becton Dickinson-specific collection kits were tested.
DNA purification.
DNA for the LightCycler PCR and relative sensitivity studies was extracted with the High Pure viral nucleic acid kit (Roche Diagnostics GmbH) according to the manufacturer's instructions. DNA was eluted with 50 μl of elution buffer and stored at −20°C.
LC-PCR.
Our in-house PCR assays were performed with the LightCycler (Roche Diagnostics, Mannheim, Germany). LC-PCR for N. gonorrhoeae was performed as described by Whiley et al. (10) with the HO1 and HO2 primer set and the NG-LC1 and NG-LC2 probes to amplify a portion of the ccpB gene present on a cryptic plasmid (4, 10). Briefly, premade reaction mixtures were obtained from the LightCycler FastStart DNA master hybridization probes kit (Roche Diagnostics). Capillaries were loaded with a reaction mixture consisting of 2 μl of 10-fold-concentrated kit master reagent (reagent 1), 0.35 pmol (4 mM) of MgCl2 (reagent 2), 2.0 pmol of primer HO1, 8.0 pmol of primer HO2, 4.0 pmol each of probes NG-LC1 and NG-LC2, and 5 μl of target DNA. All final reaction mixes were made up to 20 μl with sterile PCR-grade water (reagent 3). Each final reaction mix was subjected to an initial 10 min of incubation at 95°C, followed by 55 cycles consisting of denaturation at 95°C for 10 s, annealing at 55°C for 10 s, and extension at 72°C for 20 s.
For C. trachomatis detection by LC-PCR, we adapted the primers described earlier (6) to the SYBR Green LC-PCR format. The total reaction volume per capillary was 20 μl. Briefly each capillary was loaded with 2 μl of LightCycler FastStart reaction/enzyme mix SYBR Green 10-fold concentrated (Roche Diagnostics), 1.6 μl of 25 mM MgCl2 (3 mM), 4.0 pmol (0.2 μM) of each primer, 5 μl of purified DNA (or water as negative control) and diluted to 20 μl with molecular grade water. Initially the FastStart enzymes were activated by heating at 95°C for 10 min. This denaturation was followed by 40 amplification cycles starting with a denaturation at 95°C for 10 s, annealing at 55°C for 10 s, and elongation at 72°C for 25 s, followed by a single measurement. All transition rates were at 20°C per s. The amplification was followed by the melting curve. After a 2-s denaturation at 95°C and hybridization at 55°C for 10 s, the temperature was raised to 95°C with a transition rate of 0.2°C per s. Only those specimens that gave a clearly distinguishable peak from the primer-dimer (negative control) were considered positive. The above parameters for the C. trachomatis LC-PCR were considered optimal after performing this assay under different MgCl2 concentrations (from 1 to 5 mM) and different annealing temperatures (50 to 60°C in 2.5°C increments).
DNA precipitation.
Because the volume of specimen used by the LightCycler is normally 5 μl, we added a DNA precipitation step for all patient specimens positive by BDPT but negative by LC-PCR as a means of equalizing the amount of patient specimen used by both assays. We then retested the precipitated specimens by LC-PCR. For each precipitation, 200 μl of the specimen was added to an extraction column (see DNA purification) and eluted with 50 μl of elution buffer. For each LightCyler capillary tube, 2.5 μl of a 2.5 M sodium acetate solution (pH 5.8) was added to 25 μl of the purified DNA solution. A 10-fold volume of 100% molecular grade ice-cold ethanol was then added and placed for 30 min at −80°C. The DNA was centrifuged for 1 h at 16,000 × g in a tabletop centrifuge. The precipitates were washed once with ice-cold 70% ethanol and then air dried. The pellets were resuspended in 15 μl of an MgCl2 solution (3 mM for C. trachomatis and 4 mM for N. gonorrhoeae) and SYBR Green Fast Start Mastermix, and primers were added for C. trachomatis testing and Hybridization FastStart Mastermix and primers and probes for N. gonorrhoeae testing. The final volume for both C. trachomatis and N. gonorrhoeae testing was 20 μl.
Comparison of relative sensitivities between the BDPT strand displacement amplification and LC-PCR assays.
To compare the relative sensitivity of the LightCycler with the BDPT strand displacement amplification, we made positive control DNA from frozen stock cultures of C. trachomatis and N. gonorrhoeae (ATCC 19424). The C. trachomatis used was obtained from a College of American Pathologists Survey VR1-18 (2002) after identification from growth in McCoy cells. C. trachomatis cultures were grown in McCoy cells, and when more than 75% of cells were positive for growth of C. trachomatis by immunofluorescence, cells were harvested and placed in 200-μl aliquots for DNA purification by extraction with the High Pure viral nucleic acid kit (Roche Diagnostics) according to the manufacturer's instructions. DNA was eluted with 50 μl of elution buffer and stored at −20°C.
For N. gonorrhoeae, two to three colonies were resuspended in 500 μl of molecular grade water and boiled for 15 min; 200-μl aliquots were subjected to DNA purification as for C. trachomatis. Tenfold dilutions of purified C. trachomatis or N. gonorrhoeae DNA were made in molecular grade water, and 5 μl of each 10-fold dilution was used for LC-PCR. For BDPT, 20 μl of each 10-fold dilution was diluted to 400 μl with BDPT diluent and 100 μl was used per test. In so doing, the relative concentrations of DNA used for both the LC-PCR and the BDPT were equivalent. The detection limit was defined as the highest dilution giving a positive result.
To compare the sensitivities of the BDPT assays with the LC-PCR assays from clinical specimens, 10 patient specimens of both endocervical and urethral origin and negative by the BDPT assay were pooled. This pool was then subjected to BDPT assays for both N. gonorrhoeae and C. trachomatis and again found to be negative (MOTA score < 2000). The specimen pool was used to create separate serial dilutions for both N. gonorrhoeae DNA and C. trachomatis DNA to 10−9 of the originally spiked pool. The dilution series spiked with N. gonorrhoeae DNA was subjected to both the BDPT and LC-PCR assays for N. gonorrhoeae detection, while the dilution series spiked with C. trachomatis DNA was subjected to both the BDPT and LC-PCR assays for C. trachomatis detection.
RESULTS
C. trachomatis BDPT repeat testing and comparison with LC-PCR.
With known C. trachomatis DNA, the melting point of the C. trachomatis amplicons in the SYBR green LC-PCR was determined to be 82°C (±1°C) and was clearly distinguishable from the melting point of the primer-dimers (data not shown). Therefore, specimens that showed a distinguishable peak at 82°C (±1°C) were considered positive. Those PCRs that had no distinguishable peak near or at 82°C (±1°C) were considered negative. We then compared our LC-PCR assay with the C. trachomatis DNA dilution series we tested previously with the BDPT and found both systems to have the same relative sensitivity when DNA dilutions were performed in molecular grade water. To compare the relative sensitivity of the LC-PCR with BDPT assay from clinical specimens, actual pooled patient specimens were used to create a dilution series. Our results indicate that, for the pooled patient specimen dilution series, the LC-PCR assay was able to detect C. trachomatis DNA at the 10−7 dilution while the BDPT assay was able to detect DNA at the 10−6 dilution. Hence, in our hands, the LC-PCR was 10-fold more sensitive in detecting C. trachomatis DNA from a pooled clinical specimen than was the BDPT assay (data not shown).
Next, we randomly selected 114 BDPT C. trachomatis-positive specimens. Of these 114 specimens, 11 were found to have a MOTA score of between 2,000 and 9,999 and were retested by the BDPT assay (Table 1). Three of the 11 initially positive specimens were negative upon retesting, and only three retested again within the MOTA range of 2,000 to 9,999. The remaining five of these initially positive specimens gave repeat results with MOTA scores ranging from 10,000 to >30,000.
TABLE 1.
BDPT repeat testing results for 11 C. trachomatis-positive specimens with an initial MOTA score of between 2,000 and 9,999
| Second MOTA score | No. of specimens with initial MOTA score of:
|
|
|---|---|---|
| 2,000-4,999 | 5,000-9,999 | |
| <2,000 | 3 | 0 |
| 2,000-4,999 | 1 | 1 |
| 5,000-9,999 | 0 | 1 |
| 10,000-14,999 | 0 | 1 |
| 15,000-19,999 | 1 | 0 |
| 20,000-24,999 | 1 | 0 |
| 25,000-29,999 | 0 | 0 |
| >30,000 | 0 | 2 |
To compare the BDPT assay directly against the LC-PCR, we retested the original 114 BDPT initially C. trachomatis-positive specimens with our LC-PCR assay (Table 2). Prior to precipitation of DNA for LC-PCR analysis, 97 (85%) repeated positive and 14 (12.2%) turned out to be negative by LC-PCR. When these 14 negative specimens were subjected to precipitation and centrifugation and submitted to another round of LC-PCR testing, the comparison between the BDPT assay and the LC-PCR became even more impressive. All BDPT-retested specimens with an initial MOTA score between 2,000 and 9,999 correlated exactly with the LC-PCR assay (after precipitation) results. Further, in the high MOTA score range of >30,000, there was a 96% (79 of 82 specimens) agreement between the two systems. Overall, when precipitation is used for the LC-PCR assay, there was a 91.2% agreement between the BDPT assay and the LC-PCR assay for detection of C. trachomatis DNA and an 85% agreement when precipitation is not used for the LC-PCR assay. Three of the 114 specimens were initially positive by the BDPT assay but retested as negative by both the BDPT and the LC-PCR assay (before and after the precipitation step). The precipitation step was used so that relative concentrations of DNA tested from patient specimens would be equivalent for the LC-PCR assay compared to the larger sampling volume used by the BDPT assay.
TABLE 2.
Results comparing 114 BDPT C. trachomatis-positive specimens by MOTA score with LC-PCR assay results
| Test | No. of samples positive at MOTA score of:
|
Total | ||||
|---|---|---|---|---|---|---|
| <2000 | 2,000-9,999 | 10,000-19,999 | 20,000-29,999 | ≥30,000 | ||
| BDPT | 3a | 8a | 8 | 13 | 82 | 114 |
| LC-PCR positive | 0 | 7 | 4 | 10 | 76 | 97 |
| LC-PCR precipitation positiveb | 0 | 8 | 6 | 11 | 79 | 104 |
| % Agreementc | 100 | 100 | 75 | 85 | 96 | 91.2 |
Tested twice with the BDPT assay with the first assay giving results between 2,000 and 9,999.
Total number of LC-PCR-positive specimens (initial LC-PCR positives plus positives after precipitation of initially negative LC-PCR specimens)
Agreement between the BDPT- and LC-PCR-positive specimens (initially positive and those positive after precipitation).
N. gonorrhoeae BDPT repeat testing and comparison with LC-PCR.
To compare the relative sensitivity of the BDPT assay to the LC-PCR assay, we tested serial dilutions of DNA purified from a lysate of an N. gonorrhoeae stock culture. The LC-PCR could detect 1 log dilution more than the BDPT assay (data not shown) and therefore, its relative sensitivity is approximately 10-fold higher than that of the BDPT when dilutions were made in molecular grade water. Results of testing for relative sensitivity when a pooled clinical specimen is used to create the dilution series showed that the LC-PCR assay was able to detect N. gonorrhoeae DNA at a dilution of 10−9 while the BDPT assay was able to detect N. gonorrhoeae DNA at only the 10−7 dilution. Hence, in a pooled clinical specimen, our LC-PCR assay was 100-fold more sensitive for detection of N. gonorrhoeae than was the BDPT assay (data not shown).
Next, we randomly selected 155 BDPT N. gonorrhoeae initially positive specimens with 13 of these specimens having an initial MOTA score of between 2,000 and 9,999. On retesting these 13 specimens, we found eight to retest below a MOTA score of 2,000 while five specimens ranged in MOTA scores between 2,000 and > 30,000 (Table 3). Of the 155 BDPT initially positive specimens, 120 repeated positive on the LC-PCR and 35 were negative (Table 4). As with C. trachomatis, we were able to precipitate 32 of the 35 discrepant results between the BDPT assay and the LC-PCR. However, unlike that shown for our C. trachomatis results, precipitating negative specimens and submitting them to another round of LC-PCR testing did little to alter the initial results (only 2 of 32 initially negative LC-PCR results became positive after precipitation and retesting).
TABLE 3.
BDPT repeat testing of 13 N. gonorrhoeae-positive specimens with an initial MOTA score of between 2,000 and 9,999
| Second MOTA score | No. of positive specimens with initial MOTA score of:
|
|
|---|---|---|
| 2,000-4,999 | 5,000-9,999 | |
| <2,000 | 8 | 0 |
| 2,000-4,999 | 1 | 0 |
| 20,000-24,999 | 1 | 0 |
| 25,000-29,999 | 2 | 0 |
| >30,000 | 0 | 1 |
TABLE 4.
Results comparing BDPT N. gonorrhoeae-positive specimens by MOTA score with LightCycler assay results
| Test | No. of samples positive at MOTA score of:
|
Total | |||
|---|---|---|---|---|---|
| 2000-9,999 | 10,000-19,999 | 20,000-29,999 | ≥30,000 | ||
| BDPT | 13a | 16 | 58 | 68 | 155 |
| BDPT retest | 5 | NTb | NT | NT | NDc |
| LC-PCR positive | 1 | 6 | 49 | 64 | 120 |
| % agreementd | 7.7 | 37.5 | 84.5 | 94 | 77.4 |
See Table 3 for results of these specimens when subjected to retesting with the BDPT assay.
NT, not tested.
ND, not determined.
Agreement comparing the initial BDPT results with the LC-PCR assay.
Overall, we saw a high percentage of discrepant results between the BDPT and the LC-PCR N. gonorrhoeae assays (35 of 155, or 22.5%). It is clear from our analysis that for BDPT N. gonorrhoeae testing, discrepant results with the LC-PCR assay occur throughout the different MOTA ranges but are more concentrated in the MOTA range from 2,000 to 30,000 (only 64% agreement in this range). Of particular note is that only 5 of 13 positive specimens in the MOTA score range of 2,000 to 9,999 repeat positive by the BDPT assay, and only one specimen in this range was positive by our LC-PCR assay. Agreement between the BDPT assay and our LC-PCR assay was also poor in the MOTA score range of 10,000 to 19,999. Here 16 specimens were positive by the BDPT assay but only six of these specimens were also positive by the LC-PCR assay (Table 4).
DISCUSSION
Our laboratory has used the BDPT assay for more than 3 years, and based on the analysis of the BDPT assay package insert, we decided almost immediately to institute retesting of all specimens with an initial MOTA score range between 2,000 and 10,000. Those specimens that repeated with a MOTA score range of equal to or greater than 2,000 were reported as positive, while those specimens which repeated with a MOTA score below 2,000 were reported as indeterminate. This algorithm was applied for both C. trachomatis and N. gonorrhoeae testing. Indeterminate results, for us, are somewhat problematic because health care providers are often confused as to what indeterminate results mean. In addition, for indeterminate results we request an additional specimen which can also be problematic because it is often difficult to locate individuals for a second specimen as we test for all armed services throughout the European theater.
Therefore, our purpose, for designing an in-house LC-PCR assay, was as an additional, confirmatory test enabling us to lessen the number of or eliminate indeterminate test results. Another reason for adding confirmatory testing comes from a recent publication highlighting the lack of reproducibility of the BDPT assay for both C. trachomatis and N. gonorrhoeae testing (2). If shown to have at least equal sensitivity, a second nucleic acid amplification test assay, used judiciously and sparingly, would enable us to be more confident of our results and still allow cost containment. The confidence would come from the knowledge that our in-house LC-PCR assay is amplifying a separate and distinct DNA target for both C. trachomatis and N. gonorrhoeae DNA than of the BDPT assay. A false positive result by both assays, even at the low MOTA range for our BDPT assay, would seem highly unlikely.
To begin our study, we compared the relative sensitivity of the BDPT assay with our in-house LC-PCR assay by isolating DNA from control organisms and testing a series of dilutions on both systems. With this method, our LC-PCR assay was equivalent to the BDPT assay for C. trachomatis DNA but more sensitive, by a single 10-fold dilution, for N. gonorrhoeae DNA (data not shown). Next, we performed the LC-PCR assay on 114 BDPT-positive specimens, covering a large range of MOTA scores, for C. trachomatis. Eleven of these specimens initially tested positive by the BDPT assay with MOTA scores ranging from 2,000 to 9,999. Only 72.7% (8 of 11) were positive by BDPT on retesting giving similar results to that shown earlier (80.8%) (2) and the BDPT package insert (71.2%) when retesting initially C. trachomatis-positive specimens in the MOTA score range of 2,000 to 9,999. Three of these 11 specimens that were initially positive but retested with a MOTA score of <2000 were also negative by LC-PCR. We saw only a 50% agreement between the BDPT and the LC-PCR in the MOTA range of 10,000 to 19,999 but this improved to 75% when C. trachomatis DNA was precipitated prior to the LC-PCR assay. Similarly, in the MOTA range between 20,000 and 29,999 the agreement between the BDPT and the LC-PCR (with precipitation) improved to 85% and further improves to 96% when MOTA scores are equal to or greater than 30,000. We decided to precipitate patient specimens so that the relative amounts of DNA tested between the BDPT assay and the LC-PCR assay would be equivalent after the precipitation procedure.
Our analysis comparing the BDPT assay with the LC-PCR assay to detect the presence of N. gonorrhoeae DNA differed significantly from our comparative results for C. trachomatis detection. In contrast to BDPT C. trachomatis testing, in the MOTA score range of 2,000 to 9,999, only 5 of 13 specimens repeated positive by the BDPT N. gonorrhoeae assay (38.4%) which correlates well with earlier reproducibility results of 33% (2). Our in-house LC-PCR was able to detect N. gonorrhoeae DNA in only one of the originally 13 positive specimens. Our results for positive specimens in the MOTA score ranges from 10,000 to 19,999 were also lower than expected because we were able to achieve only a 37.5% agreement between the BDPT assay and our in-house LC-PCR assay (6 out of 16 specimens were positive by both assays). We saw significant improvement in the correlation of results between the two systems when the MOTA score ranges increased to between 20,000 and 29,999. Here we saw an 84.5% agreement which improved to 94% when MOTA scores of ≥30,000 were examined.
One argument that could explain our results is that N. gonorrhoeae DNA is lost during the purification process for our in-house LC-PCR assay. This would explain why we had a much greater correlation at the higher MOTA score range where greater DNA concentration is likely. Specimens with a small amount of initial N. gonorrhoeae DNA may be undetectable in our N. gonorrhoeae assay after our purification process. Our result, in which we analyzed a pool of 10 human specimens with spiked N. gonorrhoeae DNA, does not support this hypothesis, however, since our in-house LC-PCR assay was 100-fold more sensitive than the BDPT assay and showed that relatively minor amounts of DNA survive our purification process for the LC-PCR assay. An alternative explanation is that the primers used to generate the amplicons in our LC-PCR assay for N. gonorrhoeae hybridize to a cryptic plasmid which may not be present in all N. gonorrhoeae clinical isolates, as opposed to the BDPT assay, which detects the presence of N. gonorrhoeae genomic DNA. However, one study indicated that the cryptic plasmid known to contain the ccpB gene is present in 96% of N. gonorrhoeae isolates taken from clinical specimens (4) and that the ccpB gene can also be found chromosomally (3, 7).
Yet another consideration is that the BDPT assay itself lacks good reproducibility at the lower MOTA score range (2). Hence, a specimen which is initially positive with the BDPT assay at the MOTA score range between 2,000 and 10,000 may represent a false positive and hence the lack of correlation with our in-house LC-PCR assay for N. gonorrhoeae. Indeed, it would be highly useful to include probit analyses on the BDPT C. trachomatis and N. gonorrhoeae assays so that end users could judge these assays for reproducibility at different initial MOTA scores. Such probit analyses would be similar to that conducted by Smieja,= at al (8) for in-house C. trachomatis PCR assays.
For N. gonorrhoeae testing results in the lower MOTA score range from 2,000 to 30,000, our data did not correlate well with with data listed in the BDPT package insert. They list 6 of 24 specimens as false positives (25% false positives) in the 2,000 to 10,000 MOTA score range, while our results show only a 38.4% reproducibility (5 of 13 specimens repeated as positive) based on retesting by BDPT. We had only a 37.5% agreement between the BDPT and our in-house LC-PCR at the MOTA score range of 10,000 to 19,999 while the BDPT analysis shows a possible false positive rate of only 10.8% for this MOTA score range. However, the BDPT analysis shown in the package insert also includes results of culturing which we did not included in our analysis.
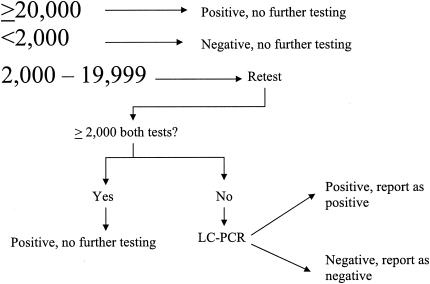
Our comparative results, between the BDPT assay and the LC-PCR for both C. trachomatis and N. gonorrhoeae testing, suggest that we repeat the BDPT assay on all initially positive specimens in the MOTA score range of 2,000 to 29,999 since above 30,000 our LC-PCR assay and the BDPT assay are in strong agreement. However, the results of Culler et al. (2) show that the BDPT assay is very reproducible at MOTA scores above 10,000 (they do not break it down further between 10,000 and 19,999 and equal to and greater than 20,000) while BDPT assay package insert results suggest that the BDPT assay is very reproducible at MOTA scores at or above 20,000. Therefore, our algorithm (Fig. 1) would be similar to our earlier testing algorithm except that we would expand our repeat range from 2,000 to 9,999 MOTA score to between 2,000 and 19,999 with repeat positives (i.e., ≥2000 MOTA score on both BDPT assays) reported as positive. Those specimens that are negative (i.e., <2,000 MOTA score) on the second BDPT assay would be considered indeterminate and would automatically be subjected to the LC-PCR assay to include the precipitation step. Those specimens positive on the third analysis by LC-PCR would be reported as positive and those that are negative would be reported negative. No indeterminate results would be reported under this algorithm.
FIG. 1.
Testing algorithm for the BDPT and LC-PCR assays for both C. trachomatis and N. gonorrhoeae. Numbers refer to MOTA scores.
Use of the LC-PCR on specimens only after two rounds of BDPT testing (initially positive followed by a negative) would substantially reduce the number of LC-PCR assays performed compared to performing the LT-PCR assay on all specimens with a MOTA score between 2,000 and 19,999. Indeed, our analysis shows that over a typical 2-month period, we tested 5,577 specimens for both C. trachomatis and N. gonorrhoeae. For C. trachomatis, 403 specimens (7.2%) were positive with 148 specimens yielding a positive result with a MOTA score between 2,000 and 19,999 (36.7% of all positives). Of these 148 specimens, 102 specimens had a MOTA score between 10,000 and 19,999 and 46 had a MOTA score between 2,000 and 9,999.
To construct a cost analysis, we estimate for C. trachomatis, based on the false positive rates proposed by the BDPT assay package insert, that approximately 15% of the specimens between 10,000 and 19,999 would likely be false positives on retesting with the BDPT assay and 30% would be negative on retesting with an initial MOTA score between 2,000 and 9,999. This would make a total of 29 C. trachomatis indeterminate specimens which would be subjected to our LC-PCR assay with our proposed testing algorithm during a typical 2-month period. At approximately US$6 per LC-PCR assay, this would add an additional US$174 to our testing costs, not counting labor. A similar analysis for N. gonorrhoeae indicates that only 32 specimens had a MOTA score between 2,000 and 19,999 during the 2-month period analyzed because our rate of positives for N. gonorrhoeae is typically less than 2% for our testing population. Therefore, the cost of retesting for N. gonorrhoeae would be relatively minor.
An argument could be made that all initial retesting should be done by LC-PCR, but our analysis indicates that while the cost per test for both the BDPT assay and the LC-PCR were approximately the same at our location, the number of samples that can be completed per day with the BDPT assay is approximately 550 and approximately 200 with the LC-PCR. Thus, when factoring in the cost of labor, the BDPT assay is a much cheaper assay to perform.
Our results and the results of Culler et al. (2) show that initial testing results for N. gonorrhoeae giving MOTA scores between 2,000 and 9,999 should not be reported as positive without further testing because of the lack of reproducibility.
The goal of our study was to see if our in-house LC-PCR could be useful in confirming both C. trachomatis and N. gonorrhoeae detection by the BDPT assay at MOTA score ranges where reproducibility or the percentage of false positives is high by BDPT's own study data. Our results indicate that in-house LC-PCR confirmatory testing is useful for resolving indeterminate results for C. trachomatis testing and for confirming results for N. gonorrhoeae testing in the MOTA score range where the BDPT assay's reproducibility is poor.
A vast majority of our specimens were taken from asymptomatic female patients as part of a yearly screen. Hence, reporting a false positive result as a positive patient result could potentially have a devastating social impact. An examination of our current prevalence rates indicates that, within our population, C. trachomatis is present in approximately 7% of our testing population while N. gonorrhoeae is present in less than 2% of our population. Therefore, we would expect the positive predictive value for our N. gonorrhoeae testing to be far less than that of our C. trachomatis testing. Confirmatory testing is therefore especially critical for N. gonorrhoeae when with the BDPT assay giving MOTA scores of less than 10,000 but should also be used as confirmatory testing for C. trachomatis as outlined in our testing algorithm.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2002. Screening tests to detect Chlamydia trachomatis and Neisseria gonorrhoeae infections—2002. Morb. Mortal. Wkly. Rep. 51:1-38. [PubMed] [Google Scholar]
- 2.Culler, E. E., A. M. Caliendo, and F. S. Nolte. 2003. Reproducibility of positive test results in the BDProbeTec ET system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 41:3911-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagblom, P., C. Korch, A. Jonsson, and S. Normack. 1986. Intragenic variation by site-specific recombination in the cryptic plasmid of Neisseria gonorrhoeae. J. Bacteriol. 167:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho, B. S. W., W. G. Feng, B. K. C. Wong, and S. I. Egglestone. 1992. Polymerase chain reaction for the detection of Neisseria gonorrhoeae in clinical samples. J. Clin. Pathol. 45:439-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little, M. C., J. Andrews, R. Moore, S. Bustos, L. Jones, C. Embres, G. Durmowicz, J. Harris, D. Berger, K. Yanson, C. Rostkowski, D. Yuris, J. Price, T. Fort, A. Walters, M. Collis, O. Llorin, J. Wood, F. Failing, C. O'Keefe, B. Scrivens, B. Pope, T. Hansen, K. Marino, K. Williams, and M. Boenisch. 1999. Strand displacement amplification and homogeneous real-time detection incorporated in a second-generation DNA probe system, BD-ProbeTec ET. Clin. Chem. 45:777-784. [PubMed] [Google Scholar]
- 6.Mahony, J. B., D. Jang, S. Chong, K. Luinstra, J. Sellors, M. Tyndall, and M. Cherneskey. 1997. Detection of Chlamydia trachomatis, Neisseria gonorrhoeae, Ureaplasma urealyticum, and Mycoplasma genitalium in first-viod urine specimens by multiplex polymerase chain reaction. Mol. Diagn. 2:161-168. [DOI] [PubMed] [Google Scholar]
- 7.Roberts, M., P. Piot, and S. Falkow. 1979. The ecology of gonococcal plasmids. J. Gen. Microbiol. 114:491-494. [DOI] [PubMed] [Google Scholar]
- 8.Smieja, M., J. B. Mahoney, C. H. Goldsmith, S. Chong, A. Petrich, and M. Cherneskey. 2001. Replicate PCR testing and probit analysis for the detection and quantification of Chlamydia pneumoniae in clinical specimens. J. Clin. Microbiol. 39:1796-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Der Pol, B., D. V. Ferrero, L. Buck-Barrington, E. Hook III, C. Lenderman, T. Quinn, C. A., Gaydos, J. Lovchik, J. Schachter, J. Moncada, G. Hall, M. J. Tuohy, and R. B. Jones. 2001. Multicenter evaluation of the BDProbeTec ET system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J. Clin. Microbiol. 39:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whiley, D. M., G. M. LeCornec, I. M. Mackay, D. J. Siebert, and T. P. Sloots. 2002. A real-time PCR assay for the detection of Neisseria gonorrhoeae by LightCycler. Diagn. Microbiol. Infect. Dis. 42:85-89. [DOI] [PubMed] [Google Scholar]