Abstract
PCR fingerprinting was used to type 177 yeast isolates obtained from two medical institutions. Candida albicans was the predominant species found, followed by C. tropicalis, C. glabrata, C. parapsilosis, C. guilliermondii, and C. krusei, which accounted for over 20% of the strains isolated. This survey represents the first study of molecular epidemiology of candidiasis in Portugal.
In the last decade, yeasts belonging to the genus Candida have emerged as major opportunistic pathogens, mainly due to the increase of immunocompromised patients (19, 26, 2, 5). Although Candida albicans is the most frequently isolated species, other species, such as C. tropicalis, C. guilliermondii, C. krusei, C. parapsilosis, and C. glabrata, have increasingly been recognized as pathogens with a wide distribution (6, 4). The significant increase in the frequency of candidiasis has promoted the study and development of a variety of molecular-based techniques aimed at the replacement of traditional methods used for identification and typing of Candida clinical isolates. Among the present molecular techniques for genotyping of yeast strains, PCR fingerprinting is in wide use for its high discriminatory power and reproducibility and because it requires very little starting material and is rapid and simple to perform. PCR fingerprinting with the primer named T3B was first developed for Streptococcus spp. identification (14), but it has been used successfully in the identification of yeast species belonging to the genus Candida (25, 1).
The aim of the present work was to study the diversity and distribution of Candida species among patients suffering from different pathologies in two medical institutions located in Braga in northern Portugal. Approximately two hundred Candida clinical isolates were analyzed by using a PCR-based methodology with primer T3B, representing the first study of the molecular epidemiology of candidiasis in this country.
Yeast clinical isolates were obtained from 123 independent patients during the year 2001 in a hospital and a health center. The yeast strains from the hospital had been previously isolated at the institution of origin and were collected from different body locations. The isolates from the health center were all collected from vaginal exudates and were isolated at the Microbiology Laboratory, University of Minho. The type cultures Candida albicans PYCC 3436 (ATCC 18804), C. parapsilosis CBS 604 (ATCC 22019), C. krusei PYCC 3343 (ATCC 6258), C. tropicalis PYCC 3097 (ATCC 750), C. guilliermondii PYCC 2730 (ATCC 6260), C. lusitaniae PYCC 2705 (ATCC 34449), C. glabrata CBS 138 (ATCC 2001), C. dubliniensis CBS 7987 (ATCC MYA-646), and C. dubliniensis CBS 7988 were used as reference strains and were supplied by the Portuguese Yeast Culture Collection (PYCC), New University of Lisbon, Portugal, except for the isolates of C. parapsilosis, C. dubliniensis, and C. glabrata, which were obtained from Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands. DNA extraction followed procedures previously described (7), and the oligonucleotide used as a single primer for arbitrary amplification was T3B (5′-AGG TCG CGG GTT CGA ATC C-3′). Amplification reactions were performed according to Thanos et al. (25), without the condensing step of the amplification products. The D1/D2 domain of 26S ribosomal DNA (rDNA) was amplified according to the procedures described by Sampaio et al. (21). Sequencing was performed with an ABI 310 Genetic Analyzer (Applied Biosystems, Inc., Foster City, Calif.) using standard protocols. Forward and reverse sequence alignments were made with MegAlign (DNAStar, Inc., Madison, Wis.) and were visually corrected.
PCR fingerprinting profiles were analyzed by using the program Bionumerics (version 2.0; Applied Maths BVBA, Sint-Martens-Latem, Belgium). Similarity coefficients were calculated with the Dice algorithm, and cluster analysis was performed by means of the unweighted paired group method using arithmetic averages (UPGMA) (23).
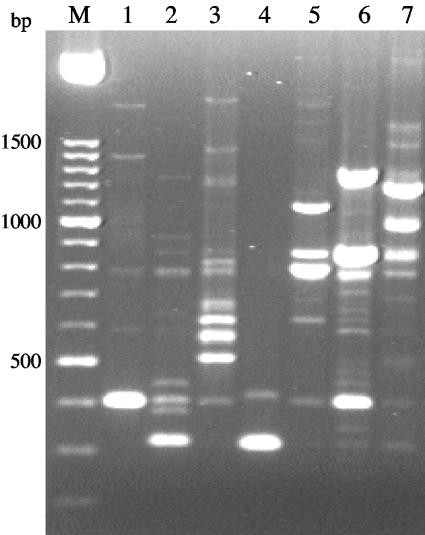
A preliminary characterization of the 177 yeast strains isolated by a rapid identification kit indicated that they all belonged to the genus Candida, although several doubts arose regarding species identity. PCR fingerprinting profiles were obtained with primer T3B for the type strains of C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. guilliermondii, C. krusei, and C. lusitaniae, the most common species found among yeast clinical isolates. Results showed that the different species tested could be clearly distinguished by their amplification patterns, because the number and size of the amplification products were characteristic for each species (Fig. 1).
FIG. 1.
PCR profiles obtained with primer T3B for C. glabrata CBS 138 (1), C. parapsilosis CBS 604 (2), C. tropicalis PYCC 3097 (3), C. krusei PYCC 3440 (4), C. guilliermondii PYCC 2730 (5), C. lusitaniae PYCC 2705 (6), and C. albicans PYCC 3436 (7). M, molecular size marker in base pairs.
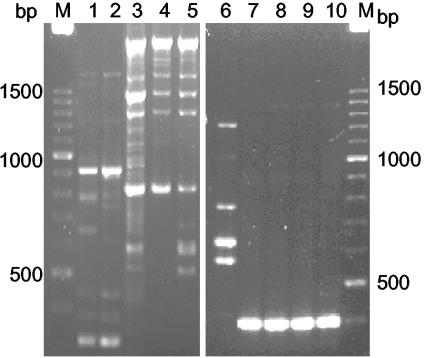
By comparing the PCR profiles of the clinical isolates with those of the reference type strains, all clinical isolates could be identified to the species level, except in the case of four of the strains that did not produce recognizable patterns. Three of them (36 M, 65 M, and 66 M) shared identical profiles, while 153 M presented a different but unique fingerprint. These strains had been preliminarily identified as C. parapsilosis and C. glabrata (Fig. 2). Although intraspecies variability was observed, PCR profiles obtained from different strains assigned to the same species were far more similar than those derived from different species. Variability was found for isolates of C. albicans, C. tropicalis, C. guilliermondii, and C. parapsilosis, with C. albicans being the species that exhibited the greatest diversity. On the contrary, no variability was observed in the profiles obtained for both C. glabrata and C. krusei isolates. Our results agree with those of Thanos et al. (25), who used the same methodology to differentiate Candida species. These authors used a condensing step of the amplification products before electrophoresis which was not performed in this study; consequently, variability within a species might have been reduced. However, as our goal was identification at the species level, the methodology without this step turned out to be less time-consuming and produced easily recognizable profiles that maintained high species discrimination. The high power of discrimination of PCR fingerprinting with primer T3B allowed the identification of over 98% of the 177 clinical isolates and the detection of misidentifications made by API 32C.
FIG. 2.
PCR profiles obtained with primer T3B for C. parapsilosis (lanes 1 and 2); Candida sp. strain 36 M (3), strain 65 M (4), strain 66 M (5), and strain 153 M (6); and C. glabrata (lanes 7 to 10). M, molecular size marker in base pairs.
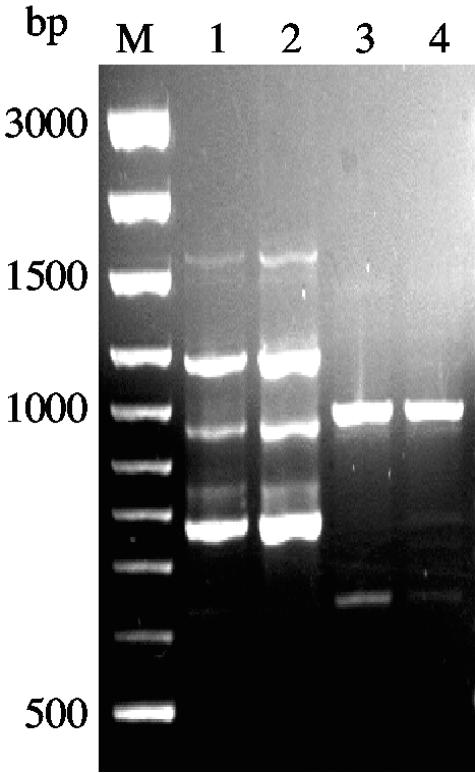
To investigate the presence of C. dubliniensis, the T3B profiles of two strains, including the type strain, were obtained and compared to the ones found for C. albicans. T3B fingerprinting clearly distinguished these closely related species, because no similarities were observed between the amplification patterns of the two species (Fig. 3). No isolates of C. dubliniensis were found, which is not surprising because this yeast species is commonly reported from oral candidiasis, mainly among human immunodeficiency virus-infected individuals (24), which are not included in this study. Previous reports also refer to the differentiation of these two species by PCR fingerprinting (16), but this was the first time that T3B fingerprints have been applied for this purpose.
FIG. 3.
PCR profiles obtained with primer T3B for C. albicans (lanes 1 and 2) and C. dubliniensis (lanes 3 and 4). M, molecular size marker in base pairs.
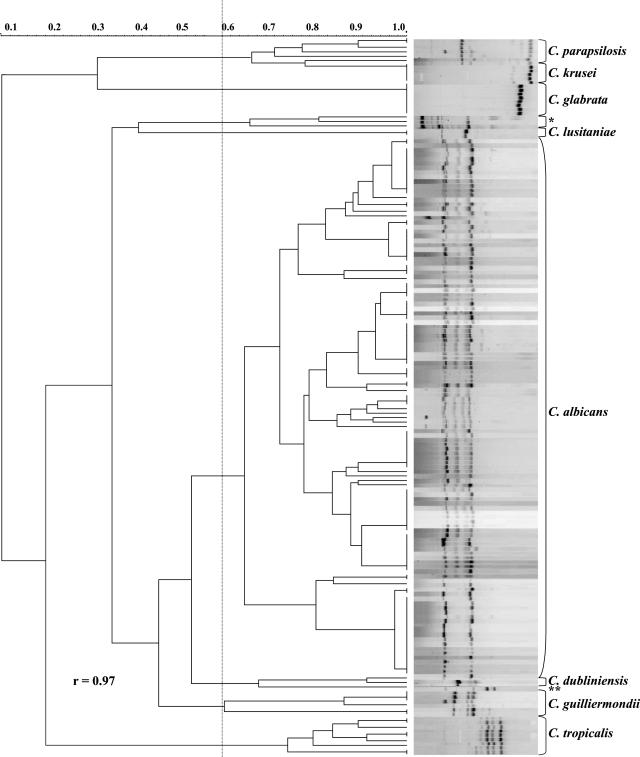
To evaluate the taxonomic resolution of T3B amplification profiles, cluster analysis was applied to the data and the dendrogram presented in Fig. 4 was produced, showing a very high correspondence between the clusters and the different Candida species. The calculated cophenetic correlation coefficient (0.97) indicated that the fit for the cluster analysis was very good. This analysis allowed the distribution of the isolates into seven major clusters corresponding to the species studied. The four isolates that did not produce recognizable T3B patterns grouped separately, and the strains that were originally misidentified grouped within the clusters corresponding to their respective T3B profile.
FIG. 4.
Dendrogram showing the degree of similarity of T3B fingerprinting profiles among the clinical Candida isolates by using the Dice coefficient and UPGMA cluster method. The unidentified strains are indicated as follows: *, 36 M, 65 M, and 66 M; **, 153 M. An arbitrary line has been drawn at 0.58 delimitating the major groups. r, cophenetic correlation coefficient.
The four strains displaying peculiar banding profiles were further investigated by sequencing the D1/D2 domain of their 26S rDNA. Sequencing results for strains 36 M, 65 M, and 66 M (GenBank accession no. AY589574) showed a 100% similarity between them and with strain Candida sp. strain NRRL Y-17456, which appears to be highly related to C. parapsilosis but has been referred to as a new species (9, 13). Furthermore, their T3B profiles did not match with the ones of other C. parapsilosis strains (Fig. 2). This species remains a source of controversy, because three genetically distinct subtypes (I, II, and III) were defined (10, 12, 11, 20) that present enough variability to justify separate species status for the three subgroups. Strain 153 M (GenBank accession no. AY589572), formerly identified as C. glabrata, also presented differences from the sequences available for C. glabrata (94% similarity). Despite seeming to be related to C. glabrata, no conclusive identification was obtained for this strain, which is, most probably, a new species based on the high number of nucleotide substitutions observed (9, 18).
Of the 177 Candida strains tested, 112 isolates were obtained from vaginal swabs, 24 were from urine, 23 were isolated from the upper respiratory system, 7 were from anal mucosa, and 11 isolates were from various sources. The number of strains belonging to each species found and their respective clinical origins are shown in Table 1. C. albicans was the predominant species (79.0%), followed by C. tropicalis (5.6%), C. glabrata (4.0%), and C. parapsilosis (3.4%), which is in accordance with several previous reports (3-6, 8). C. guilliermondii represented 2.8% of the isolates, C. krusei represented 2.3%, C. lusitaniae represented 0.6%, and the isolates whose identification was not conclusive (Candida spp.) represented 2.3% of the total. C. albicans was present in all types of clinical material except blood samples, and C. tropicalis was mainly recovered from urine and the respiratory tract. While C. albicans was predominant among vaginal isolates, the species other than C. albicans were recovered mainly from other sources. Little is known about the epidemiology of candidiasis in Portugal (17, 15, 22). Our study, despite covering a small area and only two medical institutions, is a representative survey, because patients with different pathologies were included. We show that over 20% of the infections are due to species other than C. albicans, and they may not respond to antifungal agents usually used in treatment, requiring more adequate means of therapy. Therefore, it is of the utmost importance to identify the species causing infection, and this PCR-based methodology is simple and can be implemented at relatively low cost for routine identification in hospitals and health centers. This method is more accurate in identifying species of the genus Candida than any biochemical approach presently used in clinical microbiology laboratories and, consequently, is better suited for large epidemiological surveys.
TABLE 1.
Yeast species identified, number of strains of each species, and respective sources of isolation
| Species | No. of strains | Body site |
|---|---|---|
| C. albicans | 19 | Urine |
| 16 | Respiratory tract | |
| 2 | Peritoneal fluid | |
| 95 | Vagina | |
| 6 | Anal mucosa | |
| 1 | Catheter | |
| 1 | Unknown | |
| C. tropicalis | 4 | Urine |
| 3 | Respiratory tract | |
| 1 | Pus | |
| 1 | Blood | |
| 1 | Vagina | |
| C. parapsilosis | 4 | Vagina |
| 1 | Blood | |
| 1 | Unknown | |
| C. guilliermondii | 4 | Vagina |
| 1 | Blood | |
| C. glabrata | 5 | Vagina |
| 1 | Urine | |
| 1 | Anal mucosa | |
| C. krusei | 1 | Blood |
| 1 | Unknown | |
| 2 | Vagina | |
| C. lusitaniae | 1 | Respiratory tract |
| Candida spp. | 3 | Respiratory tract |
| 1 | Vagina |
Acknowledgments
This research was supported by Fundação para a Ciência e Tecnologia (FCT) through a multiyear contract with Centro de Ciências do Ambiente (CCA), Universidade do Minho.
We thank Adelaide Alves (Hospital de S. Marcos, Braga, Portugal), Susete Polónia, and Teresa Macedo (Centro de Saúde Braga I) for providing the clinical isolates and for their overall collaboration. Magda Graça is gratefully acknowledged for operating the nucleic acid sequencer and Rogério Tenreiro for help with Bionumerics.
REFERENCES
- 1.Andrade, M. P., G. Schonian, A. Forche, L. Rosado, I. Costa, M. Muller, W. Presber, T. G. Mitchell, and H.-J. Tietz. 2000. Assessment of genetic relatedness of vaginal isolates of Candida albicans from different geographical origins. Int. J. Med. Microbiol. 290:97-104. [DOI] [PubMed] [Google Scholar]
- 2.Dean, D. A., and K. W. Burchard. 1996. Fungal infection in surgical patients. Am. J. Surg. 171:374-382. [DOI] [PubMed] [Google Scholar]
- 3.Ferrer, J. 2000. Vaginal candidosis: epidemiological and etiological factors. Int. J. Gynecol. Obstet. 71:21-27. [DOI] [PubMed] [Google Scholar]
- 4.Fidel, P. L., J. A. Vazquez, and J. D. Sobel. 1999. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hajjeh, R. A., A. N. Sofair, L. H. Harrison, G. M. Lyon, B. A. Arthington-Skaggs, S. A. Mirza, M. Phelan, J. Morgan, W. Lee-Yang, M. A. Ciblak, L. E. Benjamin, L. T. Sanza, S. Huie, S. F. Yeo, M. E. Brandt, and D. W. Warnock. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 42:1519-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazen, K. C. 1995. New and emerging yeast pathogens. Clin. Microbiol. Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 8.Kao, A. S., M. E. Brandt, W. R. Pruitt, L. A. Conn, B. A. Perkins, D. S. Stephens, W. S. Baughman, A. L. Reingold, G. A. Rothrock, M. A. Pfaller, R. W. Pinner, and R. A. Hajjeh. 1999. Population-based candidemia surveillance. Clin. Infect. Dis. 29:1164-1170. [DOI] [PubMed] [Google Scholar]
- 9.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann, P. F., D. Lin, and B. A. Lasker. 1992. Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J. Clin. Microbiol. 30:3249-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, D., L.-C. Wu, M. G. Rinaldi, and P. F. Lehmann. 1995. Three distinct genotypes within Candida parapsilosis from clinical sources. J. Clin. Microbiol. 33:1815-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lott, T. J., R. J. Kuykendal, and E. Reiss. 1993. Nucleotide sequence analysis of the 5.8S rDNA and adjacent ITS2 region of Candida albicans and related species. Yeast 9:1199-1206. [DOI] [PubMed] [Google Scholar]
- 13.Mannarelli, B. M., and C. P. Kurtzman. 1998. Rapid identification of Candida albicans and other human pathogenic yeasts by using short oligonucleotides in a PCR. J. Clin. Microbiol. 36:1634-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClelland, M., C. Petersen, and J. Welsh. 1992. Length polymorphisms in tRNA intergenic spacer detected by using the polymerase chain reaction can distinguish streptoccocal strains and species. J. Clin. Microbiol. 30:1499-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meis, J., M. Petrou, J. Bille, D. Ellis, D. Gibbs, and the Global Antifungal Surveillance Group. 2000. A global evaluation of the susceptibility of Candida species to fluconazole by disc diffusion. Diagn. Microbiol. Infect. Dis. 36:215-223. [DOI] [PubMed] [Google Scholar]
- 16.Meyer, W., K. Maszewka, and T. C. Sorrell. 2001. PCR fingerprinting: a convenient molecular tool to distinguish between Candida dubliniensis and Candida albicans. Med. Mycol. 39:185-193. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira, J. M., A. S. Cruz, A. F. Fonseca, C. P. Vaz, A. Rodrigues, F. Aurea, J. Maia, and J. A. Sousa. 1993. Prevalence of Candida albicans in vaginal fluid of asymptomatic Portuguese women. J. Reprod. Med. 38:41-42. [PubMed] [Google Scholar]
- 18.Peterson, S. W., and C. P. Kurtzman. 1991. Ribosomal RNA sequence divergence among sibling species of yeasts. Syst. Appl. Microbiol. 14:124-129. [Google Scholar]
- 19.Pfaller, M. A. 1995. Epidemiology of candidiasis. J. Hosp. Infect. 30(suppl.):329-338. [DOI] [PubMed] [Google Scholar]
- 20.Roy, B., and S. A. Meyer. 1998. Confirmation of the distinct genotype groups within the form species Candida parapsilosis. J. Clin. Microbiol. 36:216-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampaio, J. P., M. Gadanho, S. Santos, F. L. Duarte, C. Pais, A. Fonseca, and J. W. Fell. 2001. Polyphasic taxonomy of the basidiomycetous yeast genus Rhodosporidium: Rhodosporidium kratochvilovae and related anamorphic species. Int. J. Syst. Evol. Microbiol. 51:687-697. [DOI] [PubMed] [Google Scholar]
- 22.Sampaio, P., L. Gusmão, C. Alves, C. Pina-Vaz, A. Amorim, and C. Pais. 2003. Highly polymorphic microsatellite for the identification of Candida albicans strains. J. Clin. Microbiol. 41:552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. The principle and practice of numerical classification. W. H. Freeman & Co., San Francisco, Calif.
- 24.Sullivan, D., K. Haynes, J. Bille, P. Boerlin, L. Rodero, S. Lloyd, M. Henman, and D. Coleman. 1997. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 35:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thanos, M., G. Schonian, W. Meyer, C. Schweynoch, Y. Graser, T. G. Mitchell, W. Presber, and H. J. Tietz. 1996. Rapid identification of Candida species by DNA fingerprinting with PCR. J. Clin. Microbiol. 34:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wenzel, R. P. 1995. Nosocomial candidemia: risk factors and attributable mortality. Clin. Infect. Dis. 20:1531-1534. [DOI] [PubMed] [Google Scholar]