Abstract
The microtubule organizing center (MTOC) frequently polarizes to a position in front of the nucleus during cell migration, but recent work has shown conflicting evidence for the MTOC location in migratory polarized cells. Here, we show that subcellular localization of the MTOC is modulated by the extracellular matrix stiffness. Specifically, we found that ECM compliance alters the positioning of the MTOC during cell migration in scratch wound assays as well as single cell migration of mesenchymal stem cells (MSCs). In contrast to the expected polarized position in front of the nucleus, the MTOC appears randomly positioned when cells are migrating on soft matrix. The bulk of the microtubule density is also equally likely to be in front of or behind the nucleus on soft matrix, but is polarized in front of the nucleus on stiff matrix. This occurred during cell migration with cells in interphase. During cytokinesis the centrosomes polarize on either side of the chromosomes even on soft matrix, with MIIB localized in the cleavage furrow which depolarizes as cells exit cytokinesis. When cells are immobilized on micro-patterns printed on the top of substrates of different stiffness, MIIB was able to polarize if the matrix was sufficiently stiff similar to results with migrating cells. However, the MTOC was randomly positioned with respect to the nucleus independent of matrix stiffness. We deduce that cell migration is necessary to orient the MTOC in front of the nucleus and that matrix stiffness helps to drive cell polarization during migration.
Introduction
The establishment of cell polarity is critical for a many of cell functions such as division, migration, and directional transport of nutrients and chemical messengers. Microtubules (MTs) are important for establishing polarity in migrating cells (Levy and Holzbaur, 2008) and MTs stabilized by tubulin acetylation help to regulate actin polymerization necessary for extending the cell front (Kaverina and Straube, 2011). MTs are nucleated at the centrosome, one of the major microtubule organizing centers (MTOC) within the cell, and thus the position of the centrosome affects the spatial density of MTs as well as the vesicles that move along them. Although matrix stiffness has been shown to impact cell migration (Sunyer et al., 2016; Peyton and Putnam, 2005; Pelham Jr. and Wang, 1997, 1999; Stroka and Aranda-Espinoza, 2009; Ulrich et al., 2009; Lo et al., 2000; Fischer et al., 2009), division (Klein et al., 2009; Gilbert et al., 2010; Winer et al., 2009), differentiation (Tse and Engler, 2011; Engler et al., 2006), and actomyosin contractility (Beningo et al., 2006), its effects on cell polarity remain unexplored. It was, however, previously shown that the Golgi apparatus polarization was affected by matrix stiffness in wound healing assays (Ng et al., 2012), providing an inkling that matrix stiffness can influence cell polarity.
The polarization of actomyosin organization and contractile activity are strongly modulated by the effect of matrix compliance on focal adhesions (Prager-Khoutorsky et al., 2011). At the same time, there is evidence of a cross-talk between the MT system and the actomyosin cytoskeleton (Rape et al., 2011; Even-Ram et al., 2007, Rodriguez et al., 2003; Akhshi et al., 2014), and thus it is likely that the polarization of one of these may influence the other in the context of directed cell migration in various environmental contexts. To test this possibility, we decided to use human mesenchymal stem cells (MSCs) as they have been measured to migrate quite fast in vitro (Maiuri et al., 2012), and must face various spatial and mechanical environments to mobilize to sites of inflammation within the body. We find that the extracellular matrix (ECM) stiffness influences the position of the MTOC in MSCs by polarizing it in front of the nucleus only when the matrix is sufficiently stiff (≥5–6 kPa). We observe strikingly low densities of MTs in the lamellapodia of cells on soft surfaces, while on stiff matrix MTs begin to fill lamellapodia. We have previously demonstrated that ECM stiffness can change the polarity of myosin-IIB (MIIB) distribution within MSCs (Raab et al., 2012). Trying to assess whether how these changes in the actomyosin organization are coordinated with the MT cytoskeleton, we found that MIIB is localized to the cleavage furrow in cells dividing on soft gels. However, delocalization quickly occurs as the cells begin crawl away from each other. Further, using patterned matrix in the shape of polarized migrating cells, we deduce that migration is also required for this MTOC by keeping the nucleus rearward as cells polarize.
Results
In wounded monolayers, the MTOC is polarized in front of the nucleus on stiff matrix but randomly positioned on soft matrix
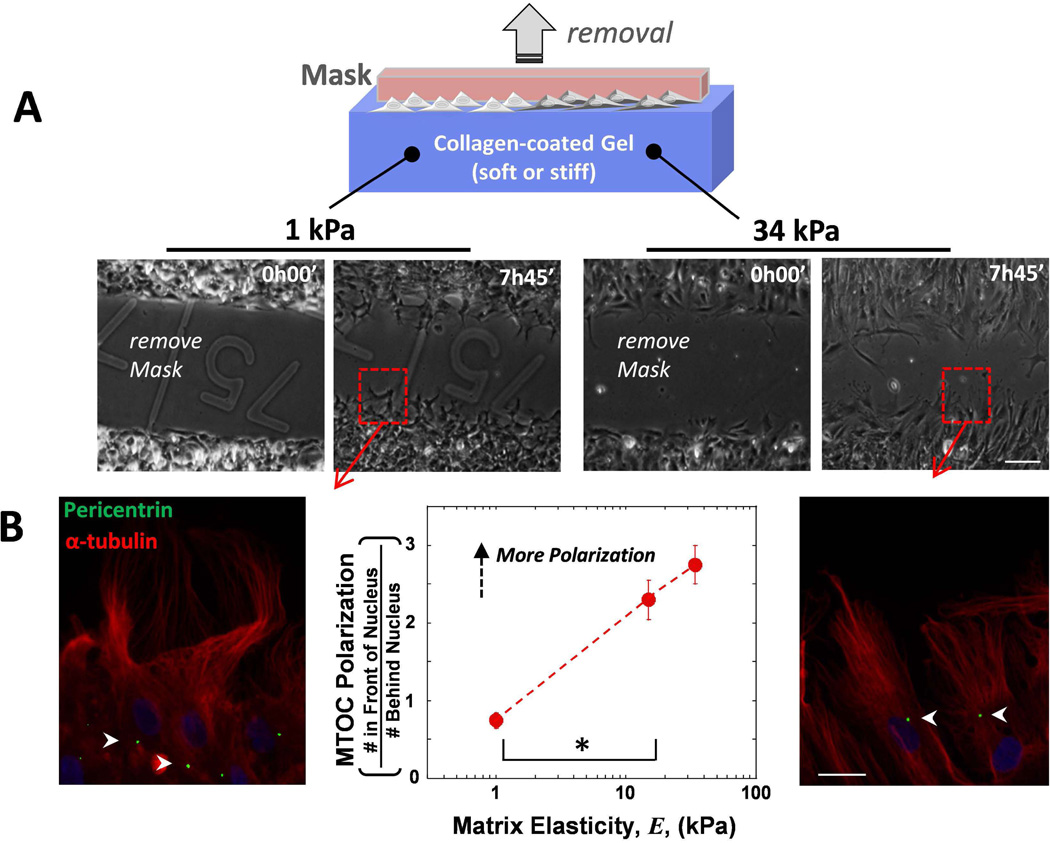
A wound assay was created on gels to induce collective cell migration of MSCs and we then tracked microtubule-organizing center (MTOC) orientation as the cells infiltrated the wound. Gels cannot be scratched without damaging the gel surface which would pose a topological barrier to cell migration. To prevent this from being an issue, a spacer was placed on the gel prior to seeding cells, and cell migration was initiated upon lifting the spacer off the gel. Despite being on soft or stiff matrices, cell sheets migrated into the ‘wound’ with only small differences in their migration rate (Fig. 1A, S1A,B), which falls in line with past results with epithelial cells (Ng et al., 2012). However, we found a striking difference with matrix stiffness in the location of the MTOC (Fig. 1B): on soft matrix, MTOCs were almost random in location with respect to the nucleus (MTOC polarization ratio ≈ 1), whereas on stiff matrix, there were 3 times more cells with the MTOC positioned in front of the nucleus (ratio = 3). Past results with other cells of mesenchymal origin migrating on scratched coverslips have also shown frontward polarization of the MTOC (Luxton et al., 2010; Gomes et al., 2005), but results shown here appear to be the first showing this is suppressed in cells migrating on soft matrix. We also found a reduction in wound closure with myosin-IIA (MIIA) depletion and a slight increase with myosin-IIB (MIIB) depletion (Fig. S1C), reminiscent of past results with single cell migration of MSCs (Raab et al., 2012). We also found that the rates of cell division of MSCs were not different between soft and stiff matrix (Fig. S2), indicating that it is not a significant factor in wound closure rates.
Fig. 1. MTOC polarization increases with increasing gel stiffness in MSCs migrating in wound assays.
(A) The mask was removed at the initial time and the Mesenchymal stem cells (MSCs) migrate into the 500 µm wide gap on gels of varying stiffness. Scale bar 100 µm (B) The MTOC was visualized with pericentrin (green), the MTs with α-tubulin (red) and DNA with Hoechst (blue) 2 hours after migration. MTOC polarization was quantified as (# In Front of the Nucleus)/(# Behind Nucleus). A ratio of 1 indicates as many cells have the MTOC in the front as the back. Arrowheads indicate location of MTOC. Scale bar 20 µm. N=3, n≥91 cells, * p<0.05. Error bars SEM.
Sparse migrating MSCs polarize MTOC in front of the nucleus only on stiff matrix
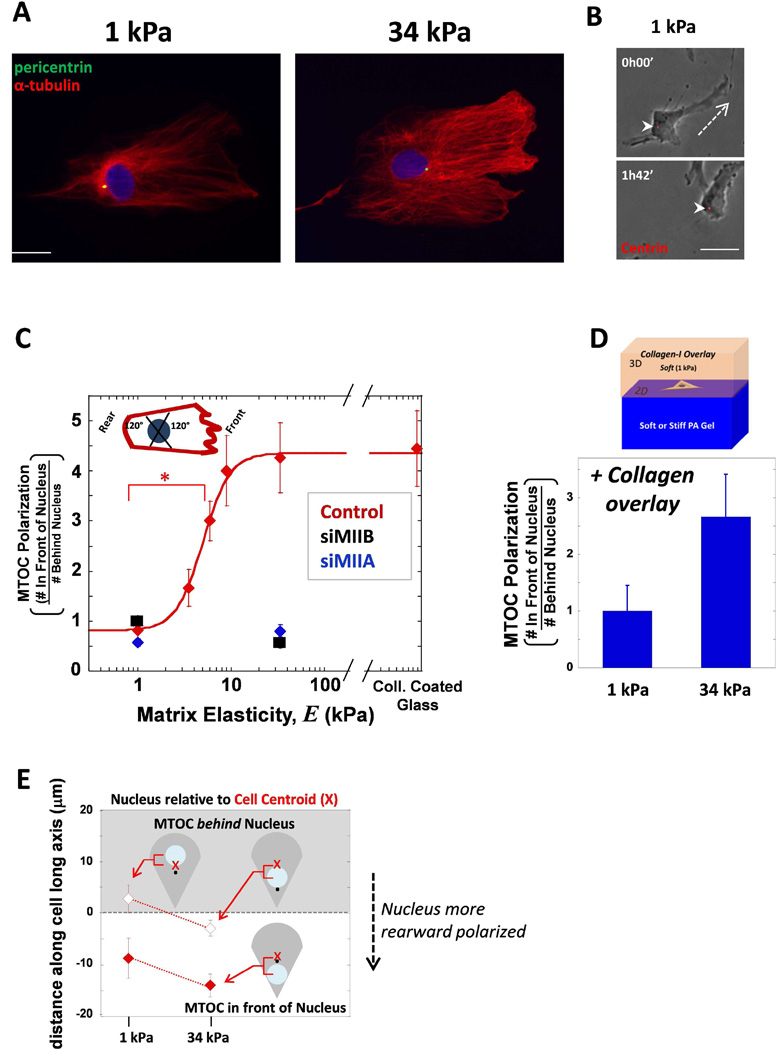
In order to assess whether the MTOC polarization phenotype above was unique to collective cell migration during wound healing, sparse cell cultures on gels were also examined. MSCs fixed and stained for localization of the MTOC and MTs typically showed their frontward positioning on soft gels but not on stiff (Fig. 2A). Tracking the position of the MTOC in MSCs expressing ds-Red-Centrin revealed MTOC’s rearward positioning during migration (Fig. 2B). MSCs displaying a polarized migrating shape were evaluated for MTOC polarization (Fig. 2C). MTOC position appeared nearly random in cells migrating on soft 1 kPa matrix (Fig. 2C), meaning that as many cells exhibited rearward positioning as frontward polarization. However, cells on high gel stiffness and also cells on collagen-coated glass, 80% of the cells (ratio = 4) exhibited polarized positioning of the MTOC. Interestingly, with increasing stiffness, the MTOC polarization ratio increases dramatically at an intermediate elasticity, with a mid-point elasticity of K ~ 5 kPa (Fig. 2C). Many tissues such as brain tissue, liver, and fat have been measured to be softer than this (Discher et al., 2005). Cells typically spread more on stiff matrix, and this value of K is similar to the matrix elasticity that yields half-maximal spreading of MSCs in a non-cooperative, sigmoidal fit of projected cell area (Rehfeldt et al., 2012). A similar K of 6.9 kPa was also obtained from a non-cooperative, sigmoidal fit of matrix elasticity dependent rearward-polarization of the minor myosin-II isoform, myosin-IIB (MIIB) – as we recently showed (Raab et al., 2012). The same study also showed that MIIB polarization depended on the activity of MIIA. We therefore knocked down both myosin isoforms separately and found that both knockdowns suppressed frontward polarization of the MTOC on stiff matrix (Fig. 2C). This means that MTOC polarization is hypersensitive to myosin levels. Perhaps even more interesting is that there is 10-fold less MIIB than MIIA in these MSCs (Raab et al., 2012) suggesting that MIIB is critical for MTOC polarization. Microtubule stabilization toward the cell front on rigid substrates has been previously reported as coupled to myosin-II activity (Even-Ram et al., 2007).
Fig. 2. MSCs polarize their MTOC on sufficiently stiff matrix during migration as single cells.
(A) Cells on either soft 1 kPa or stiff 34 kPa matrix were immunostained for pericentrin (green) and MTs (red) and DNA with hoechst (blue). Scale bar 20 µm. (B) Cell transfected with dsRed-Centrin was imaged migrating on soft 1 kPa matrix. The MTOC was located behind the nucleus during migration. Dashed line indicates direction of migration, arrowheads point to MTOC. Scale bar 20 µm (C) The ratio of the number of cells with MTOC in front of nucleus to the cells that have the MTOC behind nucleus. A schematic picture represents how the MTOC was considered in a ‘front’ or ‘rear’ position based on the migrating phenotype of a lamellipodium in the front and a tail in the rear. When MIIA (blue) or MIIB (black) is depleted, the MTOC fails to polarize to the cell front despite being on stiff matrix. Error bars SEM. N=3, n≥25, * p<0.05. (D) For PA gel stiffnesses of 1 and 34 kPa, a soft collagen was overlaid on top of the gel and the cells migrating on the surface were assessed for MTOC polarization. (E) The data from (C) was separated so that the instances the MTOC was in front of the nucleus (white background, filled shapes) was separated. The average distance between the cell centroid and the nucleus center was plotted and again the instances when the MTOC was behind the nucleus was differentiated from when it was behind. The nucleus was more rearward positioned in cells on stiffer matrix as the nucleus move closer to the rear.
Additionally, when cells on either soft or stiff gels were covered in soft collagen matrix to create a 3D culture environment, the same trend in frontal polarization of MTOC persisted (Fig. 2D); a stiffer underlying matrix still caused a higher degree of frontward MTOC positioning. The polarization on stiff 34 kPa was not as pronounced (~2.6; p < 0.05) which we attribute to cell interaction with the soft collagen surrounding the cell in addition to the stiff matrix underneath. The results are statistically the same as in collective sheet migration above (Fig. 1), suggesting that matrix stiffness has the strongest effect on polarity when cells are isolated, but when cells are in contact with each other or surrounded by very soft collagen, then the stiff gel’s effect on polarity is partially suppressed.
These results for stiff substrates appear consistent with findings for fibroblasts on rigid glass coverslips by Gundersen and coworkers (Luxton et al., 2010; Gomes et al., 2005) who described frontward polarization of the MTOC as a consequence of the nucleus being pulled backwards by myosin-II. This prior work evaluated MTOC location with respect to the cell centroid, which prompted us to examine MTOC and nuclear centroid position relative to cell centroid for the various cell populations on soft and stiff matrix (Fig. 2E). Data were segregated into 2 different populations, the first being instances in which the MTOC is positioned in front of nucleus (filled in points) and the other when the MTOC is positioned behind the nucleus (empty points). Consistent with Gundersen and coworkers (Gomes et al., 2005), on stiff matrix, the nucleus was located behind the cell centroid independent of MTOC position. On soft matrix, however, the nucleus appeared in front of the cell centroid when the MTOC was behind the nucleus (which occurred in roughly half of cells) (Fig. 2E). To summarize, on soft matrix which causes a general reduction of contractility and MIIB polarization (Raab et al., 2012), cells also fail to pull the nucleus rearward.
Microtubule density frontward polarizes on stiff matrix
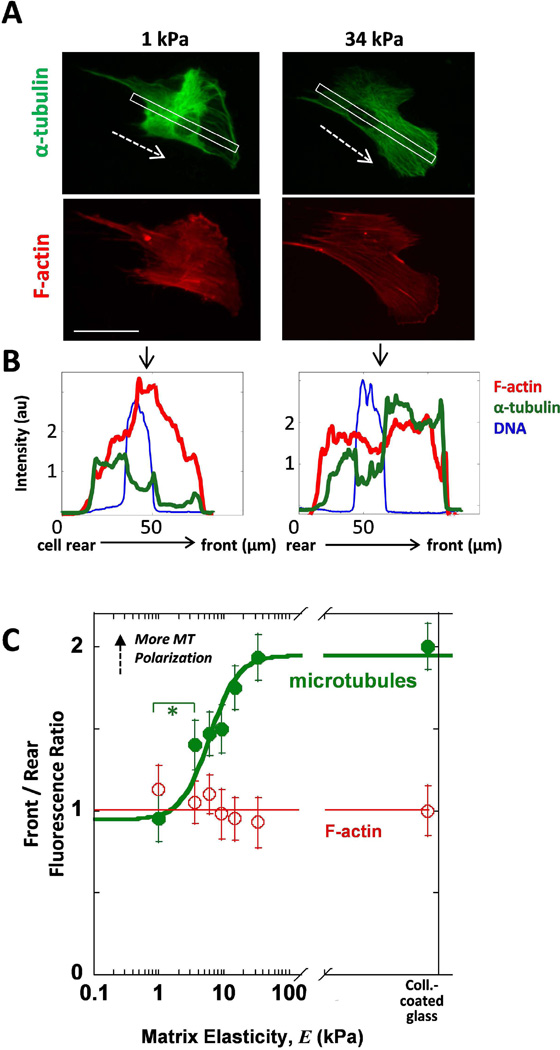
Microtubules often are reported to be frontward-polarized in cells on rigid substrates (Ezratty et al., 2009; Manneville et al., 2010), and so its importance in establishing cell polarity prompted us to check the distribution of MTs in cells on gels. Line scans were taken from the back to the front of the cell, and the results for the cells depicted in Fig. 3A are shown in Fig. 3B. The cell front was determined by the presence of a lamellipodia visualized by F-actin staining. Unique to cells on soft matrix, more than half the cells possess a lamella region that is strongly depleted of MT (Fig. 3A, S4 white arrows) rather than exhibiting the more typical enrichment on stiffer matrix(Fig. 3A, Fig. S4). Stiffer matrix promotes higher densities of MTs in front of the nucleus (Fig. 3B,C) and the data fits a sigmoidal curve similar to the MTOC data (Fig. 2C). The consistency of MT polarization on stiff matrix reflects the consistency of MTOC polarization (Fig. 1, 2). We attribute this low MT density to a thinner, more unstable lamellipodium of MSCs on soft 1 kPa matrix as is also visible in phase contrast microscopy (Fig. S1, S3, 1 kPa) which has been reported previously for fibroblasts (Pelham Jr. and Wang, 1997). A hyperbolic fit of (Rear/Front) as a function of Elasticity shows polarization of MT varies as previously shown for MIIB (Raab et al., 2012) and cell area (Rehfeldt et al., 2012) with a transition from soft to stiff matrix at a stiffness of K = 5.9 kPa, which reinforces the notion that polarization is one more aspect of a generalized, collective response to increased matrix stiffness.
Fig. 3. Stiff matrix promotes frontward distribution of MTs.
(A) Images of MSCs on soft or stiff gels show higher MT density in front of the nucleus with stiff matrix but not when on soft matrix. White outline indicates the region scanned for the line scans of fluorescence intensity. Dashed arrows indicate inferred direction of migration.. Scale bar = 50 µm. (B) Line scans taken from the back to the front of the cell show the intensity of MT density (green) as well as the nucleus (blue) and F-actin (red) as a function of distance. (C) The Front/Rear fluorescence is quantified as the ratio of the front half of the cell over the rear half on either side of the nucleus. The ratio for F-actin is plotted vs matrix elasticity and is compared to MTs. MTs polarize to the front of the cell with increasing matrix stiffness. Error bars SEM from N=3, n≥43, * p<0.05.
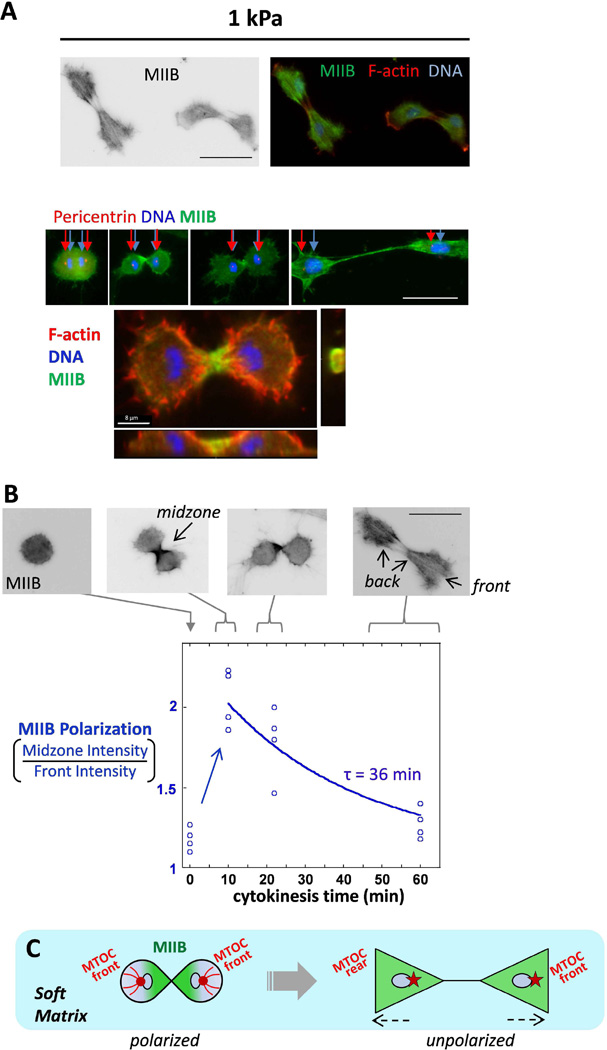
Myosin-IIB localizes to the cleavage furrow during cytokinesis and quickly depolarizes as cells pull away from each other on soft gels
We showed that MTOC polarization on stiff matrix depends on myosin (Fig. 2C). It has been shown in many cell types that myosin-IIB polarizes on stiff surfaces (Vicente-Manzanares et al., 2009) but does not polarize on soft gels during MSC migration (Raab et al., 2012). We then asked whether MIIB polarizes during cytokinesis on soft gels since it has been known that non-muscle myosin localizes to the cleavage furrow during this stage in the cell cycle (Straight et al., 2003). If there is MIIB polarization during cytokinesis on soft gels, at what point in time does the cell lose the MIIB polarization? During cytokinesis, the microtubule cytoskeleton is clearly polarized, thus we looked at polarization of myosin. The varying stages of cell division were evaluated based on the phenotype of the DNA, and these cells were evaluated for MIIB polarization (Fig. 4A). During division on soft gels, we observed MIIB polarization in the cleavage furrow, evident in the confocal image (Fig. 4A bottom), and a schematically drawn cross-sectional view of the MIIB in the cleavage furrow is shown to illustrate localization (Fig. 4A bottom, right). As cells undergo division and the chromosomes separate, there is an MTOC on either side of the set of chromosomes. After division, the 2 daughter cells begin to migrate away from each other and each new cell forms a cell front and the region where the cleavage furrow (mid-zone) had been becomes the cell rear for both cells. This is how ‘front’ and ‘rear’ of the cell are defined for the data on polarization of MTOC and MIIB (Fig. 4B).
Fig. 4. Cytokinesis on soft matrix reveals transient polarization of myosin-IIB which diminishes soon after.
(A) Cells on soft matrix which have recently divided and are migrating away from each other. The MTOC, indicated by pericentrin immunostaining, is at the front end of each daughter cell as the chromosomes are pulled apart. Z-projection of dividing daughter MSCs on 1 kPa matrix (bottom). MIIB (green) localizes to the contractile ring. (B) Cells dividing on soft matrix at progressive stages of division. The average time was approximately 10 min for a dividing cell to go from one rounded cell to the forming a cleavage furrow. In going from a single rounded cell to 2 cells migrating apart separated by about one cell length, the average time was 60 min. The extent of MIIB polarization from the midzone to the nascently formed front of the cells is plotted and fit to an exponential decay to estimate the indicated depolarization time constant. The blue empty dots indicate data for individual cells and the cytokinesis time from the fixed images was estimated based on data taken from live cell imaging of dividing MSCs in Fig. S3. (C) Schematic drawing of transient cytoskeletal polarization on soft matrix: green represents MIIB and red dot represents the MTOC. Scale bars 50 µm.
The polarization of MIIB as MSCs divide and pull away from each other on 1 kPa stiffness are quantified in Fig. 4B. In Fig. 4B (top images) sequential images of MIIB during the progression of cytokinesis on soft 1 kPa gels are shown. In the leftmost image, as the cell is initially rounded up before chromosomal separation, the MIIB is uniformly distributed throughout the cell. Next, when the DNA has separated into 2 distinct structures, there is a localization of MIIB at the cleavage furrow. As cell division progresses with time, this MIIB localization diminishes. A cytokinesis time value was set to zero when the cell was rounded up before chromosomal separation, and then the next time point was identified when there were two distinct DNA structures. The ratio of the amount of MIIB located in the midzone of the cell to the MIIB in the front halves of the cells was quantified in Fig. 4B and increased sharply after the chromosomes separate. Over the course of cytokinesis, this ratio of MIIB polarity decreases until the cells finally separate and begin to crawl away from each other when the MIIB is fully depolarized. Fitting the data with a curve of exponential decay, the time constant of depolarization τ = 36 min. Fig. 4C schematically illustrates the changes in polarity of MIIB in dividing cells on soft matrix.
Cells immobilized on patterns still exhibit stiffness dependent MIIB polarization but MTOC does not polarize
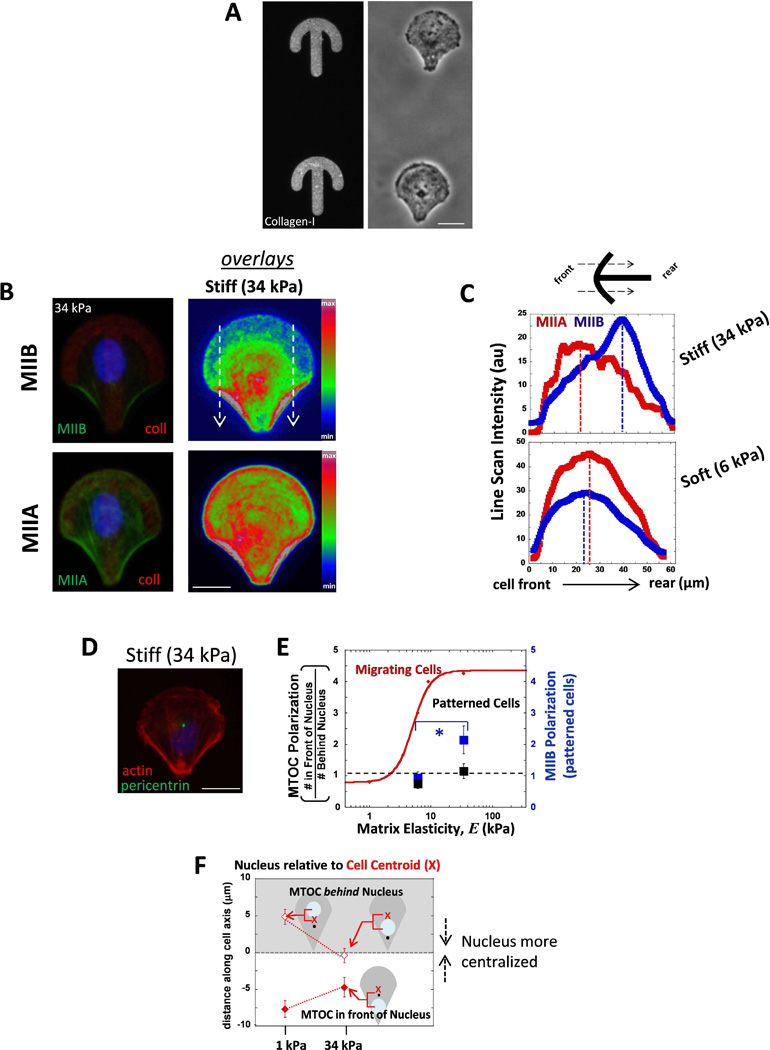
We asked whether cell migration is required for the observed cell polarization, and thus we decided to immobilize MSCs on patterned gel surfaces that forced the cells into a crossbow shape that a migrating cell will typically form. Cells were seeded on these patterns on either soft 6 kPa or stiff 34 kPa gels (Fig. 5A). Gels with stiffness less than 6 kPa were not used because they did not retain a sufficient amount collagen on the surface. After allowing several hours for cells to spread and conform into the polarized shape of the crossbow, MIIB polarized to the rear end of the cells on stiff gel patterns (Fig. 5B), whereas MIIA was more evenly distributed throughout the cell. Line scans across the entire pattern were made from front to rear to quantify the extent of MIIB polarity (Fig. 5C). Hence cells immobilized on patterns still were able to polarize their MIIB on stiff matrix even without migrating. MSCs on soft matrix patterns did not polarize their MIIB (Fig. 5C), similar to MSCs freely moving on soft unpatterned gels. It appears that MIIB polarization depends on the underlying substrate but not on migration.
Fig. 5. MSCs confined to polarized patterns exhibit myosin-IIB polarization on stiffer matrix, but MTOC polarization in front of the nucleus is not present either soft and stiff matrix.
(A) Gels with micropatterned collagen-I in polarized crossbow shapes (stained with an antibody). Phase contrast images of MSCs on the micropatterned gels. Scale bar 20 µm. (B) Sample image of cellular distributions of MIIB and MIIA in a cell on stiff (34 kPa) gels. False color represents an overlay of MIIB (top) or MIIA (bottom) staining. White arrows indicate the direction of the scan used as the region of interest in C. Scale bar 20 µm. (C) Distributions of MIIB and MIIA on stiff and soft patterned gels by taking scans of the ROI shown in B from the cell front to cell rear. MIIB is more polarized to cell rear only on stiff patterns. MIIA is not polarized to cell rear in cells on either stiffness (D) MTOC positioning in cells on patterned gels. Scale 20 µm. (E) Summary plot that compares the difference in MTOC polarization between migrating cells (red, from Fig. 2C) and patterned cells (black) and also MIIB polarization for patterned cells (blue). Patterned cells polarize MIIB with increasing matrix stiffness but MTOC polarization in front of the nucleus does not occur with increasing stiffness. * p<0.05 N=2, n≥15. (F) The positioning of the nucleus relative the cell center. The data is divided for cells that displayed MTOC behind the nucleus (open points, gray background) from cells that had the MTOC in front of the nucleus (filled in points, white background). The nucleus is more randomly positioned on soft matrix, while more centered on stiff.
Next we asked whether the polarization of the MTOC depended on stiffness when cells were confined to patterned gels. We found the MTOC centered within the shape of the pattern on both soft and stiff gels (Fig. 5D, E). Because we observed the MTOC in front of the nucleus on stiffer matrix (Fig. 2), we expected that the MTOC would similarly be in front of the nucleus for cells on stiff matrix patterns of polarized shape. However, the MTOC did not polarize significantly from the nucleus center (Fig. 5E, black squares) even on stiff gel patterns. Although the difference between the soft and stiff gel patterns was that the average distance between the nucleus and the center of the pattern was greater, meaning that the nucleus position was more centered for cells immobilized on stiff matrix (Fig. 5F).
Discussion
Many reports have documented centrosome positioning in front of the nucleus during cell migration (Burke and Roux, 2009; Vicente-Manzanares et al., 2009; Gomes et al., 2005), but some exceptions to this phenomenon have also been described (Schutze et al., 1991; Distel et al., 2010; Doyle et al., 2009). Cells migrating inside soft collagen gels (Schutze et al., 1991), decellularized matrix (Doyle et al., 2009), or in vivo within soft brain tissue (Distel et al., 2010), all show examples of an unpolarized centrosome. In all of these contexts cells are in fact migrating in soft environments: measurements made directly on these types of extracellular matrix show that collagen gels are ‘soft’ (Raab et al., 2012), as are decellularized matrix (Petrie et al., 2012) and brain tissue (Discher et al., 2005) all in the range of 1–2 kPa. Here, we show that the MTOC is positioned in front of the nucleus in cells migrating on stiff gels but by contrast when cells migrate on soft matrix the MTOC position randomizes. This stiffness-dependent positioning occurs in cells that migrate as isolated cells (Fig. 2) and also as collective sheets of interconnected cells in a wound healing model (Fig. 1). Similar results with polarization increasing with stiffer matrix using the Golgi apparatus as the marker for cell polarization were observed in the collective migration of epithelial cells (Ng et al., 2012). The Golgi and centrosome are in close proximity in polarized cells (Théry et al., 2006).
We show that knocking down either MIIA or MIIB causes a lack in MTOC polarization in isolated migrating cells (Fig. 2). In migrating cells, soft matrix inhibits polarization of MIIB (Raab et al., 2012). But during cytokinesis, a process where non-muscle myosin localizes to the cleavage furrow (Straight et al., 2003), we found that MIIB indeed localizes transiently during this process even on soft matrix (Fig. 4). A possible explanation for this polarization is that during cytokinesis the cell is in a high tension state (Lafaurie-Janvore et al., 2013; Sedzinski et al., 2011). As the cells begin to separate from each other after cytokinesis, they are likely exiting from their high tension state when on soft matrix, and thus losing their MIIB polarity.
When cells were constricted to specific polarized shapes on micropatterned gels, MIIB was polarized to the thick actin bundles in the rear of the pattern on stiff matrix but this was not the case on soft gels (Fig. 5C). This suggests that MIIB polarization does not depend on the cell’s migration process, but rather depends on the cells contractile state which increases on stiff matrix (Beningo et al., 2006; Discher et al., 2005). Moreover, any general difference in shape and size of cells between soft or stiff gels is not the reason for the difference in MIIB polarization because the patterned cells were the same in shape and size. While MIIB was able polarize in cells on stiff gel patterns, the MTOC location was centrally localized in cells on both soft and stiff gels (Fig. 5D,E). These data suggest that migration is necessary for MTOC/nucleus polarization. This falls in line with what others have reported that when cell migration in wound assays is inhibited by lack of LPA, the nucleus fails to be pulled back to polarize the MTOC in front (Luxton et al., 2010). Others have reported that the MTOC is centralized in cells on crossbow patterns (on stiff glass), and that there nucleus is slightly pulled back making the MTOC seem frontward (Théry et al., 2006). However, the nucleus in their work is ovular with long-axis left to right, seemly by the MTOC in the center pushing DNA backward. In our experiments, our nuclei remain ovular shaped in the up-down direction (Fig. 4B,D), which would explain our observed centralized nuclear position. Our results also show this centralized position of the MTOC on stiff, but tended to be farther from the pattern center on soft. However, when taking an average of many cells, the overall MTOC position was still in the center of the pattern. The reason for the central location of the MTOC could be explained by microtubule ends pushing the cell boundary, thereby forcing a centralized location of MTOC when cells are immobilized on patterns, matching the work of others in vitro on MT polymerization forces (Letort et al., 2016). Collectively, these results demonstrate that cytoskeletal polarity depends on matrix stiffness and that there is an additional dependence on cell migration for polarity of the MTOC which is not required for MIIB polarity.
Materials and Methods
Polyacrylamide Gels
25-mm circular glass coverslips were treated first with ethanol, then RCA solution (1:1:3 for 15N NH4OH:30% H2O2:dH2O), and functionalized with 1% allyltrichlorosilane and 1% triethylamine in chloroform solution. N,N′methylene-bis-acrylamide (Sigma) and the acrylamide solution (40%, Sigma) was mixed at the final concentrations in PBS. For 1 kPa gels, 3% acrylamide and 0.11% bisacrylamide was used and for 34 kPa, % acrylamide and 0.3% bisacrylamide was used. Solution was polymerized on a coverslip with 0.1% ammonium persulfate and 0.1% N,N,N′,N′-tetramethylethylenediamine. During polymerization, gels were covered with another coverslip that was pretreated with dichlorodimethylsilane. Sulfo-Sanpah (Pierce) was diluted in HEPES buffer pH 8.0 to 0.5 mg/mL and 300 uL was added to the gel and then reacted using 365 nm UV light for 10 min. Excess Sulfo-Sanpah was removed by 2 washes of buffer. Type I rat-tail collagen was diluted to 0.2 mg/mL in HEPES buffer pH 8.0 and incubated with the gel at 4°C for 4 hours. Collagen was removed and the gel was equilibrated with PBS. MSCs were plated onto gels within 24 hours of collagen attachment.
Cell Culture, Pharmacological Perturbations, and Transfections
MSCs purchased from Lonza and were used at p3–p6 for all studies and were cultured in low glucose DMEM supplemented with 10% FBS and 100 ug/ml Penicillin and 100 uM Streptomycin. To image the position of the centrosome in migrating cells, MSCs on 1 kPa gels were transfected with dsRed-centrin (Tanaka et al., 2004) using Lipofectamine LTX with Plus (Invitrogen) using 0.5 µg DNA plasmid per 200 uL optimum solution and then imaged the next day. Racemic blebbistatin (EMD Biosciences) was used at 50 µM, and cells were pretreated with mitomycin C (Sigma) for 2 hours before trypsinizing and replating onto gels.
Fixation, Immunostaining and Microscopy
Cells were fixed with 3.7% formaldehyde (Sigma) in PBS for 10 min at RT, followed by PBS washing 2× for 5 min. For microtubule staining, cells were fixed with 2.5% formaldehyde and 0.15% glutaraldehyde in PBS for 10 min at RT followed by quenching with 0.5 mg/ml sodium borohydride (Sigma) in PBS for 5 min 3×. Blocking and antibody staining was done in 1% BSA in PBS. Rabbit polyclonal ant-pericentrin antibody (Abcam) was used to stain for the MTOC at 1:1000 dilution, anti-myosin IIA antibody (Sigma) was used at 1:100, anti-myosin IIB antibody (Cell Signaling) was used at 1:150, anti-α-tubulin (Sigma) at 1:2000 and all primary antibodies were incubated at RT for 1 hour or overnight at 4°C. All secondary antibodies (Alexa dyes 488, 564, 647) were used in staining for 1 hour at RT at 1:300 dilution. TRITC-phalloidin (Sigma) was used with the donkey secondary antibodies at a concentration of 100 ng/ml and Hoechst 33342 (Invitrogen) was used to stain DNA at a concentration of 1 µg/ml for 10 min at RT. Imaging for quantification of MII levels and localization was performed using an inverted microscope (IX-71, Olympus) with a 40× LUCPlanFLN objective (NA 0.60) and a Cascade CCD camera (Photometrics). Image acquisition was performed with Image Pro software (Media Cybernetics, Inc.) and subsequently background was subtracted and image analysis was done using ImageJ.
Image Analysis
Front/Rear fluorescence polarization of F-actin, myosins, and microtubules were determined by drawing a line 6–10 µm in width (Fig. 3A) from the cell rear to front and plotting the fluorescence vs length. For microtubule measurements in Fig. 3, distances in front of the nucleus and back of the nucleus (from nuclear edge) to cell edges were cut into half. The ratio of the different halves closer to the nucleus was plotted. For dividing cells, the MIIB Polarization was calculated by taking the ratio of the average fluorescent value of the half farther from the cleavage furrow (front) to the average fluorescent value of the half closer to the cleavage furrow (midzone).
AFM Indentation
An Asylum 1-D Force Microscope (Asylum Research, Santa Barbara) was used quantify the elastic modulus of the gels. A pyramid tipped probe with spring constant 30–100 pN/nm (MCST, Veeco) was used for measuring gradient gels and the AFM head was placed over an inverted microscope (Nikon).
Time-lapse Cell Imaging
Phase contrast imaging was done at in a controlled chamber at 37°C and 5% CO2 using an inverted Olympus IX-70 microscope with a 10× or 20× phase objective, using 300 W xenon lamp illumination, and a Photometrics CoolSnap HQ high-resolution CCD camera. Deltavision Softworx software as used for acquiring images.. For centrosome tracking and cell division imaging, images were acquired in 3 min intervals for 2 hours. ImageJ was used to track the center of the nuclei.
Collagen-I Overlay
MSCs were seeded onto collagen coated polyacrylamide gels and allowed to adhere for 1–2 hours. Media was removed and 1.65 mg/ml collagen in neutral buffered solution was added to the PA gel surface and allowed to gel for 1 hour at 37°C followed by the addition of media. Cells were fixed and stained as previously described (Fischer et al., 2009). In brief, samples with fixed with 3.7% formaldehyde in PBS with 0.1% Triton X-100 for 30 min at RT and then rinsed with PBS with 0.25% Triton X-100 for 30 min. PBS with 0.5 mg/ml NaBH4 was added to reduce autofluorescence for 10 min at RT twice. Samples were subsequently blocked with PBS with 2% BSA, 1% goat serum, and 0.25% Triton X-100 overnight at 4°C. Primary antibodies were diluted in PBS with 0.25% Triton X-100 with 2% BSA and 1% goat serum at 4°C overnight. After rinsing for 30 min in PBS, secondary antibodies at 1:300 were added and incubated for 2.5 h at RT.
Micropatterned Gels
Activated micropatterns with area of 1600 µm2 (CYTOO) were incubated with 20 ug/mL rat tail collagen-I (BD Sciences) in PBS for 2 hours at room temperature, then washed 3× with PBS. Polymerizing polyacrylamide drops were placed atop glass coverslips treated with ATCS (see above) and then the micropatterned glass (excess moisture removed with a kimwipe) was placed above. To conserve micropatterns, a glass cutter was used to break the micropatterns into smaller pieces which could be used to make more micropatterned gels. Very little collagen transferred from the micropattern to the gel for 1 kPa gels, so 6 kPa was the lowest stiffness which still allowed enough transfer of collagen to the gel that was comparable to the stiff 34 kPa gel. Micropatterned gels were placed into 6 well plates and 4 mL of media with well mixed 40k MSCs was added to each well. After letting the MSCs sink to the bottom for 10 min, MSCs were incubated on the gels and were allowed to spread and polarize for 4–6 hours before fixation. Image analysis for the localization of MIIB and MTOC was done using CYTOO guidelines and their custom code in ImageJ. Collagen was immunostained using a mouse anti-collagen antibody at a 1:1000 concentration (Sigma) to label the pattern for the CYTOO code to identify the patterns.
Mask Wound Assay
Inserts (ibidi) were gently placed atop PA gels made with gridded coverslips (EMS). 70 ul of MSCs at a density of 105 per ml was added to each well, and gels were the placed in the incubator. MSCs were allowed to adhere for 2 hours and then the insert was removed and fresh media was added. MSCs were either fixed after 2 hours with 2.5% formaldehyde and 0.15% glutaraldehyde for pericentrin and microtubule staining or were imaged with a 4× phase objective every few hours for a 24 hr period to observe wound closure.
Supplementary Material
Acknowledgments
The authors would like to thank Alexis Lomakin for proofreading of the manuscript and the lab of Erika Holzbaur for kindly providing plasmids. We also thank Andrea Stout and the Cell and Developmental Biology Microscopy Core at the Perelman School of Medicine for technical support in microscopy. MR was supported by the Ashton Fellowship and this work also in part supported by the National Cancer Institute of the National Institutes of Health under PSOC Award Number U54 CA193417.
References
- Akhshi TK, Wernike D, Piekny A. Microtubules and actin crosstalk in cell migration and division. Cytoskeleton. 2014 doi: 10.1002/cm.21150. [DOI] [PubMed] [Google Scholar]
- Beningo KA, Hamao K, Dembo M, Wang YL, Hosoya H. Traction forces of fibroblasts are regulated by the Rho-dependent kinase but not by the myosin light chain kinase. Arch Biochem Biophys. 2006;456:224–231. doi: 10.1016/j.abb.2006.09.025. doi:S0003-9861(06)00369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. doi:S1534-5807(09)00440-7. [DOI] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. doi:310/5751/1139. [DOI] [PubMed] [Google Scholar]
- Distel M, Hocking JC, Volkmann K, Koster RW. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J Cell Biol. 2010;191:875–890. doi: 10.1083/jcb.201004154. doi:jcb.201004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. doi:jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. doi:S0092-8674(06)00961-5. [DOI] [PubMed] [Google Scholar]
- Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. doi:ncb1540. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Gardel M, Ma X, Adelstein RS, Waterman CM. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr Biol. 2009;19:260–265. doi: 10.1016/j.cub.2008.12.045. doi:S0960-9822(09)00538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. doi:science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. doi:S0092-8674(05)00188-1. [DOI] [PubMed] [Google Scholar]
- Kaverina I, Straube A. Regulation of cell migration by dynamic microtubules. Seminars in cell & developmental biology. 2011;22:968–974. doi: 10.1016/j.semcdb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19:1511–1518. doi: 10.1016/j.cub.2009.07.069. doi:S0960-9822(09)01548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaurie-Janvore J, Maiuri P, Wang I, Pinot M, Manneville J-B, Betz T, Balland M, Piel M. ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science (New York, N.Y.) 2013;339:1625–1629. doi: 10.1126/science.1233866. [DOI] [PubMed] [Google Scholar]
- Letort G, Nedelec F, Blanchoin L, Thery M. Centrosome centering and decentering by microtubule network rearrangement. bioRxiv. 2016;27:57968. doi: 10.1091/mbc.E16-06-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JR, Holzbaur ELF. Dynein drives nuclear rotation during forward progression of motile fibroblasts. Journal of cell science. 2008;121:3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. doi:S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science (New York, N.Y.) 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri P, Terriac E, Paul-Gilloteaux P, Vignaud T, McNally K, Onuffer J, Thorn K, Nguyen PA, Georgoulia N, Soong D, Jayo A, Beil N, Beneke J, Hong Lim JC, Pei-Ying Sim C, Chu YS, Jiménez-Dalmaroni A, Joanny JF, Thiery JP, Erfle H, Parsons M, Mitchison TJ, Lim WA, Lennon-Duménil AM, Piel M, Théry M. The first World Cell Race. Current Biology. 2012;22 doi: 10.1016/j.cub.2012.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MR, Besser A, Danuser G, Brugge JS. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. 2012;199 doi: 10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. High resolution detection of mechanical forces exerted by locomoting fibroblasts on the substrate. Mol Biol Cell. 1999;10:935–945. doi: 10.1091/mbc.10.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. The Journal of cell biology. 2012;197:439–455. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13(12):1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- Raab M, Swift J, Dingal PCDP, Shah P, Shin J-W, Discher DE. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. The Journal of Cell Biology. 2012;199:669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape A, Guo W, Wang Y. Microtubule depolymerization induces traction force increase through two distinct pathways. Journal of cell science. 2011;124:4233–4240. doi: 10.1242/jcs.090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeldt F, Brown AE, Raab M, Cai S, Zajac AL, Zemel A, Discher DE. Hyaluronic acid matrices show matrix stiffness in 2D and 3D dictates cytoskeletal order and myosin-II phosphorylation within stem cells. Integr Biol (Camb) 2012;4:422–430. doi: 10.1039/c2ib00150k. [DOI] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule–actin interactions in cell movement and morphogenesis. 2003;5 doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Schutze K, Maniotis A, Schliwa M. The position of the microtubule-organizing center in directionally migrating fibroblasts depends on the nature of the substratum. Proc Natl Acad Sci U S A. 1991;88:8367–8371. doi: 10.1073/pnas.88.19.8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Biro M, Oswald A, Tinevez J-Y, Salbreux G, Paluch E. Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature. 2011;476:462–466. doi: 10.1038/nature10286. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. doi:10.1126/science.1081412299/5613/1743 [pii] [DOI] [PubMed] [Google Scholar]
- Stroka KM, Aranda-Espinoza H. Neutrophils display biphasic relationship between migration and substrate stiffness. Cell Motil Cytoskeleton. 2009;66:328–341. doi: 10.1002/cm.20363. [DOI] [PubMed] [Google Scholar]
- Sunyer R, Conte V, Escribano J, Elosegui-Artola A, Labernadie A, Valon L, Navajas D, García-Aznar JM, Muñoz JJ, Roca-Cusachs P, Trepat X. Collective cell durotaxis emerges from long-range intercellular force transmission. Science (New York, N.Y.) 2016;353:1157–1161. doi: 10.1126/science.aaf7119. [DOI] [PubMed] [Google Scholar]
- Théry M, Racine V, Piel M, Pépin A, Dimitrov A, Chen Y, Sibarita J-B, Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse JR, Engler AJ. Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS One. 2011;6:e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. doi:0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. doi:nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer JP, Janmey PA, McCormick ME, Funaki M. Bone marrow-derived human mesenchymal stem cells become quiescent on soft substrates but remain responsive to chemical or mechanical stimuli. Tissue Eng Part A. 2009;15:147–154. doi: 10.1089/ten.tea.2007.0388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.