Abstract
The platelet-derived growth factor receptors alpha and beta (PDGFRα and PDGFRβ) mark fibroadipogenic progenitor cells/fibroblasts and pericytes in skeletal muscle, respectively. While the role that these cells play in muscle growth and development has been evaluated, it was not known whether the PDGF receptors activate signaling pathways that control transcriptional and functional changes during skeletal muscle hypertrophy. To evaluate this, we inhibited PDGFR signaling in mice subjected to a synergist ablation muscle growth procedure, and performed analyses 3 and 10 days after induction of hypertrophy. The results from this study indicate that PDGF signaling is required for fiber hypertrophy, ECM production and angiogenesis that occur during muscle growth.
Keywords: skeletal muscle, platelet-derived growth factor, extracellular matrix
Introduction
Skeletal muscle displays a remarkable ability to adapt to increased mechanical loads by undergoing hypertrophy. This adaptation involves a coordinated response of the muscle fibers that generate force, the extracellular matrix which transmits force between the skeleton and muscle fibers, and the vasculature which brings in nutrients and removes waste from the tissue [1–3]. In addition to muscle fibers, numerous other cell types, including satellite cells, fibroblasts, perivascular cells, and immune cells, among others, play important roles in the adaptation of muscle to growth signals or in its recovery following injury [1–3]. A variety of growth factors and signaling molecules direct the activities of these cells. One family of growth factors that are enriched in skeletal muscle are the platelet-derived growth factors (PDGFs) [4]. The PDGFs typically exist as hetero- or homodimers composed of four different monomers (A-D), with PDGF-BB among the most frequently occurring dimer [5]. The PDGFs activate a class of receptor tyrosine kinase transmembrane proteins, referred to as the PDGF receptors α (PDGFRα) and β (PDGFRβ) [5]. Within skeletal muscle tissue, PDGFRα is expressed on a population of cells referred to as either fibroadipogenic precursor cells or fibroblasts, while PDGFRβ is expressed on perivascular cells, which act as pluripotent stem cells [6–8]. Although PDGFs are expressed in skeletal muscle tissue, and the receptors for these proteins are located on cells that play important roles in muscle growth and remodeling, to our knowledge the specific role that PDGF signaling plays in skeletal muscle hypertrophy has not previously been explored in detail.
To investigate how PDGFR signaling impacts postnatal skeletal muscle growth, we treated mice that underwent a synergist ablation procedure to induce muscle hypertrophy with a potent and specific inhibitor of PDGFR phosphorylation, CP-673,451 [9,10]. We hypothesized that pharmacologic inhibition of PDGFR signaling would reduce the extent of muscle hypertrophy, as measured by decreased muscle mass and fiber cross-sectional area. We further hypothesized that blocking PDGFRs would reduce ECM synthesis and the formation of capillaries which typically occur during muscle hypertrophy.
Materials and Methods
Animals
All animal work was approved by the University of Michigan Institutional Animal Care & Use Committee. Wild type C57BL/6 mice, and transgenic PDGFRαEGFP/+ mice in a C57BL/6 background, were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). PDGFRαEGFP/+ mice express the H2B-eGFP reporter gene driven from the endogenous PDGFRα locus [11]. Four month old male mice were used in experiments.
Synergist Ablation
Wild type mice were randomized to 3- and 10-day groups. A tenectomy of the Achilles tendon was performed to prevent the gastrocnemius and soleus muscles from plantarflexing the talocrural joint, resulting in compensatory hypertrophy of the synergist plantaris muscle. Bilateral synergist ablation surgeries were performed under isoflurane anesthesia as previously described [12–14]. Subcutaneous buprenorphine was provided for analgesia. After surgeries, ad libitum weight-bearing and cage activity were allowed in the postoperative period. Within each time point, half of the mice were treated with vehicle or CP-673,451 (N=8; Biorbyt, Cambridge, UK), a selective inhibitor of PDGFRα and PDGFRβ [9,10], dissolved first in 2 parts dimethyl sulfoxide and then in 8 parts phosphate buffered saline. CP-673,451 was administered via intraperitoneal injection at 15 mg/kg twice daily, with the total daily dose being 30mg/kg, starting one day prior to synergist ablation and continued each day until harvest. This value was selected based on a prior study which indicated that a total daily dose of 30mg/kg of CP-673,451 was effective at blocking PDGFR phosphorylation in mice [9]. Mice were closely monitored for any adverse reactions. At harvest, mice were anesthetized with isoflurane and left plantaris muscles were collected for gene expression analysis while right plantaris muscles were used for immunohistochemistry. After the muscles were removed, mice were euthanized by cervical dislocation and induction of bilateral pneumothorax. Plantaris muscles from additional non-overloaded C57BL/6 mice were obtained as described above for gene expression analysis, and also from PDGFRαEGFP/+ mice for histology.
Quantitative RT-PCR
Gene expression was conducted as previously described [13,15]. Muscles were homogenized in QIAzol (Qiagen, Valencia, CA, USA) and RNA was isolated using a miRNeasy Micro Kit (Qiagen) supplemented with DNase I (Qiagen). RNA was reverse transcribed into cDNA with an iScript Reverse Transcription Supermix (Bio-Rad, Hercules, CA, USA). Amplification of cDNA was performed in a CFX96 real-time thermal cycler (Bio-Rad) using iTaq Universal SYBR Green Supermix (Bio-Rad). Target gene expression was normalized to the stable housekeeping gene Hspb6, and further normalized to muscles that were not subjected to synergist ablation using the 2−ddCt method. Primer sequences are provided in Supplementary Table 1.
Microarray
Microarray measurements were performed by the University of Michigan DNA Sequencing Core as previously described [16]. Equal amounts of RNA isolated from four individual muscles were pooled into a single sample for microarray analysis, and two pooled samples from each group were analyzed. RNA was pooled because gene expression from a pooled sample is similar to the average of the individual samples composing the pooled sample [17,18]. Biotinylated cDNA was prepared using the GeneChip WT PLUS Reagent Kit (Affymetrix, Santa Clara, CA, USA) and hybridized to Mouse Gene 2.1 ST Array Strips (Affymetrix). Raw microarray data were loaded into ArrayStar version 12.1 (DNASTAR, Madison, WI, USA) to calculate fold changes in gene expression. The microarray dataset is available through the NIH Gene Expression Omnibus database (accession number GSE89111).
Immunohistochemistry
Immunohistochemistry was performed as described [13,15]. Plantaris muscles were placed in a 30% sucrose solution for one hour, snap frozen in Tissue-Tek OCT Compound (Sakura Finetek, Torrance, CA, USA) and stored at −80°C. Muscles were sectioned at a thickness of 10 μm in a cryostat, and tissue sections were then fixed in methanol, permeabilized in 0.4% Triton X-100 and blocked with 5% donkey serum. For identifying cell types in muscle, slides from PDGFRαEGFP/+ mice were incubated with primary antibodies consisting of rabbit anti-PDGFRβ (1:100; SC-339, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and rat anti-laminin α-2 (1:200; sc-59854, Santa Cruz Biotechnology). To quantify muscle fiber size and blood vessel density, slides from C57BL/6 mice were incubated with rat anti-CD31 (1:100; 550274, BD Pharmingen, San Diego, CA, USA). The extracellular matrix was identified with wheat germ agglutinin (WGA) lectin conjugated to AlexaFluor 488 (1:1000; W11261, ThermoFisher, Grand Island, NY, USA). AlexaFluor-conjugated secondary antibodies (AF488, AF555, AF647, Life Technologies, Carlsbad, CA, USA), were used to detect primary antibodies. Nuclei were identified with DAPI (Life Technologies). Images were captured with a Nikon A1 confocal microscope (Nikon Instruments, Tokyo, JP). ImageJ software (NIH, Bethesda, MD) was used to quantify muscle fiber cross-sectional area and capillary density from the entire cross-section of muscles. Quantification was performed by study personnel in a blinded fashion.
Immunoblots
Protein concentration of samples were determined with a Pierce BCA Protein Assay Kit (ThermoFisher Scientific). Protein homogenates were diluted in Laemmli’s sample buffer, boiled for two minutes and 100 μg of protein was separated on a 6% SDS-PAGE gel. Proteins were transferred to 0.45-μm nitrocellulose membranes (Bio-Rad) using the Trans-Blot SD semi-dry transfer apparatus (Bio-Rad), blocked with 5% non-fat powdered milk in T-TBS solution and incubated with primary rabbit antibodies (1:1000, Cell Signaling Technology, Danvers, MA, USA) against p-PDGFRαY849/p-PDGFRβY857 (#3170), PDGFRα (#3174), PDGFRβ (#3169), p-AktT308 (#13038), p-AktS473 (#4060), Akt (#4691), p-p70S6KT389 (#9234), p-p70S6KT421/S424 (#9204), and p70S6K (#2708). β-tubulin (ab6046, rabbit, Abcam, Cambridge, MA, USA) was used as a loading control. After primary antibody incubation, membranes were rinsed and incubated with HRP-conjugated goat anti-rabbit secondary antibodies (1:10,000; ab97051, Abcam, Cambridge, MA, USA). Proteins were detected using enhanced chemiluminescent reagents (Bio-Rad) and visualized using a digital chemiluminescent documentation system (Bio-Rad).
Statistics
Results are presented as mean±SD. Prism version 7.0 (GraphPad Software, La Jolla, CA) was used to conduct analyses. A two-way ANOVA (α=0.05) followed by Holm-Sidak post hoc sorting evaluated the interaction between time after synergist ablation and CP-673,451 treatment.
Results
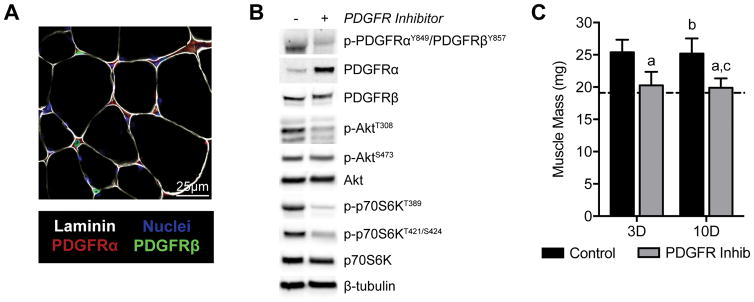
As other studies have reported, PDGFRα+ cells were observed adjacent to the muscle fiber basal lamina, while PDGFRβ+ cells existed around capillaries between muscle fibers (Figure 1A). We next sought to determine whether CP-673,451 could prevent phosphorylation of the PDGFRα and PDGFRβ receptors in vivo. Indeed, CP-673,451 reduced PDGFRα/β phosphorylation in response to synergist ablation (Figure 1B). We also evaluated activation of the Akt-p70S6K pathway, which plays a central role in regulating protein synthesis in skeletal muscle. Phosphorylation of Akt at the T308 residue was negatively affected by PDGFR inhibition, while phosphorylation at the S473 residue did not appear to be different between groups. p70S6K phosphorylation at T389 and T421/S424 were also reduced by treatment with the PDGFR inhibitor.
Figure 1. Muscle mass and inhibition of PDGFR signaling.
(A) Representative histology demonstrating PDGFRα and PDGFRβ localization in skeletal muscle tissue. (B) Representative western blots from 3D overloaded muscles demonstrating the efficacy of the PDGFR inhibitor in preventing PDGFRα, PDGFRβ, Akt, and p70S6K phosphorylation. (C) Changes in muscle mass during hypertrophy in control or PDGFR inhibitor treated mice. Values are mean±SD. N=8 per group. Dished line indicates data from non-overloaded mice. Differences tested with a two-way ANOVA followed by Holm-Sidak post-hoc sorting. a, significantly different (P<0.05) from 3D control; b, significantly different (P<0.05) from 3D PDGFR inhibitor; c, significantly different (P<0.05) from 10D control.
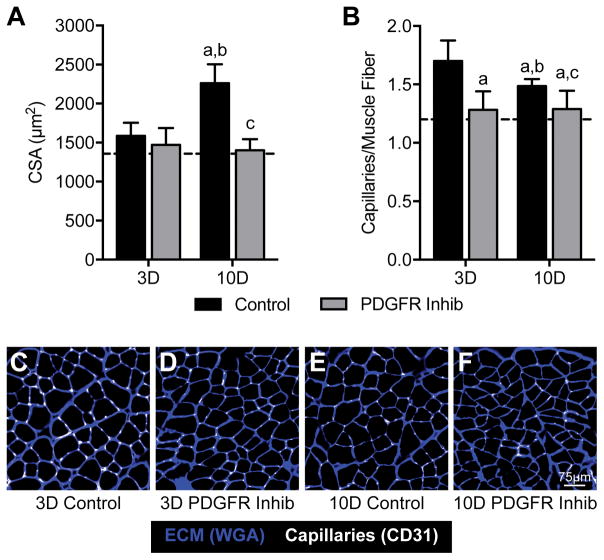
Changes in muscle mass were consistent with the observed differences in Akt/p70S6K activation. While vehicle treated mice had an approximately 25% increase in muscle mass, the PDGFR inhibitor mice did not experience an increase in muscle mass at the 3- or 10-day time points (Figure 1C). A robust 43% increase in muscle fiber CSA was observed in control mice 10 days after synergist ablation, but inhibition of PDGFR signaling prevented muscle fiber hypertrophy between 3 and 10 days (Figure 2A), which was consistent with the muscle mass findings. Capillary density followed a similar trend to muscle mass and muscle fiber CSA (Figure 2B). Representative images are shown in Figures 2C–F.
Figure 2. Muscle cross-sectional area and capillary density.
Changes in (A) muscle cross-sectional area and (B) capillary density in control or PDGFR inhibitor treated mice. (C–F) representative immunohistochemistry for extracellular matrix and capillaries is shown. Values are mean±SD. N=8 per group. Dashed line indicates data from non-overloaded mice. Differences tested with a two-way ANOVA followed by Holm-Sidak post-hoc sorting. a, significantly different (P<0.05) from 3D control; b, significantly different (P<0.05) from 3D PDGFR inhibitor; c, significantly different (P<0.05) from 10D control.
As inhibition of PDGFR signaling resulted in a muscle growth phenotype, we then sought to further explore the mechanisms behind this response by conducting microarray analysis. Microarray experiments identified that approximately one-quarter of the genome was differentially expressed in both 3-day groups, with slight differences between control and treatment animals (Figure 3). By 10 days, the overall number of differentially expressed transcripts was less than 3 days, and there was a more pronounced reduction in the PDGFR inhibited group than in control mice (Figure 3).
Figure 3. Microarray summary.
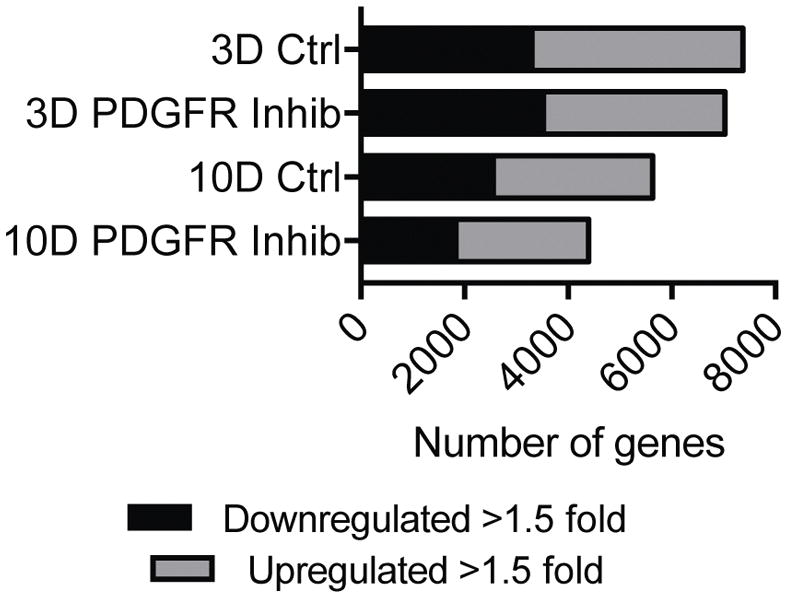
Number of genes that are downregulated greater than 1.5-fold or upregulated greater than 1.5-fold compared to non-overloaded muscles for each time point and treatment.
We then followed up on genes of interest identified from microarray results with qPCR analysis (Table 1). CD248, which is a C-type lectin domain protein important for angiogenesis, was downregulated in the PDGF inhibitor group by 63% at 3 days and 52% at 10 days. For genes involved with atrophy and myogenesis, there was a 33% increase in the E3-ubiquitin ligase Atrogin-1 in the 3-day treatment group, but this was downregulated by 40% at 10 days. The myoblast fusion gene myomaker (Tmem8c) was 61% lower in treated muscles at 3 days. Numerous extracellular matrix synthesis and remodeling genes were affected by PDGFR inhibitor treatment. Type I collagen expression was reduced by over 97% in treated mice compared to controls at each time point, while type III collagen was reduced by 71% in the 10 day treatment mice. The basement membrane collagens, type IV–VI, were at least 70% lower in the 10 day treatment mice compared to 10 day controls, and elastin expression was also reduced by 52% in these mice. MMP8, which is a collagenase, was upregulated 4-fold at 3 days and by 18-fold at 10 days in inhibitor treated mice. The gelatinase MMP9 was also elevated by 6-fold at 10 days in treated mice, while the membrane tethered MMP14 was downregulated by 78% in these mice. FSP-1, which is a marker of fibroblasts, was reduced by 62% in 3-day inhibitor treated mice, while FAP, which is expressed in activated fibroblasts, was downregulated by 53% in 10 day inhibitor treated mice. Finally, for markers of immune cells, CD11b which is expressed in M1 macrophages, and CD206 which is expressed in M2 macrophages, were downregulated by approximately 50% in PDGFR inhibitor treated mice at 3 days compared to vehicle treated controls.
Table 1. Gene expression values from vehicle and PDGFR inhibitor treated muscles.
| Gene | 3 day control | 3 day treatment | 10 day control | 10 day treatment |
|---|---|---|---|---|
| Angiogenesis | ||||
| Cd248 | 4.3±2.0 | 1.6±0.6a | 3.1±1.3b | 1.5±0.2a,c |
| Pecam1 | 1.5±0.4 | 1.3±0.4 | 1.6±0.5 | 1.2±0.2 |
| Tsp1 | 9.6±4.3 | 9.4±3.3 | 2.5±0.7a,b | 3.4±1.0a,b |
| Vegfa | 0.6±0.2 | 0.6±0.3 | 0.7±0.6 | 0.4±0.2 |
| Atrophy and Myogenesis | ||||
| Fboxo30 | 1.2±0.4 | 1.1±0.3 | 1.2±0.4 | 0.8±0.1 |
| Fboxo32 | 0.3±0.1 | 0.4±0.2a | 0.5±0.1a | 0.3±0.1b,c |
| Gdf11 | 1.1±0.1 | 0.7±0.1 | 2.3±0.7a,b | 1.0±0.2c |
| Tmem8c | 38.3±10.6 | 15.1±15.0a | 14.4±7.1a | 5.3±2.4a |
| Trim63 | 1.2±0.3 | 1.0±0.2 | 0.9±0.3 | 0.6±0.1a,b |
| Extracellular Matrix | ||||
| Col1a1 | 8.6±4.6 | 0.2±0.4a | 14.6±10.1b | 0.4±0.8a,c |
| Col3a1 | 7.6±3.6 | 3.4±1.5 | 8.7±5.4b | 2.5±0.6a,c |
| Col4a1 | 3.1±1.2 | 1.9±0.7 | 3.6±1.4b | 1.1±0.2a,c |
| Col5a1 | 7.8±4.3 | 3.1±1.5a | 8.2±5.0b | 1.7±0.4a,c |
| Col6a1 | 3.7±1.6 | 1.6±0.8 | 5.1±2.8b | 1.2±0.2a,c |
| Eln | 3.6±0.7 | 2.9±1.4 | 4.6±1.8 | 2.2±0.4c |
| Has1 | 47.5±40.4 | 52.4±33.7 | 8.1±7.1a,b | 3.7±1.3a,b |
| Has2 | 4.9±4.0 | 4.1±1.9 | 2.7±1.6 | 1.1±0.2a |
| Hyal1 | 0.9±0.3 | 0.7±0.2 | 1.8±0.5a,b | 0.8±0.1c |
| Lox | 26.8±13.9 | 12.6±6.3a | 6.0±4.3a | 3.0±0.4a |
| Mmp2 | 1.2±0.4 | 0.9±0.2 | 4.1±2.3a,b | 1.4±0.3c |
| Mmp3 | 13.6±7.1 | 9.8±6.9 | 19.5±11.8 | 17.9±4.3 |
| Mmp8 | 5.6±5.3 | 23.9±10.2a | 1.2±0.5b | 22.2±5.0a,c |
| Mmp9 | 4.6±5.4 | 4.4±1.4 | 1.0±0.4 | 6.0±0.9c |
| Mmp13 | 5.1±3.6 | 2.2±1.0 | 4.1±2.4 | 2.5±1.6 |
| Mmp14 | 6.0±3.6 | 2.8±1.2 | 11.7±10.2b | 2.6±0.8c |
| Timp1 | 143.0±75.1 | 65.6±41.6a | 14.1±8.7a | 7.7±1.2a |
| Timp2 | 2.0±0.9 | 1.2±0.4 | 2.8±1.3b | 1.2±0.3c |
| Fibroblast Markers | ||||
| Fap | 1.2±0.4 | 0.9±0.2 | 1.7±0.6b | 0.8±0.1c |
| Fsp1 | 11.7±6.4 | 4.4±2.4a | 5.0±2.4a | 2.5±0.9a |
| Immune Cell Markers | ||||
| Cd11b | 15.9±7.3 | 8.3±4.8a | 3.0±1.1a | 1.9±0.4a,b |
| Cd68 | 23.3±7.3 | 24.8±5.1 | 3.1±1.6a,b | 1.7±0.4a,b |
| Cd163 | 4.9±2.3 | 3.3±1.4 | 2.5±1.2a | 1.5±0.2a |
| Cd206 | 12.1±5.5 | 6.8±3.9a | 4.2±2.0a | 2.0±0.5a |
| F480 | 0.9±0.1 | 0.8±0.1 | 1.3±0.3a,b | 1.0±0.1 |
| Ly6c | 1.3±0.4 | 1.1±0.2 | 1.6±0.6 | 1.0±0.1 |
Target gene expression was normalized to the stable housekeeping gene Hspb6, and further normalized to muscles that were not subjected to synergist ablation. Values are mean±SD, N = 6–8 muscles for each group. Differences between groups were tested using a two-way ANOVA (α = 0.05) followed by Holm-Sidak post hoc sorting:
different from 3 day control;
different from 3 day PDGFR inhibitor treatment;
different from 10 day control.
Discussion
The PDGFRs, α and β, are expressed on cells that play important roles in muscle hypertrophy and regeneration. PDGFRα+ cells are thought to be involved in extracellular matrix synthesis and remodeling [6,8,19,20], while PDGFRβ+ cells function as multipotent progenitor cells that are enriched in the vasculature [7,21]. There are also multiple spliced isoforms of PDGFRα with different polyadenylation sites, including a variant that encodes an isoform containing a truncated kinase domain which acts as a decoy to inhibit PDGF signaling and inhibit fibroblast activity [22]. Ligands for the PDGFRs, the PDGFs, are present in skeletal muscle and appear to be induced during muscle regeneration [4,23,24]. While the expression of the two PDGFRs marks two distinct cell types that are critical for muscle growth and regeneration, the role that PDGF signaling played in skeletal muscle hypertrophy was not known. By using a specific inhibitor of the PDGFRs in a mouse model of skeletal muscle growth, we report that PDGFR signaling is important for proper skeletal muscle tissue hypertrophy.
The rodent synergist ablation model has frequently been used in the study of skeletal muscle hypertrophy. In this model, by removing the contribution of the gastrocnemius and soleus muscles to ankle plantarflexion, the synergist plantaris muscle undergoes compensatory growth. This growth is typically accompanied by increased muscle mass and fiber CSA, ECM accumulation, and growth of new capillaries [12,14,17,25,26]. The PDGFRα+ cell population, which also express the transcription factor Tcf4, plays an important role in muscle growth by regulating the activity of satellite cells and the synthesis of new myofibrillar proteins in muscle fibers [27]. Ablation of this cell population also prevents muscle fiber hypertrophy, likely through the inhibition of satellite cell expansion and the induction of premature myoblast differentiation [20]. In the current study, inhibition of PDGFR signaling prevented muscle hypertrophy and ECM accumulation, which is similar to the observed changes in muscles that lack PDGFRα+ cells. PDGFRβ+ cells exist in capillaries and provide a source of progenitor cells for capillary growth and for other cell populations, including myogenic cells [21,28,29]. CD248, also known as endosialin, is expressed on pericytes and is positively associated with angiogenesis in skeletal muscle [30], and was markedly reduced by blocking PDGFR signaling. The reduced capillary density values and smaller muscle fibers observed in the PDGFR inhibited treatment group are consistent with a role of reduced PDGFRβ+ cell activity that was observed in previous studies. While PDGFR inhibition did not have a striking effect on many genes involved with myogenesis and atrophy, myomaker, which is a protein that promotes myoblast fusion [31], was downregulated in PDGF treated muscles. PDGFs have also been posited to be involved in the recruitment of immune cells to muscle to help participate in muscle regeneration [32]. Although some modest effects were observed for macrophage markers, we did not observe a substantial outcome of PDGFR inhibition on the expression of markers of different types of immune cells in overloaded muscles. Combined, it appears not only that various cell populations in skeletal muscle express PDGFRs, activation of these receptors is required for proper skeletal muscle growth.
The Akt-p70S6K pathway plays a central role in regulating skeletal muscle growth in large party by activating mRNA translation [33]. In the current study, inhibition of PDGFR signaling blocked phosphorylation of important regulatory sites on Akt and p70S6K. The T308 site in Akt is phosphorylated by PDK1, while the S473 site is phosphorylated by the mTORC2 complex, and phosphorylation of both sites is required for the full activation of Akt and subsequent activation of mTOR [34]. p70S6K, which phosphorylates several proteins involved in the initiation of translation, is activated downstream of Akt, chiefly via mTOR-mediated phosphorylation of the T389 residue of p70S6K [35]. Various upstream kinases can also phosphorylate the T421 and S424 residues, which lead to conformational changes that enhance the activation of p70S6K [35], and p70S6K activation is required for proper skeletal muscle hypertrophy [36]. The inhibition of muscle growth in the PDGFR inhibitor treated animals corresponds well with observed changes in phosphorylation of Akt and p70S6K, and the observed effects of PDGFR inhibition on Akt activation appear to be mediated by PDK1. Several receptor tyrosine kinases, including the PDGFRs, as well as mechanical stimuli, can activate the Akt-p70S6K pathway [37,38]. However, the PDGFRs are not expressed in skeletal muscle fibers, and based on the size and abundance of muscle fibers compared to PDGFRα+ and PDGFRβ+ cells, the vast majority of proteins in the Akt-p70S6K pathway in muscle tissue would be found within fibers. The effect of PDGFR inhibition on activation of the Akt-p70S6K pathway in muscle fibers is therefore likely to be indirect. The Tcf4+/PDGFRα+ population of fibroadipogenic precursor cells play an important role in muscle hypertrophy by indirectly regulating the synthesis of sarcomeric proteins [27], and although this is speculative, PDGFR signaling may be important role in fibroadipogenic precursor cell-mediated muscle hypertrophy.
There are several limitations to this short report. We used a pharmacological inhibitor of both PDGFRα and PDGFRβ, and were not able to selectively inhibit each receptor. While muscle morphology was assessed, muscle contractility and other functional assays were not measured. Histology sections were taken from the midportion of muscles, but quantifying capillaries in cross-section may not be reflective of changes throughout the muscle. We only analyzed two time points after synergist ablation, and additional insight could be gained from longer term experiments. As microarray data were pooled, and the number of pooled groups was low, we only calculated fold-change values in our analysis of array data. Finally, although we measured changes in the transcriptome using microarrays and qPCR, other than measuring protein phosphorylation we did not directly measure protein abundance, and changes in the transcriptome might not be reflected in the proteome.
Skeletal muscle hypertrophy is a complex process that involves the coordinated activity of muscle fibers, fibroblasts, stem cells, and immune cells to adapt to increased mechanical loads placed upon the tissue. Several elegant studies have demonstrated the requirement for cells that express PDGFRα or PDGFRβ in muscle growth and regeneration [7,8,19,20]. This study provided the first evidence that PDGFR signaling is important for muscle fiber hypertrophy and ECM production in growing skeletal muscle tissue in vivo. Future studies which use targeted inactivation of each PDGFR in specific cell types would provide additional insight into the role of PDGFR signaling in skeletal muscle hypertrophy. Additionally, small molecule inhibitors of PDGFR signaling are used for the treatment of acute myeloid leukemia and other types of other cancer [39], and the work from this study suggests that these drugs might have unwanted side effects in skeletal muscle tissue.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIAMS grants R01-AR063649 and F32-AR067086.
References
- 1.Gumucio JP, Sugg KB, Mendias CL. TGF-β Superfamily Signaling in Muscle and Tendon Adaptation to Resistance Exercise. Exerc Sport Sci Rev. 2015;43:93–99. doi: 10.1249/JES.0000000000000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mounier R, Chrétien F, Chazaud B. Blood vessels and the satellite cell niche. Curr Top Dev Biol. 2011;96:121–138. doi: 10.1016/B978-0-12-385940-2.00005-X. [DOI] [PubMed] [Google Scholar]
- 3.Kjaer M, Magnusson P, Krogsgaard M, Boysen Møller J, Olesen J, Heinemeier K, Hansen M, Haraldsson B, Koskinen S, Esmarck B, Langberg H. Extracellular matrix adaptation of tendon and skeletal muscle to exercise. J Anat. 2006;208:445–450. doi: 10.1111/j.1469-7580.2006.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen G, Birnbaum RS, Yablonka-Reuveni Z, Quinn LS. Separation of mouse crushed muscle extract into distinct mitogenic activities by heparin affinity chromatography. J Cell Physiol. 1994;160:563–572. doi: 10.1002/jcp.1041600320. [DOI] [PubMed] [Google Scholar]
- 5.Breitkopf K, Roeyen CV, Sawitza I, Wickert L, Floege J, Gressner AM. Expression patterns of PDGF-A, -B, -C and -D and the PDGF-receptors alpha and beta in activated rat hepatic stellate cells (HSC) Cytokine. 2005;31:349–357. doi: 10.1016/j.cyto.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Joe AWB, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FMV. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birbrair A, Zhang T, Wang Z-M, Messi ML, Mintz A, Delbono O. Pericytes at the intersection between tissue regeneration and pathology. Clin Sci. 2015;128:81–93. doi: 10.1042/CS20140278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uezumi A, Fukada S, Yamamoto N, Ikemoto-Uezumi M, Nakatani M, Morita M, Yamaguchi A, Yamada H, Nishino I, Hamada Y, Tsuchida K. Identification and characterization of PDGFRα+ mesenchymal progenitors in human skeletal muscle. Cell Death Dis. 2014;5:e1186. doi: 10.1038/cddis.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts WG, Whalen PM, Soderstrom E, Moraski G, Lyssikatos JP, Wang H-F, Cooper B, Baker DA, Savage D, Dalvie D, Atherton JA, Ralston S, Szewc R, Kath JC, Lin J, Soderstrom C, Tkalcevic G, Cohen BD, Pollack V, Barth W, Hungerford W, Ung E. Antiangiogenic and antitumor activity of a selective PDGFR tyrosine kinase inhibitor, CP-673,451. Cancer Res. 2005;65:957–966. [PubMed] [Google Scholar]
- 10.Ehnman M, Missiaglia E, Folestad E, Selfe J, Strell C, Thway K, Brodin B, Pietras K, Shipley J, Östman A, Eriksson U. Distinct effects of ligand-induced PDGFRα and PDGFRβ signaling in the human rhabdomyosarcoma tumor cell and stroma cell compartments. Cancer Res. 2013;73:2139–2149. doi: 10.1158/0008-5472.CAN-12-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calve S, Isaac J, Gumucio JP, Mendias CL. Hyaluronic acid, HAS1, and HAS2 are significantly upregulated during muscle hypertrophy. AJP - Cell Physiology. 2012;303:C577–88. doi: 10.1152/ajpcell.00057.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gumucio JP, Phan AC, Ruehlmann DG, Noah AC, Mendias CL. Synergist ablation induces rapid tendon growth through the synthesis of a neotendon matrix. J Appl Physiol. 2014;117:1287–1291. doi: 10.1152/japplphysiol.00720.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendias CL, Schwartz AJ, Grekin JA, Gumucio JP, Sugg KB. Changes in muscle fiber contractility and extracellular matrix production during skeletal muscle hypertrophy. J Appl Physiol. 2016 doi: 10.1152/japplphysiol.00719.2016. jap.00719.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gumucio JP, Flood MD, Phan AC, Brooks SV, Mendias CL. Targeted inhibition of TGF-β results in an initial improvement but long-term deficit in force production after contraction-induced skeletal muscle injury. J Appl Physiol. 2013;115:539–545. doi: 10.1152/japplphysiol.00374.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudgens JL, Sugg KB, Grekin JA, Gumucio JP, Bedi A, Mendias CL. Platelet-Rich Plasma Activates Proinflammatory Signaling Pathways and Induces Oxidative Stress in Tendon Fibroblasts. Am J Sports Med. 2016;44:1931–1940. doi: 10.1177/0363546516637176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaillou T, Jackson JR, England JH, Kirby TJ, Richards-White J, Esser KA, Dupont-Versteegden EE, Mccarthy JJ. Identification of a conserved set of upregulated genes in mouse skeletal muscle hypertrophy and regrowth. J Appl Physiol. 2015;118:86–97. doi: 10.1152/japplphysiol.00351.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uezumi A, Fukada S-I, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 20.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birbrair A, Zhang T, Wang Z-M, Messi ML, Enikolopov GN, Mintz A, Delbono O. Role of Pericytes in Skeletal Muscle Regeneration and Fat Accumulation. Stem Cells Dev. 2013;22:2298–2314. doi: 10.1089/scd.2012.0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller AA, van Velthoven CT, Fukumoto KD, Cheung TH, Rando TA. Intronic polyadenylation of PDGFRα in resident stem cells attenuates muscle fibrosis. Nature. 2016;540:276–279. doi: 10.1038/nature20160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duehrkop C, Rieben R. Refinement of tourniquet-induced peripheral ischemia/reperfusion injury in rats: comparison of 2 h vs 24 h reperfusion. Lab Anim. 2014;48:143–154. doi: 10.1177/0023677213516313. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F, Hu EC, Gerzenshtein J, Lei M-P, Lineaweaver WC. The expression of proinflammatory cytokines in the rat muscle flap with ischemia-reperfusion injury. Ann Plast Surg. 2005;54:313–317. [PubMed] [Google Scholar]
- 25.Mccarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, Srikuea R, Lawson BA, Grimes B, Keller C, Van Zant G, Campbell KS, Esser KA, Dupont-Versteegden EE, Peterson CA. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138:3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamaki T, Uchiyama Y, Okada Y, Tono K, Nitta M, Hoshi A, Akatsuka A. Multiple stimulations for muscle-nerve-blood vessel unit in compensatory hypertrophied skeletal muscle of rat surgical ablation model. Histochem Cell Biol. 2009;132:59–70. doi: 10.1007/s00418-009-0585-1. [DOI] [PubMed] [Google Scholar]
- 27.Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA, Hansen MS, Angus-Hill M, Kardon G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138:371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birbrair A, Zhang T, Wang Z-M, Messi ML, Enikolopov GN, Mintz A, Delbono O. Skeletal muscle pericyte subtypes differ in their differentiation potential. Stem Cell Res. 2013;10:67–84. doi: 10.1016/j.scr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, Innocenzi A, Galvez BG, Messina G, Morosetti R, Li S, Belicchi M, Peretti G, Chamberlain JS, Wright WE, Torrente Y, Ferrari S, Bianco P, Cossu G. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 30.Naylor AJ, McGettrick HM, Maynard WD, May P, Barone F, Croft AP, Egginton S, Buckley CD. A differential role for CD248 (Endosialin) in PDGF-mediated skeletal muscle angiogenesis. PLoS ONE. 2014;9:e107146. doi: 10.1371/journal.pone.0107146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millay DP, O’Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499:301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tidball JG. Inflammatory cell response to acute muscle injury. Medicine and science in sports and exercise. 1995;27:1022–1032. doi: 10.1249/00005768-199507000-00011. [DOI] [PubMed] [Google Scholar]
- 33.Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda, Md) 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 34.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavares MR, Pavan ICB, Amaral CL, Meneguello L, Luchessi AD, Simabuco FM. The S6K protein family in health and disease. Life Sci. 2015;131:1–10. doi: 10.1016/j.lfs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Marabita M, Baraldo M, Solagna F, Ceelen JJM, Sartori R, Nolte H, Nemazanyy I, Pyronnet S, Kruger M, Pende M, Blaauw B. S6K1 Is Required for Increasing Skeletal Muscle Force during Hypertrophy. Cell Rep. 2016;17:501–513. doi: 10.1016/j.celrep.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 37.Moritz A, Li Y, Guo A, Villén J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, Ren JM, Hornbeck P, Cantley LC, Gygi SP, Rush J, Comb MJ. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Science signaling. 2010;3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornberger TA. Mechanotransduction and the regulation of mTORC1 signaling in skeletal muscle. Int J Biochem Cell Biol. 2011;43:1267–1276. doi: 10.1016/j.biocel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmerman EI, Turner DC, Buaboonnam J, Hu S, Orwick S, Roberts MS, Janke LJ, Ramachandran A, Stewart CF, Inaba H, Baker SD. Crenolanib is active against models of drug-resistant FLT3-ITD-positive acute myeloid leukemia. Blood. 2013;122:3607–3615. doi: 10.1182/blood-2013-07-513044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.