Abstract
Methane monooxygenase (MMO) enzymes activate O2 for oxidation of methane. Two distinct MMOs exist in nature, a soluble form that uses a diiron active site (sMMO) and a membrane-bound form with a catalytic copper center (pMMO). Understanding the reaction mechanisms of these enzymes is of fundamental importance to biologists and chemists, and is also relevant to the development of new biocatalysts. The sMMO catalytic cycle has been elucidated in detail, including O2 activation intermediates and the nature of the methane-oxidizing species. By contrast, many aspects of pMMO catalysis remain unclear, most notably the nuclearity and molecular details of the copper active site. Here, we review the current state of knowledge for both enzymes, and consider pMMO O2 activation intermediates suggested by computational and synthetic studies in the context of existing biochemical data. Further work is needed on all fronts, with the ultimate goal of understanding how these two remarkable enzymes catalyze a reaction not readily achieved by any other metalloenzyme or biomimetic compound.
Keywords: Methanotroph, Methane monooxygenase, Dioxygen activation, Diiron, Copper
Introduction
Humans are reliant on a dwindling supply of fossil fuels to maintain our current standard of living, often at the expense of ecological homeostasis [1, 2]. Consequently, greenhouse gas emissions are intensely scrutinized, and minimizing climate change is a primary issue in global policy [3]. New technologies to sequester highly potent greenhouse gases while providing alternative uses for them are, thus, valued tremendously. Methane is ~84 times more potent than carbon dioxide as a greenhouse gas, and is present in deposits worldwide [4, 5]. It is routinely used as feedstock in the production of liquid fuels at Fischer–Tropsch facilities [6], but current techniques for producing fuels from methane require large capital expenditure and risk, along with environmental and anthropological perturbation [6, 7]. The desire for environmentally benign and cost-effective methane utilization has prompted researchers to search for alternative catalysts. One avenue for this pursuit is the study of methanotrophic bacteria, a diverse collection of organisms capable of utilizing methane as their sole carbon and energy source. Methanotrophs have been studied intensely over the ~110 years since Kaserer and Söhngen independently identified bacteria capable of methane oxidation [8–10]. The diversity among methanotrophs is a reflection of the distinct environments in which they are found; they can live in temperature, salt, and pH extremes, and survive either facultatively or obligately on C-1 compounds [11–13].
Classically, aerobic methanotrophs have been divided into three categories based on carbon assimilation pathways, intracytoplasmic membrane morphology, and phospholipid composition, as well as other physiological and genetic traits [13–15]. The type I (Gammaproteobacterial) methanotrophs utilize the ribulose monophosphate (RuMP) pathway for carbon assimilation whereas the type II (Alphaproteobacterial) methanotrophs couple methane oxidation to serine formation through the serine pathway. The type X methanotrophs, sometimes categorized as a subset of type I [13, 16], utilize the RuMP pathway, but express ribulose-bisphosphate carboxylase, an enzyme used in the serine pathway. These classifications do not include the comparatively recently discovered Verrucomicrobia [17–19], which seemingly fit neither category [20], and may have a distinct mechanism of methane oxidation [15].
The first step in methanotroph metabolism, oxidation of methane to methanol, is catalyzed by methane monooxygenases (MMOs). Methanotrophs produce two genetically unrelated MMOs: soluble MMO (sMMO) expressed by a subset of methanotrophs and membrane-bound, particulate MMO (pMMO) expressed by nearly all methanotrophs [13]. These enzymes can activate the very strong C–H bond of methane (105 kcal/mol) [21] at ambient temperature and pressure. A pMMO homolog expressed by ammonia oxidizers, ammonia monooxygenase (AMO) [22, 23], is also capable of methane oxidation. In organisms that have genes for both sMMO and pMMO, expression levels are coupled to intracellular copper levels in a mechanism known as the “copper switch”, wherein sMMO is produced at low copper concentrations while pMMO expression is mildly upregulated and sMMO expression is downregulated when copper is available [24–28]. The enzyme active sites reflect this metalloregulation: sMMO contains a non-heme diiron active site and pMMO has a copper active site [29–31]. Whereas many aspects of sMMO function are established, the state of knowledge regarding pMMO is far less advanced. In this minireview, we summarize what is known about O2 activation by both enzymes, with an emphasis on prevailing hypotheses and unresolved questions surrounding pMMO. More detailed reviews of MMO structure, kinetics, efficiency, and biotechnological viability have been published elsewhere [32–34].
Soluble methane monooxygenase
Overall structure and active site
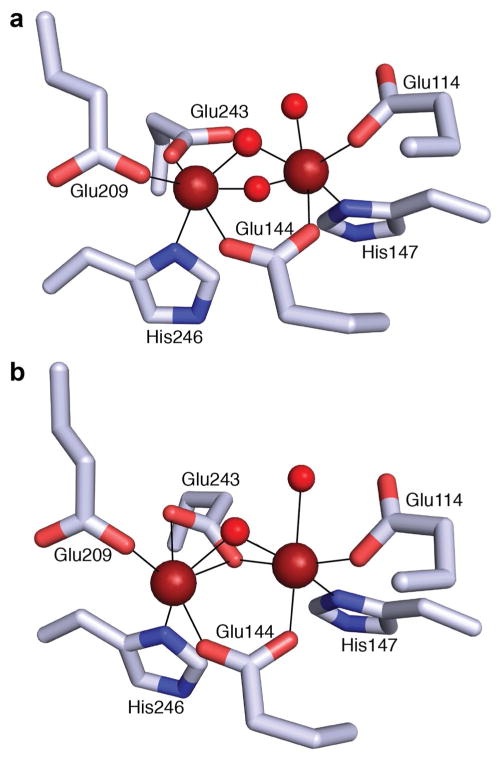
sMMO, which belongs to the larger bacterial multicomponent monooxygenase (BMM) family, requires three protein components for maximal catalytic activity: the hydroxylase (MMOH), the reductase (MMOR), and the regulatory protein (MMOB) [32, 35]. MMOH is a 251 kDa α2β2γ2 homodimer, composed of mostly α helices (Fig. 1a). The diiron active site of each homodimer is located in the α subunit, and no other metal centers are present [29]. The resting state active site (MMOHox) consists of two Fe(III) ions coordinated by Glu114, His147, and a solvent molecule (Fe1), and Glu209, Glu243, and His246 (Fe2). The iron ions are 3.1 Å apart, coordinated in pseudooctahedral fashion and bridged by two solvent molecules as well as Glu144 [29, 36] (Fig. 2a). The two electron reduction of MMOHox to the diiron(II) state (MMOHred) increases the Fe–Fe distance to ~3.3 Å, and a bridging solvent molecule is displaced by Glu243 in a rearrangement known as a carboxylate shift [36] (Fig. 2b).
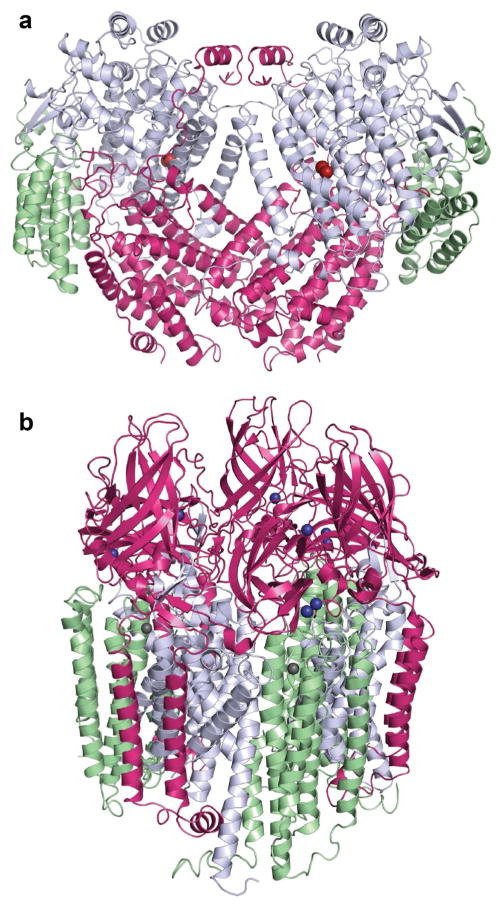
Fig. 1.
Overall structures of MMOs. a M. capsulatus (Bath) sMMO (PDB accession code 1MTY) showing the α subunits in gray, β subunits in magenta, and γ subunits in green. Iron ions are colored red–brown. b M. capsulatus (Bath) pMMO (PDB accession code 3RGB) showing the PmoC subunits in green, PmoB subunits in magenta, and PmoA subunits in gray. Copper ions are dark blue and zinc ions are gray
Fig. 2.
The diiron active site of MMOH in the a oxidized (PDB accession code 1MTY) and b reduced (PDB accession code 1FYZ) forms, demonstrating the carboxylate shift of Glu243, displacing a solvent ligand (colored red). Iron ions are colored red–brown
The active site is ~12 Å from the bulk solvent and can be accessed via a hydrophilic region known as the “pore” that is gated by residues Thr213, Asn214, and Glu240 [37, 38]. The egress from this pore leads to the “canyon” region, a large surface at the MMOH dimer interface that is the site of MMOR and MMOB binding. There are three hydrophobic pockets [37] that can be occupied in structures of MMOH and the related BMM toluene/o-xylene monooxygenase with bound substrate or product analogs [29, 39–42]. The roles of the pore and these cavities in transport of substrates, products, protons, and electrons have not been established definitively. In one model, the hydrophobic cavities constitute the pathway for substrate access (O2 and methane) and/or product exit to and from the active site, and the pore provides the entrance for protons and electrons as well as the exit for water, facilitated by residue Glu240 [37, 38]. However, substrate access via the 36–40 Å pathway connecting the cavities is not consistent with kinetic data showing that reaction of substrates with the oxidizing intermediate Q (vide infra) has a linear concentration dependence [43, 44]. Successive occupancy of these sites would be expected to yield zero order kinetics with respect to substrate. Alternatively, substrates could access the active site via the pore. In support of this second model, mutation of MMOB residues adjacent to the pore [38] significantly affects the reaction rate with intermediate Q, and does so in a substrate-dependent fashion [45, 46], which would be consistent with the pore functioning as the substrate entry site.
MMOR is a ~40 kDa protein composed of three domains: a [2Fe-2S] ferredoxin domain, an FAD-binding domain, and an NADH-binding domain [47–49]. Structures of the isolated reduced ferredoxin domain and reduced FAD- and NADH-binding domains have been determined via NMR [50, 51]. Interactions between the ferredoxin and hydroxylase components have been observed crystallographically for the BMM toluene 4-monooxygenase, in which the reduced ferredoxin domain binds in the hydroxylase canyon region atop the pore [52]. This same binding site was proposed for sMMO based on hydrogen–deuterium exchange coupled mass spectrometry (HDX-MS) and computational docking [53]. Additionally, chemical crosslinking studies indicate that MMOR interacts with the α [54] and β [55] subunits of MMOH. The redox potential of MMOH increases in the presence of MMOR, indicating a higher affinity of MMOR for MMOHred [56].
The 16 kDa regulatory component MMOB contains no cofactors. MMOB binding to MMOH affects the structure of MMOH [38] and the geometry of the diiron center as manifested by alterations of the EPR, CD, and MCD spectra [55, 57, 58]. These changes increase methane oxidation activity by accelerating reaction intermediate formation as well as O2 and proton delivery to the active site [47, 55, 59]. Specifically, complex formation repositions MMOH residue Phe188, which gates the hydrophobic cavities [38] and closes the pore via movement of Glu240 (suggested to reposition for proton donation to the sMMO active site), perhaps preventing electrons from prematurely quenching reactive intermediates [37, 38]. Fluorescence anisotropy and double electron–electron resonance (DEER) studies suggest that spin-labeled MMOB binds MMOHred with higher affinity than MMOHox and that the N-terminal tail of MMOB, truncation of which severely impacts MMOH activity [38], participates in an ordered interaction only with MMOHred [60]. These observations are not compatible with previous work showing that the redox potential of MMOH decreases in the presence of MMOB [61, 62], which requires that MMOB bind MMOHox with a higher affinity than MMOHred [56]. In addition, fluorescence titrations using labeled MMOB indicate a higher affinity for MMOHox than MMOHred [63].
O2 activation intermediates
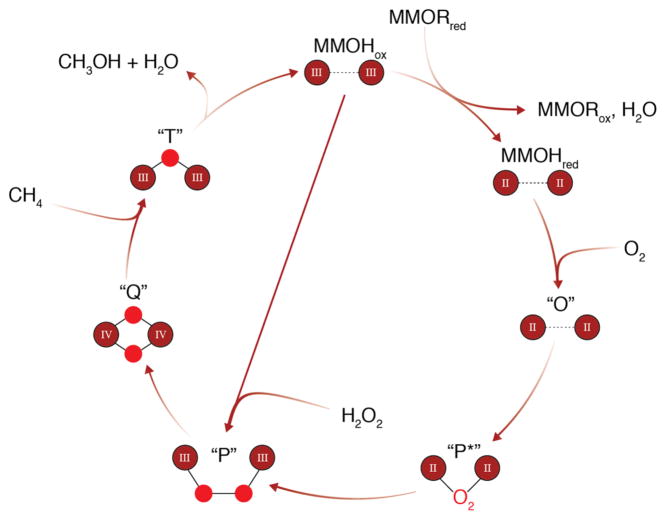
The intermediates involved in O2 activation by sMMO have been largely characterized (Fig. 3), although exactly how MMOR and MMOB regulate activity has not been fully elucidated. The antiferromagnetically coupled, high-spin site in MMOHox [29, 64–67] is reduced by two electrons to the weakly ferromagnetically coupled, high-spin state, MMOHred [68]; this change is correlated with the carboxylate shift of residue Glu243 (Fig. 2). The electrons come from reduction of the MMOR FAD cofactor by NADH, followed by transfer to the MMOR [2Fe-2S] cluster, and then sequential electron transfer to the MMOH diiron site [69]. The Glu243 carboxylate shift upon conversion to MMOHred provides an open coordination site for O2 binding [36]. After formation of MMOHred, O2 associates with the MMOHred-2MMOB complex, in which MMOB has been proposed to displace MMOR [37, 53]. Whether MMOB and MMOR bind in the same location remains controversial. MMOB inhibits MMOR binding to MMOH [53, 55], which would be consistent with a shared binding site. However, MMOB and MMOR are also known to form a protein–protein complex [55], which could explain inhibition of MMOR binding by MMOB without invoking a common binding site. Moreover, MMOR decreases the rate of O2 reactivity of the MMOHred-2MMOB complex, indicating that a ternary complex can form [56], and kinetic data are best modeled with the formation of a complex between all three components [55, 70]. The high affinities for both MMOB and MMOR for MMOH are not consistent with a model in which electron transfer is simply controlled by competitive binding of the two components [37], and further consideration of this issue is warranted.
Fig. 3.
Catalytic cycle of sMMO, with iron ions shown in red–brown, and their oxidation states designated. Oxygen atoms are represented as red circles, except for intermediate P*, for which the binding mode of O2 to the active site is unknown. Fe–Fe, Fe–O, and O–O distances and bond angles through the catalytic cycle are not implied. Intermediate P can also be generated by reaction of MMOHox with H2O2 in a mechanism known as the peroxide shunt
The interaction of MMOHred with O2 produces intermediate O, which has been hypothesized to be a complex of MMOHred with O2 at a location other than the diiron site [59, 71, 72]. Intermediate O is converted to intermediates P* and then P [73]. It has been suggested that P* and P are electronically equivalent as a site, with protonation of either a coordinating ligand or solvent molecule and minor structural changes producing P from P* [74]. However, recent Mössbauer data indicate that P* is an center [75]. P is believed to be a μ-1,2 peroxo-bridged species, formed from P* in a pH-dependent manner, indicating that proton transfer is involved in the rate-determining step [71, 73, 76, 77]. Intermediate P can alternatively be generated by mixing MMOHox with excess H2O2, a mechanism known as the peroxide shunt [78, 79].
Proton transfer and homolytic O–O bond cleavage then produce compound Q, the methane-oxidizing species [43, 80–82]. The structure of Q was elucidated via time resolved resonance Raman spectroscopy as a diamond di(μ-oxo) core [83]. Intermediate Q reacts with methane to insert a single oxygen atom derived from the bound O2 via an unknown mechanism. This critical step has been studied computationally [84] as well as experimentally; radical and/or cationic mechanisms [85–91] as well as concerted reaction mechanisms have been proposed [92–94]. Experiments using chiral ethane as a substrate suggest that hydrogen abstraction by intermediate Q yields a radical intermediate [95, 96], but one that is best explained by formation of a “bound-radical” intermediate in which methyl radical rotation is constrained by interaction with the diiron center [97]. This model also includes a second nonsynchronous concerted pathway, with the reaction proposed to involve a mixture of the two pathways [97, 98]. Extensive studies utilizing radical clock probes are consistent with this proposal [92]. It should be noted that MMOH can oxidize a large range of substrates, including C1–C8 linear alkanes, alkenes, cyclic and aromatic carbon compounds [99–101], and that understanding C–H activation may be complicated by the use of various probe substrates that may react by different or multiple mechanisms [102]. Finally, compound T forms after Q reaction with methane, and is assigned as a mono(μ-oxo) bridged site [83].
Particulate methane monooxygenase
Overall structure and metal centers
The only known protein component of pMMO is the hydroxylase, referred to simply as pMMO. pMMO from four methanotrophs has been characterized crystallographically, and in all instances the oligomerization state is a ~300 kDa α3β3γ3 trimer composed of PmoA (24 kDa) and PmoC (22 kDa), almost entirely transmembrane domains, and PmoB (42 kDa), a primarily periplasmic subunit comprising two cupredoxin-like β-barrels joined by two trans-membrane helices [30, 103–105] (Fig. 1b). Additionally, a helix of unknown sequence and function associates with PmoC in all pMMO structures except that from Methylococcus (M.) capsulatus (Bath) and is likely of biological, if not mechanistic, significance [32]. The physiological relevance of the trimeric state and general subunit assembly observed in the crystal structures has been validated by electron microscopic structural characterization of an active M. capsulatus (Bath) pMMO [106, 107].
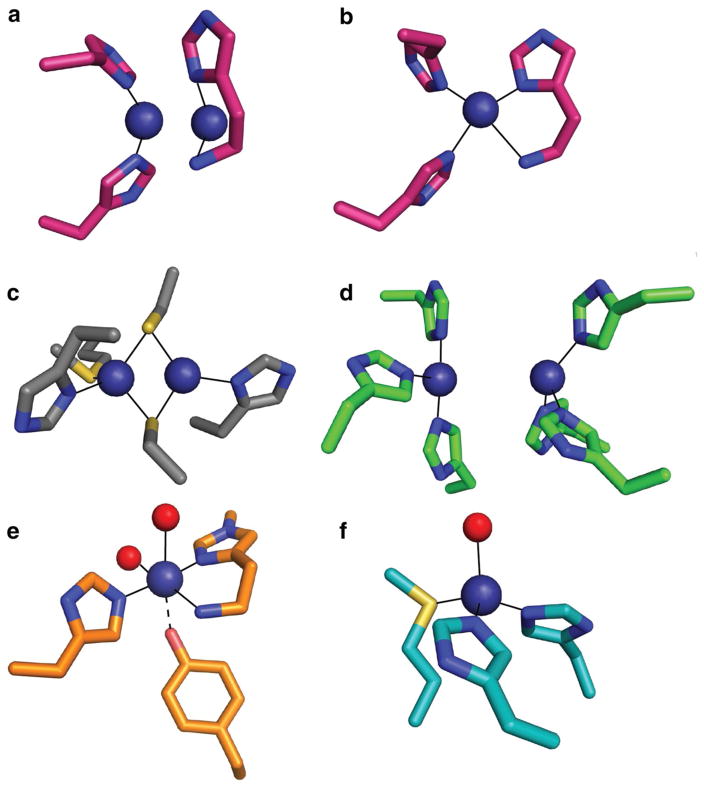
Unlike sMMO, structures of pMMO have not unambiguously identified the active site: the first structure, obtained for M. capsulatus (Bath) pMMO, revealed several metal centers. These sites include (1) a monocopper site coordinated by nonconserved residue His48 and residue His72 from the N-terminal subdomain of PmoB, (2) a dicopper site in the same subdomain with a Cu–Cu distance of ~2.5 Å, ligated by the N-terminal amino group and side chain of His33 (Cu1) as well as His137 and His139 (Cu2) (Fig. 4a), and (3) a zinc ion in PmoC ~20 Å away from the PmoB dicopper site and attributed to the crystallization solution [30, 108]. The dicopper and zinc site ligands are conserved across all pMMOs and AMOs except for the distantly related Verrucomicrobial pMMOs, which lack the residues corresponding to His33, His137, and His139 [20, 109, 110]. With the highest resolution pMMO structure with two copper ions modeled in this site solved to 2.68 Å [103], the copper ions are not observable as discrete atoms. Thus, assignment of nuclearity has relied on extended X-ray absorption fine structure (EXAFS) data, which are consistently best fit with Cu–Cu distances of ~2.5 Å for the resting state and ~2.65 Å for chemically reduced samples [103, 108]. The three metal sites have been variably occupied in later pMMO structures, with the monocopper center not observed in any other pMMO, the PmoC site occupied by either copper or zinc depending on the crystallization conditions, and the dicopper site often modeled with only a single copper ion [103–105] (Fig. 4b). The crystal structure of a soluble AmoB domain also contains only a single copper ion in the dicopper site, although the amino terminal histidine was not visible in the structure and this protein fragment did not exhibit methane oxidation activity [109]. Despite these issues, a dicopper center at this location remains the leading hypothesis for the pMMO active site (vide infra).
Fig. 4.
Comparison of pMMO dicopper site occupied with a two copper ions or b one copper ion from Methylocystis species strain M (PDB accession code 3RFR) to c the cytochrome c oxidase CuA site (PDB accession code 1AR1), d the deoxytyrosinase dicopper site (PDB accession code 2Y9X), e the monocopper active site of LPMO (PDB accession code 5ACG), and f the CuB/CuM site of PHM (PDB accession code 1PHM). Solvent ligands are shown in red and copper ions are in dark blue
Attempts to cocrystallize pMMO with substrate to identify the active site and assess pathways for hydrocarbon access and product egress have not been successful. One possible methane entryway is through the membranes [15], favored in part because of the higher solubility of methane in lipid bilayers relative to solution [111]. A hydrophobic pocket in PmoA was proposed to provide such access, justified by steric constraints consistent with pMMO substrate size limitations [112], but a pathway from this site to the PmoB copper active site has not been established. Unique pathways for smaller (C1–C2) versus larger (C3–C5) hydrocarbons have also been proposed, a notion somewhat corroborated by the dramatically different affinity of pMMO for propane compared to methane [113, 114]. If hydrocarbons do access the pMMO active site through the membranes, navigation to the active site would not necessitate solvent occlusion through extensive protein channels as proposed for MMOH. Thus, an adventitious pathway for larger hydrocarbon substrates could exist, leading to the different affinities. Hydrophobic cavities identified between PmoC and PmoB may also play roles in directing substrate to the pMMO active site [33, 106, 107].
The copper active site
Understanding O2 activation by pMMO has been hindered by uncertainty regarding the metal composition of the active site. It is currently accepted that pMMO uses copper to oxidize methane, despite earlier suggestions of an iron active site [32, 115]. The dependence of pMMO activity on copper was established by metal reconstitution experiments showing that activity of metal-depleted, membrane-bound pMMO can be restored by copper and not by iron [31, 105]. In addition, much of the available information indicates that the active site is located at the site of the dicopper center in PmoB. First, the His48 ligand to the monocopper center observed in the M. capsulatus (Bath) pMMO structure is not conserved and this site is not present in any other pMMO structures [103–105]. Second and most important, a recombinantly expressed fragment of PmoB is capable of methane oxidation [31]. This protein, denoted as spmoB, consists of the two soluble PmoB subdomains joined by a flexible linker region, and exhibits methane oxidation activity dependent on binding of approximately two copper equivalents. Copper presumably binds at the location of the crystallographically modeled dicopper active site since mutation of the histidine ligands diminishes metal binding and completely abrogates activity [31], and EXAFS data indicate the presence of a short Cu–Cu distance as observed in pMMO [31]. These data also rule out the PmoC metal center and a proposed tricopper center in PmoA [116, 117] as the active site since these subunits are not present in spmoB.
There are several potential discrepancies with this active site model. First, mutation of the His139 equivalent residue in the C2–C4 alkane-oxidizing pMMO homolog hydrocarbon monooxygenase (HMO) from Mycobacterium NBB4 does not completely abrogate its ability to consume butane, although it is severely reduced [118]. It is possible that this mutant HMO active site might still bind one copper ion and be capable of activating the comparatively weaker C–H bond in C2–C4 alkanes [21]. Second, the histidine residues involved in dicopper site coordination are not conserved in the distantly related Verrucomicrobia PmoB subunit [20]. However, the extreme growth conditions of these Verrucomicrobial methanotrophs (capable of growth below pH 1 and up to 65 °C) may very well require a distinct active site and mechanism from that of the Proteobacterial pMMOs [15, 32]. Finally, pMMO/AMO studies with radiolabeled acetylene, a suicide substrate, suggest involvement of PmoA in the active site [119–122]. Since PmoA houses no identified metal centers, either conformational changes during catalysis or product migration after reaction at the active site may explain these data. Since these discrepancies do not strongly support an alternative location for the active site, the copper site at the N-terminus of PmoB remains the most viable candidate.
Assuming that O2 activation occurs at this site, the question of nuclearity is critical. As mentioned above, EXAFS data consistently indicate a short Cu–Cu interaction [103, 108], but crystallographic data are ambiguous and often best fit with a single copper ion [103–105] (Fig. 4b). The situation is further complicated by the loss of enzymatic activity and copper upon purification and crystallization of some pMMO samples [104, 105] as well as the consistent observation that two copper ions are required for maximal activity recovery in reconstitution experiments [31, 105]. These two copper ions could both be located within the putative dicopper center or there could be an alternative copper-requiring site, such as that in PmoC. While spmoB does not contain additional sites, its activity is significantly less than that of pMMO, suggesting that the rest of the enzyme, possibly including other metal centers, plays an important role in activity. Analysis of the M. capsulatus (Bath) pMMO and spmoB resting state EPR spectroscopic features suggest the presence of a valence-localized, scrambled, CuICuII dicopper active site, an additional CuI occupying the nonconserved monocopper site, and no CuII in the PmoC site [123]. Since no evidence for strong dipolar coupling, electron delocalization over the copper ions, or an (anti)ferromagnetically coupled ground state (except for studies claiming to observe the trinuclear copper cluster signature [116, 124, 125]) has been obtained, a CuICuII resting state is the only consistent assignment if the site is indeed dinuclear and not CuICuI.
Understanding the pMMO copper active site has also been hindered by the lack of similar sites in well-characterized copper proteins [126]. The proposed dicopper center shares some features with the dinuclear CuA centers, which exhibit a nearly identical Cu–Cu distance to that in pMMO [127] (Fig. 4c) and with the type 3 dinuclear copper centers of tyrosinase, hemocyanin, and multicopper oxidases (MCOs), which coordinate copper using exclusively histidine ligands [128] (Fig. 4d). However, the CuA site is defined by two bridging cysteine sulfur ligands, and EPR spectroscopic characterization indicates complete electron delocalization over the two copper ions [128]. By contrast, pMMO consistently exhibits a type 2 copper EPR signal, typically observed for mononuclear CuII [123]. Although a fully delocalized CuA center can give an apparent mononuclear localized EPR signature [129], no Cu–S scattering has ever been observed via EXAFS for pMMO (PmoB does not in fact contain any cysteine residues), and no bridging ligands are observed by crystallography. The type 3 copper sites differ from pMMO in that they feature an EPR-silent, antiferromagnetically coupled CuIICuII oxy ground state and also have significantly longer Cu–Cu distances [128, 130].
If the site is actually mononuclear, it resembles that in lytic polysaccharide monooxygenases (LPMOs), in which an N-terminal histidine also coordinates a copper ion in a bidentate fashion, a mode termed the histidine brace [131–133] (Fig. 4e). pMMO contains a third histidine ligand not present in LPMO, however. In some, but not all, LPMO structures, the histidine is methylated; there is no evidence for this occurring in pMMO. In addition, a tyrosine side chain oxygen is within 2.6 Å of the LPMO copper ion. In pMMO, there are tyrosines as close as 5 Å to the copper ion, but not coordinating. Another relevant example is the CuB/CuM site in peptidylglycine α-hydroxylating monooxygenase (PHM) and dopamine β-hydroxylase (DβH), in which O2 activation occurs at a single copper ion coordinated by two histidines and a methionine that is important for activity [134, 135] (Fig. 4f). However, there are no conserved methionines in the vicinity of the pMMO site and no evidence from EXAFS for Cu–S interactions [103, 108].
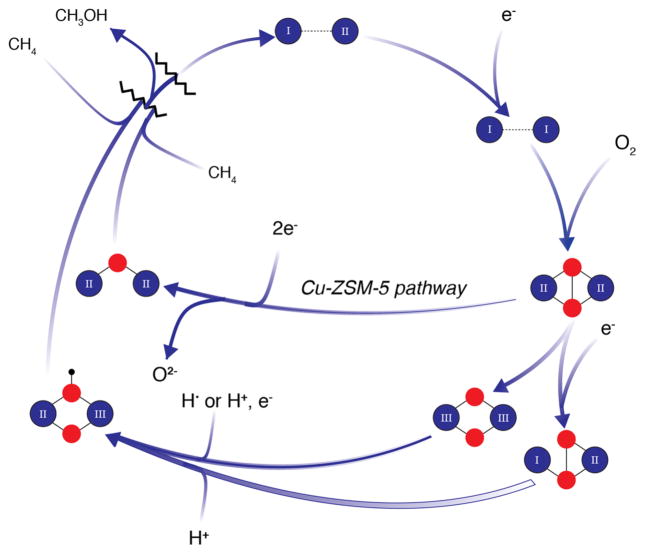
Possible O2 activation intermediates
Given the uncertainty surrounding the active site nuclearity, it is necessary to consider how O2 activation might occur at both mono- and dicopper centers. The monocopper site in LPMOs (Fig. 4e) cleaves glycosidic C–H bonds on the order of 95 kcal/mol [132], and the PHM and DβH monocopper sites (Fig. 4f) activate O2 to hydroxylate substrates with C–H bond strengths of 87 and 85 kcal/mol, respectively [136]. Thus, a key question is whether a somewhat similar monocopper site can cleave the methane 105 kcal/mol C–H bond. Density functional theory (DFT) and quantum mechanics/molecular mechanics (QM/MM) calculations using the M. capsulatus (Bath) pMMO structure as a starting model have suggested that a mononuclear copper active site may be viable, proceeding through a CuIII-oxo (CuII-O·) species [137]. However, this study used the nonconserved monocopper site ligated by His48 and His72 as a starting model, and it would be worthwhile to reinvestigate the calculations using a monocopper site coordinated by the residues equivalent to M. capsulatus (Bath) His33, His137, and His139 (Fig. 4b).
Although the computational work deemed the mono-nuclear CuIII-oxo species sufficiently reactive for methane oxidation, the formation of a dicopper–oxygen species is more favorable. In this pathway, also calculated for M. capsulatus (Bath) pMMO, a μ-η2:η2-peroxo- or μ-η1:η2-peroxo-CuICuII species is converted to a reactive di(μ-oxo) CuIICuIII or (μ-oxo)(μ-hydroxo)CuIICuIII species capable of methane oxidation [137–139]. In these calculated species, one copper ion is coordinated by the side chain nitrogens of His137 and His139 while the other is coordinated by the side chain of His33, and instead of the amino terminal nitrogen (Fig. 4a), an oxygen from the side chain of Glu35. Residue Glu35 is included in the starting models for these calculations to maintain charge neutrality, but is not within coordinating distance in the crystal structures. The (μ-oxo) (μ-hydroxo)CuIICuIII species is proposed to form via an H-atom transfer or proton-coupled electron transfer (PCET) mechanism in which homolytic cleavage of the O–H bond of a second-sphere tyrosine residue, Tyr374, and transfer to the μ-η2:η2-peroxo- produces a μ-η1:η2hydroperoxo-CuICuII species, which is converted to a reactive (μ-oxo) (μ-hydroxo)-CuIICuIII species [138] (Fig. 5). One caveat to this proposed mechanism is CuIII has not been observed in biological systems [140–142]. While the invoked tyrosine residue is not in the same position in every pMMO, there is always one present, including Tyr341, Tyr352, and Tyr341 in the structures of pMMO from Methylocystis species strain M, Methylosinus trichosporium OB3b, and Methylocystis species strain Rockwell, respectively, all with a Cu–Tyr hydroxyl distances of ~5 Å. The functional relevance of these tyrosines has not yet been addressed experimentally.
Fig. 5.
Possible O2 activation intermediates for pMMO. Copper ions with oxidation states designated are shown in dark blue, oxygen atoms in red, and hydrogen atoms in black
There is some experimental support for the formation of dicopper–oxygen species in pMMO. Upon reaction of reduced pMMO with H2O2, a species with an absorbance feature at 345 nm appears and was tentatively assigned as a μ-η2:η2-peroxo- species. This species disappears upon incubation with methane and some methanol product is detected [32, 143]. The feature was not produced when pMMO was incubated with O2-saturated buffer, however. This may be due to difficulty in supplying sufficient O2 to the pMMO active site since the feature was generated in the presumably more solvent-accessible active site of spmoB with either O2-saturated buffer or H2O2 [143]. This notion is consistent with studies suggesting that a hemerythrin found natively in the membranes and cytoplasm of M. capsulatus (Bath) [144] may facilitate O2 delivery to the pMMO active site [145]. Computational studies suggest that a μ-η2:η2-peroxo- species formed in the pMMO active site is not planar, but adopts a bent butterfly structure [138, 146, 147]. The calculated absorption spectrum of this species exhibits a peak at 370 nm, and the difference between 345 and 370 nm was attributed to the calculation algorithm [146]. It is also possible that the 345 nm feature formed by pMMO is not due to a μ-η2:η2-peroxo- species, and further spectroscopic characterization is necessary.
Finally, some insight into O2 activation by the pMMO copper active site can be derived from work on non-biological systems. The copper zeolite Cu-ZSM-5 oxidizes methane to methanol under relatively mild conditions using a bent mono(μ-oxo) reactive species [148] that forms from a μ-η2:η2-peroxo- adduct [149] (Fig. 5), suggesting that such a species could participate in methane oxidation by pMMO. Similarly, dicopper complexes assembled using either primary amines or histamine ligands to yield a planar bis(μ-oxide) coordination exhibit short Cu–Cu distances and strong Cu–N bonding, and are able to oxidize hydrocarbons with C–H bond strengths of ~76 kcal/mol [150, 151]. These compounds demonstrate that histidine imidazoles can stabilize CuIII ligation under the conditions employed, and may provide a chemical rationale for the presence of an N-terminal histidine ligand in pMMO (as well as LPMO). Model compounds provide an excellent foundation for eventual interpretation of spectroscopic features of pMMO reaction intermediates, but it should be noted that no isolated dicopper complex has been shown to oxidize methane. Methane to methanol conversion by tricopper complexes and zeolites containing tricopper species has also been reported [152–156]. However, no copper has been observed at the site of the proposed PmoA tricopper active site [116, 117], and the exact electronic structure of the species giving rise to an EPR signal assigned to a tricopper site [116, 124, 125] remains unclear [106, 157–159].
Finally, some effort has been made to probe possible mechanisms of C–H activation by pMMO. Chiral ethane hydroxylation proceeds with enantioselectivity, favoring a concerted pentacoordinate C–Cu or C–O hydrocarbon intermediate preceding oxygen insertion and C–H bond cleavage rather than a radical or cationic mechanism [160]. These findings have been recapitulated with other substrates and point toward a binding cavity that orients substrate specifically [113, 161, 162]. However, a radical or cationic mechanism could be rationalized if pMMO substantially slows the rate of hydrocarbon C-C bond rotation through binding constraints [33, 160]. Due to the more limited substrate specificity of pMMO (C1–C5 linear alkanes and alkenes) [99, 163], radical clock substrates cannot be utilized as they were for sMMO.
Reduction of the pMMO active site
A final piece of the pMMO puzzle that requires discussion is the nature of the physiological reductant. Whereas reduction of MMOH by MMOR is well understood, the source of electrons for pMMO remains under debate. Activity assays on membrane-bound pMMO routinely utilize NADH or duroquinol as reductant (succinate has been shown to work as well), while only duroquinol and to a lesser extent, other quinols, are effective for solubilized and purified samples [164, 165]. Theories for the native reductant have traditionally fallen into two camps, active site reduction via metallocofactor or via endogenous quinols, which may be mimicked by duroquinol [164]. Biological electron transfer through metallocofactors such as those in type 1 Cu and CuA proteins and c-type cytochromes is well established [166–168]. An as yet unidentified complementary reductase component may house such a cofactor, possibly associating with conserved acidic residues near the copper active site [30]. Alternatively, electrons from methanol and/or formaldehyde oxidation could be recycled to reduce a cytochrome c which would then reduce the pMMO active site [169–172]. This latter model is consistent with protein–protein interactions detected between pMMO and methanol dehydrogenase (MDH) [173]. The PmoB C-terminal subdomain, which does not bind metal, might play a role in this interaction as suggested for a purported pMMO/MDH supercomplex [171], although it has not been possible to isolate this complex in a stable form [173]. Notably, recent metabolic reconstructions suggest that methanol oxidation could indeed support methane oxidation [174].
In favor of the quinol model, the inability of NADH to function as a reductant for purified pMMO may be a consequence of removing a type 2 NADH:quinone oxidoreductase (NDH-2) complex during the purification process, implying that in vivo, pMMO is reduced by membrane-available quinols generated by the NDH-2. NDH-2 involvement is supported by an increase in pMMO activity upon adding duroquinone, NADH, and NDH-2, as compared to direct duroquinol addition [175, 176]. There is no evidence that pMMO and NDH-2 form a stable complex, however. No quinol binding site has been identified in pMMO, but one possibility is that quinols bind near the PmoC metal binding site. The inhibitory effect of zinc on pMMO has been attributed to binding at this site, which may interfere with delivery of protons to the active site [105], and could also affect electron transfer. The potential importance of this site is underscored by the complete conservation of metal binding residues, as well as studies in which mutation of residues comprising the analogous site in HMO completely abolished activity [118]. Assuming direct tunneling electron transfer, which is limited to ~14 Å [177], the ~20 Å distance from the PmoC metal site to the copper active site is not too far outside of this range, supporting the notion that quinols could bind nearby. A clearer understanding of the initial reduction step of the pMMO active site could enhance activity in vitro and thus facilitate future identification of O2 activation intermediates.
Conclusions and outlook
MMOs are unique in their ability to generate an appropriately potent oxidant for methane, and are, therefore, of central interest to the O2 activation field. It is remarkable that two completely different biological solutions to this challenging reaction have evolved, and in some cases, exist in the same organism despite divergence in other metabolic processes. sMMO utilizes three protein components to orchestrate conformational and electronic changes in MMOH that enable efficient methane oxidation at its diiron active site. The exact binding sites and timing of MMOR and MMOB association and dissociation with MMOH remain under discussion, however. pMMO accomplishes the same reaction using copper, but the molecular details of its active site have not been established. In the case of pMMO, questions also remain regarding its native reductant and the possible involvement of additional protein components. The sMMO structures suggest several routes for substrate to access the active site; the existence of such pathways in pMMO remains more speculative.
Understanding of O2 activation by sMMO is advanced, including identification of the oxidizing species Q as a diamond di(μ-oxo) diiron(IV) core. Future computational and spectroscopic studies will likely provide additional insight into this species. By contrast, O2 activation by pMMO must still be considered in the context of both mono- and dicopper sites, with calculations and biochemical studies currently favoring dicopper–oxygen intermediates. Further computational work utilizing more recent pMMO crystal structures may be informative. However, given that the current picture of the pMMO active site may not be complete, calculations and studies of model complexes should be interpreted with some caution. It is particularly intriguing that several lines of investigation suggest the involvement of a CuIII species in methane oxidation by pMMO. An overarching consideration is that neither other enzymes with diiron, dicopper, or monocopper sites nor biomimetic synthetic complexes can oxidize methane. Thus, further characterization of sMMO intermediate Q and identification of the pMMO oxidizing species must be accompanied by detailed investigations of how the two distinct protein scaffolds precisely control the oxidation chemistry.
Acknowledgments
Work in the Rosenzweig laboratory on biological methane oxidation is supported by National Institutes of Health Grant GM118035 (A. C. R.). M. O. R. was supported in part by National Institutes of Health Grant 5T32GM008382.
Abbreviations
- AMO
Ammonia monooxygenase
- BMM
Bacterial multicomponent monooxygenase
- DβH
Dopamine β-hydroxylase
- EPR
Electron paramagnetic resonance
- EXAFS
Extended X-ray absorption fine structure
- HMO
Hydrocarbon monooxygenase
- MDH
Methanol dehydrogenase
- MMO
Methane monooxygenase
- MMOB
sMMO regulatory protein
- MMOH
sMMO hydroxylase
- MMOHox
MMOH with a diiron(III) site
- MMOHred
MMOH with a diiron(II) site
- MMOR
sMMO reductase
- NDH-2
Type 2 NADH:quinone oxidoreductase
- PHM
Peptidylglycine α-hydroxylating monooxygenase
- pMMO
Particulate MMO
- sMMO
Soluble MMO
References
- 1.Covert T, Greenstone M, Knittel CR. J Econ Perspect. 2016;30:117–137. [Google Scholar]
- 2.Heinberg R. Afterburn: society beyond fossil fuels. New Society Publishers; Gabriola Island: 2015. [Google Scholar]
- 3.Girod B, van Vuuren DP, Hertwich EG. Glob Environ Chang. 2014;25:5–15. [Google Scholar]
- 4.Myhre G, Shindell D, Bréon F-M, Collins W, Fuglestvedt J, Huang J, Koch D, Lamarque J-F, Lee D, Mendoza B. Climate change 2013: the physical science basis. In: Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM, editors. Contribution of working Group I to the fifth assessment report of the intergovernmental panel on climate change. Cambridge University Press; Cambridge and New York: 2013. pp. 659–740. [Google Scholar]
- 5.Bibler CJ, Marshall JS, Pilcher RC. Int J Coal Geol. 1998;35:283–310. [Google Scholar]
- 6.Dry ME. Catal Today. 2002;71:227–241. [Google Scholar]
- 7.Cartwright E. In: Cultures of energy: power, practices, technologies. Strauss S, Rupp S, Love T, editors. Left Coast Press, Inc; Walnut Creek: 2013. pp. 201–212. [Google Scholar]
- 8.Hutton WE, ZoBell CE. J Bacteriol. 1949;58:463–473. doi: 10.1128/jb.58.4.463-473.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaserer H. Z landv Versuchsw Deut Oesterr. 1905;8:789. [Google Scholar]
- 10.Söhngen NL. Zentr Bakt Parasitenk. 1906;15:513–517. [Google Scholar]
- 11.Dedysh SN, Dunfield PF. Methods Enzymol. 2011;495:31–44. doi: 10.1016/B978-0-12-386905-0.00003-6. [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Chen Y, Jiang P, Zhang C, Smith TJ, Murrell JC, Xing X-H. Biochem Eng J. 2010;49:277–288. [Google Scholar]
- 13.Semrau JD, DiSpirito AA, Yoon S. FEMS Microbiol Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 14.Hanson RS, Hanson TE. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culpepper MA, Rosenzweig AC. Crit Rev Biochem Mol Biol. 2012;47:483–492. doi: 10.3109/10409238.2012.697865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowman JP, Sly LI, Nichols PD, Hayward AC. Int J Syst Evol Microbiol. 1993;43:735–753. [Google Scholar]
- 17.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, Op den Camp HJM. Nature. 2007;450:874–878. doi: 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 18.Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M. Nature. 2007;450:879–882. doi: 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- 19.Islam T, Jensen S, Reigstad LJ, Larsen Ø, Birkeland N-K. Proc Natl Acad Sci. 2008;105:300–304. doi: 10.1073/pnas.0704162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A, Dunfield PF. Environ Microbiol Rep. 2009;1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 21.Blanksby SJ, Ellison GB. Acc Chem Res. 2003;36:255–263. doi: 10.1021/ar020230d. [DOI] [PubMed] [Google Scholar]
- 22.Arp DJ, Sayavedra-Soto LA, Hommes NG. Arch Microbiol. 2002;178:250–255. doi: 10.1007/s00203-002-0452-0. [DOI] [PubMed] [Google Scholar]
- 23.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen AK, Gerdes K, Murrell JC. Mol Microbiol. 1997;25:399–409. doi: 10.1046/j.1365-2958.1997.4801846.x. [DOI] [PubMed] [Google Scholar]
- 25.Kenney GE, Sadek M, Rosenzweig AC. Metallomics. 2016;8:931–940. doi: 10.1039/c5mt00289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murrell JC, McDonald IR, Gilbert B. Trends Microbiol. 2000;8:221–225. doi: 10.1016/s0966-842x(00)01739-x. [DOI] [PubMed] [Google Scholar]
- 27.Prior SD, Dalton H. Microbiology. 1985;131:155–163. [Google Scholar]
- 28.Stanley S, Prior S, Leak D, Dalton H. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- 29.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 30.Lieberman RL, Rosenzweig AC. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 31.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirajuddin S, Rosenzweig AC. Biochemistry. 2015;54:2283–2294. doi: 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sazinsky MH, Lippard SJ. Met Ions Life Sci. 2015;15:205–256. doi: 10.1007/978-3-319-12415-5_6. [DOI] [PubMed] [Google Scholar]
- 34.Lawton TJ, Rosenzweig AC. J Am Chem Soc. 2016;138:9327–9340. doi: 10.1021/jacs.6b04568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Müller J, Lippard SJ. Angew Chem Int Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 36.Rosenzweig AC, Nordlund P, Takahara PM, Frederick CA, Lippard SJ. Chem Biol. 1995;2:409–418. [PubMed] [Google Scholar]
- 37.Wang W, Liang AD, Lippard SJ. Acc Chem Res. 2015;48:2632–2639. doi: 10.1021/acs.accounts.5b00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SJ, McCormick MS, Lippard SJ, Cho U-S. Nature. 2013;494:380–384. doi: 10.1038/nature11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sazinsky MH, Lippard SJ. Acc Chem Res. 2006;39:558–566. doi: 10.1021/ar030204v. [DOI] [PubMed] [Google Scholar]
- 40.Sazinsky MH, Bard J, Di Donato A, Lippard SJ. J Biol Chem. 2004;279:30600–30610. doi: 10.1074/jbc.M400710200. [DOI] [PubMed] [Google Scholar]
- 41.Sazinsky MH, Lippard SJ. J Am Chem Soc. 2005;127:5814–5825. doi: 10.1021/ja044099b. [DOI] [PubMed] [Google Scholar]
- 42.Whittington DA, Rosenzweig AC, Frederick CA, Lippard SJ. Biochemistry. 2001;40:3476–3482. doi: 10.1021/bi0022487. [DOI] [PubMed] [Google Scholar]
- 43.Lee S-K, Nesheim JC, Lipscomb JD. J Biol Chem. 1993;268:21569–21577. [PubMed] [Google Scholar]
- 44.Tinberg CE, Lippard SJ. Biochemistry. 2010;49:7902–7912. doi: 10.1021/bi1009375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallar BJ, Lipscomb JD. Biochemistry. 2001;40:2220–2233. doi: 10.1021/bi002298b. [DOI] [PubMed] [Google Scholar]
- 46.Brazeau BJ, Lipscomb JD. Biochemistry. 2003;42:5618–5631. doi: 10.1021/bi027429i. [DOI] [PubMed] [Google Scholar]
- 47.Fox BG, Froland WA, Dege JE, Lipscomb JD. J Biol Chem. 1989;264:10023–10033. [PubMed] [Google Scholar]
- 48.Colby J, Dalton H. Biochem J. 1978;171:461–468. doi: 10.1042/bj1710461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colby J, Dalton H. Biochem J. 1979;177:903–908. doi: 10.1042/bj1770903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller J, Lugovskoy AA, Wagner G, Lippard SJ. Biochemistry. 2002;41:42–51. doi: 10.1021/bi015668k. [DOI] [PubMed] [Google Scholar]
- 51.Chatwood LL, Müller J, Gross JD, Wagner G, Lippard SJ. Biochemistry. 2004;43:11983–11991. doi: 10.1021/bi049066n. [DOI] [PubMed] [Google Scholar]
- 52.Acheson JF, Bailey LJ, Elsen NL, Fox BG. Nat Commun. 2014;5:5009. doi: 10.1038/ncomms6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang W, Iacob RE, Luoh RP, Engen JR, Lippard SJ. J Am Chem Soc. 2014;136:9754–9762. doi: 10.1021/ja504688z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kopp DA, Berg EA, Costello CE, Lippard SJ. J Biol Chem. 2003;278:20939–20945. doi: 10.1074/jbc.M301581200. [DOI] [PubMed] [Google Scholar]
- 55.Fox BG, Liu Y, Dege JE, Lipscomb JD. J Biol Chem. 1991;266:540–550. [PubMed] [Google Scholar]
- 56.Liu Y, Nesheim JC, Paulsen KE, Stankovich MT, Lipscomb JD. Biochemistry. 1997;36:5223–5233. doi: 10.1021/bi962743w. [DOI] [PubMed] [Google Scholar]
- 57.Pulver SC, Froland WA, Lipscomb JD, Solomon EI. J Am Chem Soc. 1997;119:387–395. [Google Scholar]
- 58.Pulver SC, Tong WH, Bollinger JM, Jr, Stubbe J, Solomon EI. J Am Chem Soc. 1995;117:12664–12678. [Google Scholar]
- 59.Liu Y, Nesheim JC, Lee SK, Lipscomb JD. J Biol Chem. 1995;270:24662–24665. doi: 10.1074/jbc.270.42.24662. [DOI] [PubMed] [Google Scholar]
- 60.Wang W, Lippard SJ. J Am Chem Soc. 2014;136:2244–2247. doi: 10.1021/ja412351b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paulsen KE, Liu Y, Fox BG, Lipscomb JD, Münck E, Stankovich MT. Biochemistry. 1994;33:713–722. doi: 10.1021/bi00169a013. [DOI] [PubMed] [Google Scholar]
- 62.Liu KE, Lippard SJ. J Biol Chem. 1991;266(12836–12839):24859. [PubMed] [Google Scholar]
- 63.Brazeau BJ, Wallar BJ, Lipscomb JD. Biochem Biophys Res Commun. 2003;312:143–148. doi: 10.1016/j.bbrc.2003.09.242. [DOI] [PubMed] [Google Scholar]
- 64.Davydov R, Valentine AM, Komar-Panicucci S, Hoffman BM, Lippard SJ. Biochemistry. 1999;38:4188–4197. doi: 10.1021/bi982391o. [DOI] [PubMed] [Google Scholar]
- 65.DeWitt JG, Bentsen JG, Rosenzweig AC, Hedman B, Green J, Pilkington S, Papaefthymiou GC, Dalton H, Hodgson KO, Lippard SJ. J Am Chem Soc. 1991;113:9219–9235. [Google Scholar]
- 66.Fox BG, Surerus KK, Münck E, Lipscomb JD. J Biol Chem. 1988;263:10553–10556. [PubMed] [Google Scholar]
- 67.Fox BG, Hendrich MP, Surerus KK, Andersson KK, Froland WA, Lipscomb JD, Munck E. J Am Chem Soc. 1993;115:3688–3701. [Google Scholar]
- 68.Hendrich MP, Munck E, Fox BG, Lipscomb JD. J Am Chem Soc. 1990;112:5861–5865. [Google Scholar]
- 69.Lund J, Woodland MP, Dalton H. Eur J Biochem. 1985;147:297–305. doi: 10.1111/j.1432-1033.1985.tb08750.x. [DOI] [PubMed] [Google Scholar]
- 70.Gassner GT, Lippard SJ. Biochemistry. 1999;38:12768–12785. doi: 10.1021/bi990841m. [DOI] [PubMed] [Google Scholar]
- 71.Tinberg CE, Lippard SJ. Acc Chem Res. 2011;44:280–288. doi: 10.1021/ar1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brazeau BJ, Lipscomb JD. Biochemistry. 2000;39:13503–13515. doi: 10.1021/bi001473l. [DOI] [PubMed] [Google Scholar]
- 73.Lee S-K, Lipscomb JD. Biochemistry. 1999;38:4423–4432. doi: 10.1021/bi982712w. [DOI] [PubMed] [Google Scholar]
- 74.Tinberg CE, Lippard SJ. Biochemistry. 2009;48:12145–12158. doi: 10.1021/bi901672n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Banerjee R, Meier KK, Münck E, Lipscomb JD. Biochemistry. 2013;52:4331–4342. doi: 10.1021/bi400182y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skulan AJ, Brunold TC, Baldwin J, Saleh L, Bollinger JM, Solomon EI. J Am Chem Soc. 2004;126:8842–8855. doi: 10.1021/ja049106a. [DOI] [PubMed] [Google Scholar]
- 77.Han W-G, Noodleman L. Inorg Chem. 2008;47:2975–2986. doi: 10.1021/ic701194b. [DOI] [PubMed] [Google Scholar]
- 78.Andersson KK, Froland WA, Lee S-K, Lipscomb JD. New J Chem. 1991;15:411–415. [Google Scholar]
- 79.Jiang Y, Wilkins PC, Dalton H. Biochem Biophys Res Commun. 1993;1163:105–112. doi: 10.1016/0167-4838(93)90285-y. [DOI] [PubMed] [Google Scholar]
- 80.Lee S-K, Fox BG, Froland WA, Lipscomb JD, Münck E. J Am Chem Soc. 1993;115:6450–6451. [Google Scholar]
- 81.Liu KE, Valentine AM, Wang D, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. J Am Chem Soc. 1995;117:10174–10185. [Google Scholar]
- 82.Liu KE, Wang D, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. J Am Chem Soc. 1994;116:7465–7466. [Google Scholar]
- 83.Banerjee R, Proshlyakov Y, Lipscomb JD, Proshlyakov DA. Nature. 2015;518:431–434. doi: 10.1038/nature14160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang S-P, Shiota Y, Yoshizawa K. Dalton Trans. 2013;42:1011–1023. doi: 10.1039/c2dt31304a. [DOI] [PubMed] [Google Scholar]
- 85.Deighton N, Podmore ID, Symons MC, Wilkins PC, Dalton H. J Chem Soc Chem Commun. 1991;16:1086–1088. [Google Scholar]
- 86.Brazeau BJ, Austin RN, Tarr C, Groves JT, Lipscomb JD. J Am Chem Soc. 2001;123:11831–11837. doi: 10.1021/ja016376+. [DOI] [PubMed] [Google Scholar]
- 87.Wilkins PC, Dalton H, Podmore ID, Deighton N, Symons MCR. Eur J Biochem. 1992;210:67–72. doi: 10.1111/j.1432-1033.1992.tb17391.x. [DOI] [PubMed] [Google Scholar]
- 88.Dalton H, Wilkins PC, Deighton N, Podmore ID, Symons MCR. Faraday Discuss. 1992;93:163–171. [Google Scholar]
- 89.Jin Y, Lipscomb JD. Biochemistry. 1999;38:6178–6186. doi: 10.1021/bi990068v. [DOI] [PubMed] [Google Scholar]
- 90.Liu A, Jin Y, Zhang J, Brazeau BJ, Lipscomb JD. Biochem Biophys Res Commun. 2005;338:254–261. doi: 10.1016/j.bbrc.2005.08.216. [DOI] [PubMed] [Google Scholar]
- 91.Choi S-Y, Eaton PE, Kopp DA, Lippard SJ, Newcomb M, Shen R. J Am Chem Soc. 1999;121:12198–12199. [Google Scholar]
- 92.Baik M-H, Newcomb M, Friesner RA, Lippard SJ. Chem Rev. 2003;103:2385–2419. doi: 10.1021/cr950244f. [DOI] [PubMed] [Google Scholar]
- 93.Liu KE, Johnson CC, Newcomb M, Lippard SJ. J Am Chem Soc. 1993;115:939–947. [Google Scholar]
- 94.Valentine AM, LeTadic-Biadatti M-H, Toy PH, Newcomb M, Lippard SJ. J Biol Chem. 1999;274:10771–10776. doi: 10.1074/jbc.274.16.10771. [DOI] [PubMed] [Google Scholar]
- 95.Priestley ND, Floss HG, Froland WA, Lipscomb JD, Williams PG, Morimoto H. J Am Chem Soc. 1992;114:7561–7562. [Google Scholar]
- 96.Valentine AM, Wilkinson B, Liu KE, Komar-Panicucci S, Priestley ND, Williams PG, Morimoto H, Floss HG, Lippard SJ. J Am Chem Soc. 1997;119:1818–1827. [Google Scholar]
- 97.Gherman BF, Dunietz BD, Whittington DA, Lippard SJ, Friesner RA. J Am Chem Soc. 2001;123:3836–3837. doi: 10.1021/ja0055108. [DOI] [PubMed] [Google Scholar]
- 98.Guallar V, Gherman BF, Miller WH, Lippard SJ, Friesner RA. J Am Chem Soc. 2002;124:3377–3384. doi: 10.1021/ja0167248. [DOI] [PubMed] [Google Scholar]
- 99.Burrows KJ, Cornish A, Scott D, Higgins IJ. Microbiology. 1984;130:3327–3333. [Google Scholar]
- 100.Green J, Dalton H. J Biol Chem. 1989;264:17698–17703. [PubMed] [Google Scholar]
- 101.Colby J, Stirling DI, Dalton H. Biochem J. 1977;165:395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kopp DA, Lippard SJ. Curr Opin Chem Biol. 2002;6:568–576. doi: 10.1016/s1367-5931(02)00366-6. [DOI] [PubMed] [Google Scholar]
- 103.Smith SM, Rawat S, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. Biochemistry. 2011;50:10231–10240. doi: 10.1021/bi200801z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hakemian AS, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. Biochemistry. 2008;47:6793–6801. doi: 10.1021/bi800598h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sirajuddin S, Barupala D, Helling S, Marcus K, Stemmler TL, Rosenzweig AC. J Biol Chem. 2014;289:21782–21794. doi: 10.1074/jbc.M114.581363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Balasubramanian R, Rosenzweig AC. Acc Chem Res. 2007;40:573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- 107.Kitmitto A, Myronova N, Basu P, Dalton H. Biochemistry. 2005;44:10954–10965. doi: 10.1021/bi050820u. [DOI] [PubMed] [Google Scholar]
- 108.Lieberman RL, Kondapalli KC, Shrestha DB, Hakemian AS, Smith SM, Telser J, Kuzelka J, Gupta R, Borovik AS, Lippard SJ, Hoffman BM, Rosenzweig AC, Stemmler TL. Inorg Chem. 2006;45:8372–8381. doi: 10.1021/ic060739v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lawton TJ, Ham J, Sun T, Rosenzweig AC. Proteins Struct Funct Bioinf. 2014;82:2263–2267. doi: 10.1002/prot.24535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hakemian AS, Rosenzweig AC. Annu Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 111.Miller KW, Hammond L, Porter EG. Chem Phys Lipids. 1977;20:229–241. [Google Scholar]
- 112.Ng K-Y, Tu L-C, Wang Y-S, Chan SI, Yu SSF. ChemBio-Chem. 2008;9:1116–1123. doi: 10.1002/cbic.200700628. [DOI] [PubMed] [Google Scholar]
- 113.Miyaji A, Miyoshi T, Motokura K, Baba T. Biotechnol Lett. 2011;33:2241–2246. doi: 10.1007/s10529-011-0688-3. [DOI] [PubMed] [Google Scholar]
- 114.Sugimori D, Ando R, Okura I. Appl Biochem Biotechnol. 1995;53:199–205. [Google Scholar]
- 115.Martinho M, Choi DW, DiSpirito AA, Antholine WE, Semrau JD, Münck E. J Am Chem Soc. 2007;129:15783–15785. doi: 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chan SI, Wang VCC, Lai JCH, Yu SSF, Chen PPY, Chen KHC, Chen C-L, Chan MK. Angew Chem Int Ed. 2007;46:1992–1994. doi: 10.1002/anie.200604647. [DOI] [PubMed] [Google Scholar]
- 117.Pham MD, Lin YP, Vuong QV, Nagababu P, Chang BTA, Ng KY, Chen CH, Han CC, Chen CH, Li MS, Yu SSF, Chan SI. Biochim Biophys Acta. 2015;1854:1842–1852. doi: 10.1016/j.bbapap.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 118.Liew EF, Tong D, Coleman NV, Holmes AJ. Microbiology. 2014;160:1267–1277. doi: 10.1099/mic.0.078584-0. [DOI] [PubMed] [Google Scholar]
- 119.Zahn JA, DiSpirito AA. J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gilch S, Vogel M, Lorenz MW, Meyer O, Schmidt I. Microbiology. 2009;155:279–284. doi: 10.1099/mic.0.023721-0. [DOI] [PubMed] [Google Scholar]
- 121.Cook SA, Shiemke AK. J Inorg Biochem. 1996;63:273–284. [Google Scholar]
- 122.Prior SD, Dalton H. FEMS Microbiol Lett. 1985;29:105–109. [Google Scholar]
- 123.Culpepper MA, Cutsail GE, III, Gunderson WA, Hoffman BM, Rosenzweig AC. J Am Chem Soc. 2014;136:11767–11775. doi: 10.1021/ja5053126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nguyen HH, Shiemke AK, Jacobs SJ, Hales BJ, Lidstrom ME, Chan SI. J Biol Chem. 1994;269:14995–15005. [PubMed] [Google Scholar]
- 125.Chan SI, Chen KHC, Yu SSF, Chen C-L, Kuo SSJ. Biochemistry. 2004;43:4421–4430. doi: 10.1021/bi0497603. [DOI] [PubMed] [Google Scholar]
- 126.Lieberman RL, Rosenzweig AC. Dalton Trans. 2005;21:3390–3396. doi: 10.1039/b506651d. [DOI] [PubMed] [Google Scholar]
- 127.Blackburn NJ, Barr ME, Woodruff WH, van der Ooost J, de Vries S. Biochemistry. 1994;33:10401–10407. doi: 10.1021/bi00200a022. [DOI] [PubMed] [Google Scholar]
- 128.Solomon EI, Sundaram UM, Machonkin TE. Chem Rev. 1996;96:2563–2606. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 129.Xie X, Gorelsky SI, Sarangi R, Garner DK, Hwang HJ, Hodgson KO, Hedman B, Lu Y, Solomon EI. J Am Chem Soc. 2008;130:5194–5205. doi: 10.1021/ja7102668. [DOI] [PubMed] [Google Scholar]
- 130.Woolery G, Powers L, Peisach J, Spiro T. Biochemistry. 1984;23:3428–3434. doi: 10.1021/bi00310a008. [DOI] [PubMed] [Google Scholar]
- 131.Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen J-CN, Johansen KS, Krogh KBRM, Jørgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. Proc Natl Acad Sci. 2011;108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frandsen KEH, Simmons TJ, Dupree P, Poulsen J-CN, Hemsworth GR, Ciano L, Johnston EM, Tovborg M, Johansen KS, von Freiesleben P, Marmuse L, Fort S, Cottaz S, Driguez H, Henrissat B, Lenfant N, Tuna F, Baldansuren A, Davies GJ, Lo Leggio L, Walton PH. Nat Chem Biol. 2016;12:298–303. doi: 10.1038/nchembio.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Walton PH, Davies GJ. Curr Opin Chem Biol. 2016;31:195–207. doi: 10.1016/j.cbpa.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 134.Bauman AT, Broers BA, Kline CD, Blackburn NJ. Biochemistry. 2011;50:10819–10828. doi: 10.1021/bi201193j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Prigge ST, Eipper BA, Mains RE, Amzel LM. Science. 2004;304:864–867. doi: 10.1126/science.1094583. [DOI] [PubMed] [Google Scholar]
- 136.Klinman JP. J Biol Chem. 2006;281:3013–3016. doi: 10.1074/jbc.R500011200. [DOI] [PubMed] [Google Scholar]
- 137.Yoshizawa K, Shiota Y. J Am Chem Soc. 2006;128:9873–9881. doi: 10.1021/ja061604r. [DOI] [PubMed] [Google Scholar]
- 138.Shiota Y, Juhasz G, Yoshizawa K. Inorg Chem. 2013;52:7907–7917. doi: 10.1021/ic400417d. [DOI] [PubMed] [Google Scholar]
- 139.Shiota Y, Yoshizawa K. Inorg Chem. 2009;48:838–845. doi: 10.1021/ic8003933. [DOI] [PubMed] [Google Scholar]
- 140.Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber-Emmons MT, Kjaergaard CH, Hadt RG, Tian L. Chem Rev. 2014;114:3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kaim W, Rall J. Angew Chem Int Ed. 1996;35:43–60. [Google Scholar]
- 142.Crichton RR, Pierre J-L. Biometals. 2001;14:99–112. doi: 10.1023/a:1016710810701. [DOI] [PubMed] [Google Scholar]
- 143.Culpepper MA, Cutsail GE, III, Hoffman BM, Rosenzweig AC. J Am Chem Soc. 2012;134:7640–7643. doi: 10.1021/ja302195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kao W-C, Chen Y-R, Yi EC, Lee H, Tian Q, Wu K-M, Tsai S-F, Yu SS-F, Chen Y-J, Aebersold R, Chan SI. J Biol Chem. 2004;279:51554–51560. doi: 10.1074/jbc.M408013200. [DOI] [PubMed] [Google Scholar]
- 145.Chen KHC, Wu H-H, Ke S-F, Rao Y-T, Tu C-M, Chen Y-P, Kuei K-H, Chen Y-S, Wang VCC, Kao W-C, Chan SI. J Inorg Biochem. 2012;111:10–17. doi: 10.1016/j.jinorgbio.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 146.Itoyama S, Doitomi K, Kamachi T, Shiota Y, Yoshizawa K. Inorg Chem. 2016;55:2771–2775. doi: 10.1021/acs.inorgchem.5b02603. [DOI] [PubMed] [Google Scholar]
- 147.Blackburn NJ, Strange RW, Farooq A, Haka MS, Karlin KD. J Am Chem Soc. 1988;110:4263–4272. [Google Scholar]
- 148.Woertink JS, Smeets PJ, Groothaert MH, Vance MA, Sels BF, Schoonheydt RA, Solomon EI. Proc Natl Acad Sci. 2009;106:18908–18913. doi: 10.1073/pnas.0910461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Smeets PJ, Hadt RG, Woertink JS, Vanelderen P, Schoonheydt RA, Sels BF, Solomon EI. J Am Chem Soc. 2010;132:14736–14738. doi: 10.1021/ja106283u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Citek C, Gary JB, Wasinger EC, Stack TDP. J Am Chem Soc. 2015;137:6991–6994. doi: 10.1021/jacs.5b02157. [DOI] [PubMed] [Google Scholar]
- 151.Citek C, Lin B-L, Phelps TE, Wasinger EC, Stack TDP. J Am Chem Soc. 2014;136:14405–14408. doi: 10.1021/ja508630d. [DOI] [PubMed] [Google Scholar]
- 152.Chan SI, Lu YJ, Nagababu P, Maji S, Hung MC, Lee MM, Hsu IJ, Minh PD, Lai JCH, Ng KY. Angew Chem Int Ed. 2013;52:3731–3735. doi: 10.1002/anie.201209846. [DOI] [PubMed] [Google Scholar]
- 153.Grundner S, Markovits MAC, Li G, Tromp M, Pidko EA, Hensen EJM, Jentys A, Sanchez-Sanchez M, Lercher JA. Nat Commun. 2015;6:7546. doi: 10.1038/ncomms8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chan SI, Chien CYC, Yu CSC, Nagababu P, Maji S, Chen PPY. J Catal. 2012;293:186–194. [Google Scholar]
- 155.Nagababu P, Maji S, Kumar MP, Chen PPY, Yu SSF, Chan SI. Adv Synth Catal. 2012;354:3275–3282. [Google Scholar]
- 156.Liu CC, Mou CY, Yu SSF, Chan SI. Energy Environ Sci. 2016;9:1361–1374. [Google Scholar]
- 157.Basu P, Katterle B, Andersson KK, Dalton H. Biochem J. 2003;369:417–427. doi: 10.1042/BJ20020823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yuan H, Collins MLP, Antholine WE. J Am Chem Soc. 1997;119:5073–5074. [Google Scholar]
- 159.Yuan H, Collins MLP, Antholine WE. J Inorg Biochem. 1998;72:179–185. doi: 10.1016/s0162-0134(98)10078-8. [DOI] [PubMed] [Google Scholar]
- 160.Wilkinson B, Zhu M, Priestley ND, Nguyen HHT, Morimoto H, Williams PG, Chan SI, Floss HG. J Am Chem Soc. 1996;118:921–922. [Google Scholar]
- 161.Elliott SJ, Zhu M, Tso L, Nguyen HHT, Yip JHK, Chan SI. J Am Chem Soc. 1997;119:9949–9955. [Google Scholar]
- 162.Ono M, Okura I. J Mol Catal. 1990;61:113–122. [Google Scholar]
- 163.Drummond S, Dalton H. Eur J Biochem. 1989;182:667–671. doi: 10.1111/j.1432-1033.1989.tb14877.x. [DOI] [PubMed] [Google Scholar]
- 164.Shiemke AK, Cook SA, Miley T, Singleton P. Arch Biochem Biophys. 1995;321:421–428. doi: 10.1006/abbi.1995.1413. [DOI] [PubMed] [Google Scholar]
- 165.Cornish A, MacDonald J, Burrows KJ, King TS, Scott D, Higgins IJ. Biotechnol Lett. 1985;7:319–324. [Google Scholar]
- 166.Farrar JA, Neese F, Lappalainen P, Kroneck PMH, Saraste M, Zumft WG, Thomson AJ. J Am Chem Soc. 1996;118:11501–11514. [Google Scholar]
- 167.Suzuki S, Kohzuma T, Deligeer A, Yamaguchi K, Nakamura N, Shidara S, Kobayashi K, Tagawa S. J Am Chem Soc. 1994;116:11145–11146. [Google Scholar]
- 168.Pelletier H, Kraut J. Science. 1992;258:1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 169.Tonge GM, Harrison DEF, Knowles CJ, Higgins IJ. FEBS Lett. 1975;58:293–299. doi: 10.1016/0014-5793(75)80282-1. [DOI] [PubMed] [Google Scholar]
- 170.Leak DJ, Dalton H. Appl Microbiol Biotechnol. 1986;23:477–481. [Google Scholar]
- 171.Myronova N, Kitmitto A, Collins RF, Miyaji A, Dalton H. Biochemistry. 2006;45:11905–11914. doi: 10.1021/bi061294p. [DOI] [PubMed] [Google Scholar]
- 172.Anthony C. The biochemistry of methylotrophs. Academic Press; New York: 1982. [Google Scholar]
- 173.Culpepper MA, Rosenzweig AC. Biochemistry. 2014;53:6211–6219. doi: 10.1021/bi500850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.de la Torre A, Metivier A, Chu F, Laurens LML, Beck DAC, Pienkos PT, Lidstrom ME, Kalyuzhnaya MG. Microb Cell Fact. 2015;14:188. doi: 10.1186/s12934-015-0377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cook SA, Shiemke AK. Arch Biochem Biophys. 2002;398:32–40. doi: 10.1006/abbi.2001.2628. [DOI] [PubMed] [Google Scholar]
- 176.Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han JI, Zahn JA, Boyd JM, de la Mora AM, DiSpirito AA. J Bacteriol. 2003;185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Page CC, Moser CC, Chen X, Dutton PL. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]