Abstract
Major advances in genetic and epigenetic profiling of acute lymphoblastic leukaemia (ALL) have enhanced the understanding of key biological subsets of de novo and relapsed ALL, which has led to improved risk stratification of patients. These achievements have further defined critical leukaemia-associated pathways and somatic alterations that may be preferentially sensitive to treatment with kinase inhibitors, epigenetic therapy or other novel agents. Therapeutic success in childhood ALL currently relies upon refined risk stratification of patients based on (1) underlying biological and clinical characteristics and (2) depth of initial treatment response with appropriate modulation of chemotherapy intensity. This review describes the current mutational landscape of childhood ALL and discusses opportunities for substantial improvements in survival with implementation of molecularly targeted therapies.
Keywords: acute lymphoblastic leukaemia, genomics, paediatrics, precision medicine, therapy
INTRODUCTION
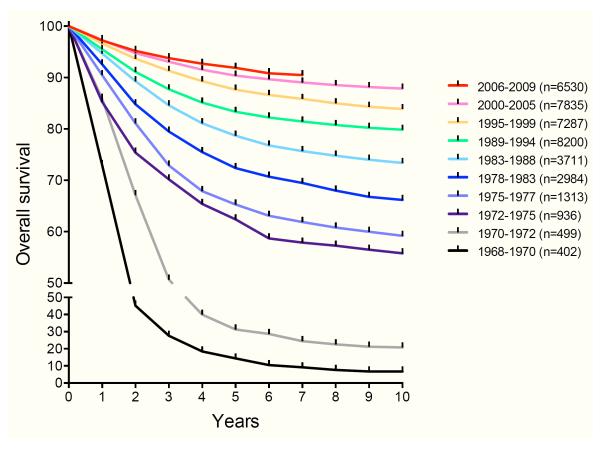
Remarkable advances have been made during the past 60 years in the treatment of patients with acute lymphoblastic leukaemia (ALL), the most common childhood cancer. In the 1960s, fewer than 10% of children with ALL were long-term survivors. With contemporary therapy, 5-year event-free survival (EFS) and overall survival (OS) rates now approach or exceed 85% and 90%, respectively (Hunger, et al 2012) (Figure 1). Key factors contributing to this dramatic improvement in outcomes include integration of central nervous system (CNS)-directed treatment and development of risk-adapted multi-agent chemotherapy regimens. Enhanced supportive care has also decreased morbidity and mortality and contributed to improved survival (Pui, et al 2015a).
Figure 1. Improved overall survival of children with ALL treated on cooperative group trials in North America.
Parenthetical numbers indicate number of subjects analysed for each treatment era. Longer-term follow-up is available for some populations (not shown). Data provided by the Children’s Oncology Group.
Serial improvements in both EFS and OS first occurred with identification of superior therapeutic strategies via phase 3 clinical trials conducted by paediatric oncology consortia in Europe and in North America (Hunger, et al 2012, Pui, et al 2015a). These cooperative group trials have demonstrated that age and initial white blood cell (WBC) count are consistent predictors of outcome. Older children and/or those with higher WBC counts fare less well than younger children and/or those with lower WBC counts, probably partly due to inherent biological differences of these leukaemias (Hunger, et al 2012, Pui, et al 2015a). These key diagnostic factors were used to delineate standard risk and high risk subtypes of ALL by the National Cancer Institute (NCI)-Rome criteria (Smith, et al 1996). NCI-Rome risk classification is used by many paediatric oncology consortia for stratification of children with B-lymphoblastic leukaemia (B-ALL), but is not prognostic in T-lymphoblastic leukaemia (T-ALL) (Pui, et al 2015a).
Risk stratification of children with ALL has been further refined by development and clinical implementation of sensitive, highly reproducible minimal residual disease (MRD) response monitoring techniques. Children with MRD positivity above specific pre-defined thresholds (typically 0.01%) at the end of induction therapy have a significantly higher risk of treatment failure and/or relapse regardless of underlying leukaemia-associated alterations (Borowitz, et al 2015, Pui, et al 2015b). Recent studies have further demonstrated particularly dismal outcomes for children with B-ALL and T-ALL who remain MRD positive approximately 3 months after starting therapy (Borowitz, et al 2015, Schrappe, et al 2011).
Numerous germline genetic variants and somatic alterations have been identified in de novo and relapsed childhood ALL (Pui, et al 2015a), which may also have prognostic implications. Efforts are now ongoing to characterize the epigenetic, biochemical, and other functional sequelae of these mutations that may provide therapeutic vulnerabilities. Ultimately, tailored therapeutic strategies to target ALL-associated driver lesions and pathways may increase anti-leukaemia efficacy and decrease relapse, as well as reduce undesirable off-target toxicities.
B-CELL ACUTE LYMPHOBLASTIC LEUKAEMIA
Approximately 80-85% of paediatric ALL is of B cell origin, resulting from arrest at an immature B-precursor cell stage. Most B-ALL cases appear to arise spontaneously and are classified by the presence of recurrent somatic cytogenetic or molecular alterations (Hunger and Mullighan 2015). The underlying aetiologies of most cases of childhood ALL remain largely unknown, although various environmental, ethnic, immunological, infectious, socioeconomic and other epidemiological factors have been rigorously evaluated as potential contributors to leukaemogenesis (Wiemels 2012). Childhood ALL is also associated with uncommon constitutional leukaemia predisposition syndromes, such as trisomy 21 and TP53 mutations (Li-Fraumeni syndrome) (Stieglitz and Loh 2013). Rare germline ETV6 and PAX5 mutations and ARID5B, CEBPE, GATA3, and IKZF1 polymorphisms have also been linked to increased ALL occurrence (Perez-Andreu, et al 2015, Shah, et al 2013, Zhang, et al 2015).
Somatic Chromosomal Gains and Losses
Hyperdiploidy
Cytogenetic and/or fluorescence in situ hybridization (FISH) assays are routinely used to identify structural chromosomal gains or losses within leukaemia cells (Table I). High hyperdiploidy (51-67 chromosomes per leukaemia cell, most often with +4, +6, +10, +14, +17, +18, +21 and +X) occurs in approximately 25% of childhood ALL and is more common in younger children (Paulsson, et al 2015). Children with high-hyperdiploid B-ALL have excellent outcomes and may be candidates for reduced-intensity chemotherapy, which has been demonstrated to minimize treatment toxicities without compromising survival (Vora, et al 2013). The inferior clinical outcomes previously associated with low-hyperdiploidy (47-50 chromosomes) appear to be ameliorated with contemporary therapy regimens (Hunger and Mullighan 2015, Pui, et al 2015a).
Table I. Common genetic alterations in childhood ALL.
Note that percentages may total more than 100% due to co-occurrence of genetic lesions. Data are summarized from those delineated in the main text and updated from Tasian et al (2015)
| Genetic subtype | Common alterations | Frequency in ALL |
Prognosis | Comment |
|---|---|---|---|---|
| B-ALL | ||||
| Abnomalities in Chromosome Number | ||||
| High hyperdiploidy (51-67 chromosomes) |
25% | Favourable | ||
| Low hyperdiploidy (47-50 chromosomes) |
14% | Previously unfavourable, now intermediate |
||
| Hypodiploidy (<44 chromosomes) | Near-haploidy (24-31 chromosomes), low- hypodiploidy (32-39 chromosomes) |
1-2% | Unfavourable | Association with TP53 mutations, IKZF2 and IKZF3 deletions, and RAS and PI3K pathway mutations |
|
Recurrent Chromosomal
Translocations |
||||
| t(12;21)(p13;q22) | ETV6-RUNX1 (TEL-AML1) | 20% | Favourable | |
| t(1;19)(q23;p13.1) | TCF3-PBX1 (E2A-PBX1) | 4% | Intermediate | |
| t(17;19)(q22;p13) | TCF3-HLF | <0.5% | Unfavourable | |
| KMT2A (MLL) rearrangements | 5-6% | Unfavourable (infants), intermediate (non- infants) |
Highest frequency in younger infants (80%); associated with FLT3 overexpression and epigenetic dysregulation |
|
| t(1;11)(q21;q23) | KMT2A-MLLT11 | Less unfavourable | Rare | |
| t(4;11)(q21;q23) | KMT2A-AFF1 (AF4) | Particularly unfavourable |
Comprises 50% of infant KMT2A- rearranged ALL |
|
| t(9;11)(p22;q23) | KMT2A-MLLT3 (AF9) | Comprises 15% of infant KMT2A- rearranged ALL |
||
| t(10;11)(p12;q23) | KMT2A-MLLT10 (AF10) | Comprises 5% of infant KMT2A- rearranged ALL |
||
| t(11;19)(q23;p13.3) | KMT2A-MLLT1 (ENL) | Comprises 20-25% of infant KMT2A- rearranged ALL |
||
| Other fusion partners | ||||
| t(9;22)(q34;q11.2) | BCR-ABL1 | 3-5% | Unfavourable prior to TKI therapy, intermediate with TKI therapy? |
Associated with IKZF1 deletions |
| Other | ||||
| Ph-like |
IGH-CRLF2, P2RY8- CRLF2 |
7-8% | Unfavourable | 50% of Ph-like; associated with JAK1 and JAK2 mutations, CDKN2A/B deletions, IKZF1 deletions; increasing incidence with older age; possibly targetable with TKIs |
|
ABL1, ABL2, CSF1R, PDGFRB rearrangements |
5-6% | Unfavourable | 10-20% of Ph-like; potentially targetable with TKIs |
|
|
EPOR, JAK2 rearrangements |
2% | Unfavourable | 10% of Ph-like; potentially targetable with TKIs |
|
| Trisomy 21-associated ALL |
P2RY8-CRLF2, JAK2 mutations |
50-60% of DS-ALL |
Intermediate | |
| iAMP21 | Multiple copies of RUNX1 | 2% | Unfavourable | Rare rob(15;21)(q10;q10)c associated with greatly increased risk of iAMP21 ALL |
| DUX4 rearrangements | IGH-DUX4, ERG-DUX4 | 3-7% | Favourable | Associated with IKZF1 deletions, ERG dysregulation, aberrant CD2 expression |
| MEF2D rearrangements |
MEF2D-BCL9, MEF2D-
HNRNPUL1 |
3-6% | Unfavourable | Multiple fusion partners, possible role for epigenetic therapies |
| ZNF384 rearrangements | EP300-ZNF384 | 4% | Intermediate | Multiple fusion partners, possible role for JAK inhibitors |
|
| ||||
| T-ALL | ||||
|
Recurrent Chromosomal
Translocations |
||||
| t(10;14)(q24;q11) | TLX1 (HOX11) fusions | 5-10% of T- ALL |
Favourable | Associated with PHF6 mutations |
| t(7;19)(q34;p13) | LYL1 fusions | 10% of T- ALL |
Unfavourable | |
| t(1;14)(p32;q11), t(1;7)(p32;q34), t(11;14)(p15;q11), t(11;14)(p13;q11) |
TAL1, LMO1, LMO2 fusions |
50-60% of T- ALL |
Unfavourable | Associated with PHF6 mutations |
| t(11;14)(p15;q11), t(5;14)(q35;q32) | TLX3 (HOX11L2) fusions | 20-25% of T- ALL |
Unfavourable (some studies), intermediate (some studies), favourable (some studies) |
Associated with PHF6 mutations |
| t(8;14)(q24;q11) | TRA-MYC, TRC-MYC | 1% of T-ALL | Probably unfavourable |
Associated with MYC activation and aggressive phenotype |
| 7p15 translocations |
HOXA10, HOXA9 overexpression |
3% of T-ALL | Unfavourable | |
| KMT2A (11q23) rearrangements |
KMT2A-AFF1, KMT2A- MLLT1 |
5% of T-ALL | Possibly favourable |
|
| t(10;11)(p13;q21) |
PICALM-MLLT10 (CALM-
AF10) |
5-10% of T- ALL |
Unfavourable (some studies), intermediate (other studies) |
Associated with EZH2 alterations |
| t(9;14)(q34;q32) | NUP214-ABL1 | 5-15% of T- ALL |
Unfavourable (some studies), intermediate (other studies) |
Associated with TLX1 and TLX3 (HOX11L2) overexpression |
| Other | ||||
| NOTCH1 mutations | 50-60% of T- ALL |
Favourable | Associated with CDKN2A and FBXW7 deletions |
|
| ETP | 10-15% of T- ALL |
Unfavourable (some studies), intermediate (other studies) |
Associated with Ras pathway mutations; characteristic immunophenotype (CD1a-, CD8-, CD5- or CD5-dim with co- expression of myeloid or stem cell markers) |
|
| FBXW7 mutation | 15% of T- ALL |
Associated with NOTCH1 activation via impairment of proteasomal degradation of NOTCH1 |
||
| Other T-ALL | 6% of T-ALL | |||
|
| ||||
| Relapsed ALL | Associated with chemotherapy resistance | |||
| CREBBP mutation | 20% of relapsed ALL |
Probably confers resistance to glucocorticoids |
||
| NT5C2 mutation | 20% of relapsed ALL |
Probably confers resistance to nucleoside analogues |
||
| PRPS1 mutation | 7% of relapsed ALL |
Probably confers resistance to nucleoside analogues |
||
| MSH6 deletion | ||||
| NR3C1 deletion | ||||
| SETD2 mutation | 12% of relapsed ALL |
Suggests possible role for epigenetic therapies |
||
| KDM6A mutation | ||||
| KMT2D (MLL2) mutation | ||||
| RAS pathway mutations | 30-50% of relapsed ALL |
|||
ALL = acute lymphoblastic leukaemia; B-ALL = B cell acute lymphoblastic leukaemia; DS-ALL = Down syndrome-associated ALL; iAMP21 = intrachromosomal amplification of chromosome 21; T-ALL = T cell acute lymphoblastic leukaemia; TKI = tyrosine kinase inhibitor.
Hypodiploidy
Conversely, hypodiploid B-ALL (<44 chromosomes) comprises 1-2% of childhood ALL and is associated with inferior survival, particularly in those with end-of-induction MRD positivity. While many patients with hypodiploid ALL empirically undergo haematopoietic stem cell transplantation (HSCT) in first complete remission given their unfavourable prognoses, it is not clear that HSCT is necessary or will improve outcomes for all patients (Mehta, et al 2015, Mullighan, et al 2015). TP53 mutations occur commonly in children with low-hypodiploid (30-39 chromosomes) ALL, many of whom have germline TP53 mutations consistent with Li-Fraumeni syndrome (Holmfeldt, et al 2013, Safavi, et al 2015). Preclinical studies suggest potential therapeutic activity of phosphatidylinositol-3-kinase (PI3K) or mitogen/extracellular signal-regulated kinase (MEK) inhibitors in hypodiploid ALL (Holmfeldt, et al 2013, Safavi, et al 2015).
Recurrent Chromosomal Translocations
Sentinel chromosomal translocations occur in nearly all of childhood B-ALL, many of which have prognostic significance. Most recurrent ALL-associated translocations appear to be very early or initiating events that drive leukaemogenesis. Some rearrangements are not readily detectable by routine cytogenetic analyses of metaphase chromosomes, but can be identified via reverse-transcription polymerase chain reaction (RT-PCR) amplification of fusion genes created by these translocations or via FISH assays.
ETV6-RUNX1 Rearrangement
Approximately 25% of standard risk childhood B-ALL cases have a cryptic t(12;21)(p13;q22), which results in ETV6-RUNX1 (TEL-AML1) fusion. While detection of ETV6-RUNX1 fusions in preserved blood spots from children who subsequently develop ALL implicates potential prenatal origin of leukaemogenesis, these fusions are also detectable in children who do not develop ALL. These latter data suggest that ETV6-RUNX1 translocations cooperate with additional necessary mutations to contribute to ALL pathogenesis (Greaves 2009). In general, children with standard risk B-ALL with ETV6-RUNX1 fusion have extremely favourable outcomes with standard therapy(Hunger and Mullighan 2015). As in high hyperdiploid ALL, successful treatment with lower-intensity regimens may also be feasible for some patients with ETV6-RUNX1 ALL (Vora, et al 2013).
Very recently, a small number of “ETV6-RUNX1-like” B-ALL cases have been reported, which lack classic ETV6-RUNX1 rearrangement, but are associated with other ETV6 fusions and with IKZF1 deletions. The clinical outcomes of such rare patients remain incompletely elucidated, although relapse does not appear to be common (Lilljebjorn, et al 2016).
TCF3 Rearrangement
Other recurrent chromosomal translocations in B-ALL include t(1;19)(q23;p13.3) resulting in TCF3-PBX1 (E2A-PBX1) fusion. While previously associated with inferior prognosis, TCF3-PBX1 rearrangement is no longer considered a poor risk factor with contemporary treatment (Felice, et al 2011). Although rare (<0.5% of children with B-ALL), the TCF3-HLF fusion resulting from t(17;19)(q22;p13.3) is associated with extremely poor outcomes (Mullighan 2012). Some preclinical studies have demonstrated activity of the tyrosine kinase inhibitor (TKI) dasatinib in TCF3-rearranged ALL (Lenz, et al 2014).
KMT2A (MLL) Rearrangement
Somatic translocations of KMT2A (lysine methyltransferase 2A; formerly MLL, mixed lineage leukaemia) occur in approximately 75% of infants with B-ALL, particularly in those <6 months of age. Interestingly, infant KMT2A-rearranged (KMT2A-R) ALL has a remarkable paucity of other genetic alterations (Andersson, et al 2015, Bernt and Armstrong 2011). KMT2A translocations also occur in about 2% of older children, adolescents and adults with ALL, with >100 fusion partners identified to date (Bernt and Armstrong 2011). Clinical prognoses vary somewhat based upon the specific KMT2A translocation, although outcomes of infants and children with KMT2A-R ALL are generally inferior to those of patients with non-KMT2A-R ALL. Outcomes are particularly dismal for infants diagnosed at <90 days of age, raising the question of HSCT in first complete remission for this highest-risk population, although data suggest that other infants with ALL do not benefit from HSCT (Mann, et al 2010).
Research to characterize dysregulated signalling and transcriptional pathways in KMT2A-R ALL is ongoing with a goal of identifying potential new therapeutic targets (Andersson, et al 2015). FLT3 (fms-related tyrosine kinase 3 receptor) overexpression is common in KMT2A-R infant ALL (Brown, et al 2005). Unfortunately, addition of the FLT3 inhibitor (FLT3i) lestaurtinib to chemotherapy in infants with KMT2A-R ALL did not improve EFS versus chemotherapy alone (Brown, et al 2016). Similarly, no appreciable clinical responses were observed in infants with relapsed KMT2A-R ALL treated on a phase 1 trial with the selective FLT3i quizartinib and chemotherapy (Cooper, et al 2016). Additional investigation is thus needed to determine potential efficacy of FLT3-targeted therapies (perhaps with more potent or selective FLT3 inhibitors) in KMT2A-R ALL.
Another potential therapeutic approach for infant ALL relies upon the discovery of frequent epigenetic dysregulation in KMT2A-R leukaemias (Bernt and Armstrong 2011). Drugs targeting histone deacetylases (e.g., vorinostat, panobinostat, bromodomain inhibitors) or methyltransferases (e.g., decitabine, 5-azacytidine, disruptor of telomeric silencing 1-like histone H3K79 methyltransferase [DOT1L] inhibitors) are under early-phase clinical evaluation in adults and children with KMT2A-R leukaemias (NCT02141828, NCT01483690, NCT01321346, NCT02828358).
BCR-ABL1 Rearrangement
BCR-ABL1 fusion resulting from t(9;22)(q34;q11.2) (the Philadelphia chromosome, Ph+) occurs in nearly all patients with chronic myeloid leukaemia (CML) and a subset of patients with ALL, including 3-5% of childhood B-ALL. BCR–ABL1 fusion results in constitutive activation of ABL1 kinase and associated downstream signalling. Most Ph+ ALL cases also have deletions in transcription factors that regulate B-cell development, including IKZF1 (Ikaros) and PAX5 (paired box 5) (Mullighan, et al 2009a), which has been associated with poor outcomes in both Ph+ and Ph− ALL.
Prior to the 21st century, children and adults with Ph+ ALL had dismal clinical outcomes despite maximal intensity of conventional chemotherapy and frequent utilization of HSCT (Arico, et al 2010). Pivotal studies conducted in the early 2000s demonstrated remarkable clinical efficacy of the ABL TKI, imatinib, in adults with CML, resulting in major cytogenetic and molecular remissions. Childhood leukaemia cooperative group trials subsequently demonstrated safety of combining imatinib with intensive chemotherapy in children with Ph+ ALL, as well as dramatic improvements in survival (Biondi, et al 2012, Schultz, et al 2009). Mature clinical trial data now show that most children with Ph+ ALL can be successfully treated with imatinib and chemotherapy without the need for HSCT, and the 10-year OS for this population now approaches 80% (Schultz, et al 2014). These remarkable clinical responses further establish BCR-ABL1 as a driver oncogene in Ph+ ALL and provides a precedent for successful precision medicine therapies in childhood ALL.
Selective pressure of imatinib therapy over time can lead to development of ABL tyrosine kinase domain (TKD) point mutations that confer reduced TKI sensitivity or overt therapeutic resistance. Limited available data suggest that BCR-ABL1 TKD mutations occur in very small number of children with Ph+ ALL who relapse after imatinib and chemotherapy, presumably because combination therapy significantly overcomes selective pressure of TKI monotherapy (Cazzaniga, et al 2015, Chang, et al 2012).
Second- and third-generation ABL TKIs (e.g., dasatinib, nilotinib, bosutinib, ponatinib, bafetinib) have been developed to overcome therapeutic resistance (Leoni and Biondi 2015). Many of these drugs also inhibit SRC kinases and have improved CNS penetration. While superiority of one TKI has not been definitively proven in Ph+ leukaemias (Leoni and Biondi 2015), comparably excellent remission rates and outcomes with dasatinib and imatinib have been demonstrated in adults with Ph+ ALL (Foa, et al 2011). In paediatric trials, treatment with combined dasatinib and intensive multi-agent chemotherapy also resulted in outstanding 3-year EFS and OS, which were analogous to outcomes of children with Ph+ ALL treated with continuous imatinib and chemotherapy (Schultz, et al 2014, Slayton, et al 2015). Additional trials of ABL1-targeting TKIs in children with Ph+ ALL are ongoing or in development.
BCR-ABL1-Like or Philadelphia Chromosome-Like ALL
Philadelphia chromosome-like (Ph-like) or BCR-ABL1-like ALL is a recently-described subset of B-ALL defined by an activated kinase gene expression profile similar to that of Ph+ ALL and associated with a diverse range of genetic alterations that activate cytokine receptor signalling pathways (Den Boer, et al 2009, Mullighan, et al 2009a, Roberts, et al 2014). In paediatrics, the Ph-like subtype comprises approximately 10% of NCI standard risk and 13% of NCI high risk ALL cases. The incidence of Ph-like ALL further increases with age, accounting for 21% and 27% of B-ALL cases in adolescents and younger adults (<40 years), respectively (Roberts, et al 2014).
As in Ph+ ALL, deletions and inactivating mutations of IKZF1 and other lymphoid-associated transcription factors genes are common in Ph-like ALL (Den Boer, et al 2009, Mullighan, et al 2009a). In North America, CRLF2 (cytokine receptor-like factor 2) rearrangement with resulting gene overexpression occur in 50% of Ph-like ALL cases, including P2RY8 (purinergic receptor P2Y, G-protein coupled, 8)-CRLF2 fusions and IGH (immunoglobulin heavy locus)-CRLF2 translocations (Mullighan, et al 2009b, Russell, et al 2009). CRLF2 rearrangements occur more commonly in older adolescents and young adults (AYAs) and in patients of Native American and Hispanic/Latino genetic ancestry, which may contribute to differences in Ph-like ALL incidence reported in European and North American studies (Attarbaschi, et al 2012). Concomitant JAK2 or JAK1 point mutations occur in about half of CRLF2-rearranged ALL cases (Mullighan, et al 2009b, Russell, et al 2009).
An additional 15-20% of Ph-like ALL cases (without CRLF2 rearrangement) activate kinase signalling via cryptic translocations involving “ABL class” genes ABL1 (Abelson kinase 1), ABL2 (Abelson kinase 2), CSF1R (colony stimulating factor 1 receptor) or PDGFRB (platelet-derived growth factor receptor beta) (Roberts, et al 2014). These rearrangements involve a diverse variety of translocation partners and encode fusion proteins that activate receptor tyrosine kinase (RTK) and non-RTK intracellular signalling. Another 10-15% of Ph-like ALL harbour JAK2 fusions or truncating rearrangements of EPOR, resulting in activated JAK/STAT signalling (Iacobucci, et al 2016, Roberts, et al 2014). Other rare fusions potentially sensitive to kinase inhibitors have also been identified in Ph-like ALL (Roberts, et al 2014).
Importantly, children and adolescents with Ph-like ALL have high rates of treatment failure, relapse and death when treated with conventional cytotoxic chemotherapy (Den Boer, et al 2009, Mullighan, et al 2009a). Given these poor clinical outcomes and robust preclinical data demonstrating constitutive activation of kinase signalling in Ph-like ALL (Roberts, et al 2014), development of new therapeutic approaches is imperative. Various preclinical studies have reported in vitro and/or in vivo sensitivity of models of Ph-like ALL with ABL class fusions to imatinib and dasatinib and of CRLF2-, JAK2-, and EPOR-rearranged ALL models to the JAK inhibitor ruxolitinib (Iacobucci, et al 2016, Maude, et al 2012, Roberts, et al 2014).
Anecdotal reports of remarkable responses of children with PDGFRB-rearranged ALL with poor early responses to chemotherapy when imatinib or dasatinib was added to chemotherapy have now been published (Roberts, et al 2014). It is now clear that PDGFRB rearrangements occur frequently in patients with induction failure (defined as ≥25% residual leukaemia after induction therapy) (Schwab, et al 2016). Clinical testing for PDGFRB alterations is thus recommended for such patients.
Taken together, these data strongly suggest potential clinical efficacy of kinase inhibitors in specific subsets of patients with Ph-like ALL and raise the possibility of combinatorial therapy, either via addition of a kinase inhibitor to a chemotherapy backbone or via dual inhibitor therapy targeting discrete signalling pathways. Clinical trials are now underway (NCT01406756, NCT02723994) or under active development by paediatric oncology cooperative groups to test the efficacy of addition of dasatinib or ruxolitinib to chemotherapy for patients with Ph-like ALL harbouring ABL-class fusions or JAK pathway alterations, respectively. Such efforts have required extensive multi-disciplinary collaboration to develop testing algorithms capable of identifying the diverse milieu of Ph-like ALL genomic alterations in relative real time. Ideally, rapid identification of children and AYA with Ph-like ALL via comprehensive genomic testing will occur during induction therapy. Patients can then be allocated to clinical trials testing the efficacy of combined kinase inhibition and chemotherapy, as has been the therapeutic paradigm for Ph+ ALL.
Trisomy 21-Associated ALL
Children with trisomy 21 (Down Syndrome) have a 10-fold increased risk of developing B-ALL (DS-ALL), although the contribution of germline trisomy 21 to leukaemogenesis remains incompletely understood. Interestingly, DS-associated ALL is almost always B-lineage and has a lower incidence of hyperdiploidy and fewer recurrent cytogenetic translocations than in non-DS-ALL. Children with DS-ALL also have increased risk of chemotherapy-related toxicity and inferior survival (Buitenkamp, et al 2014).
CRLF2 rearrangements are prevalent in children with DS-ALL, occurring in 50-60% of patients. The P2RY8-CRLF2 fusion, resulting from interstitial deletion of the pseudoautosomal region of the sex chromosomes, is most common (Russell, et al 2009). Concomitant JAK mutations occur in approximately 50% of CRLF2-rearranged DS-ALL cases, most frequently a JAK2 R683G point mutation (Bercovich, et al 2008). To date, JAK inhibitors have not been formally evaluated in children with DS-ALL.
Intrachromosomal Amplification of Chromosome 21
Intrachromosomal amplification of chromosome 21 (iAMP21) was initially discovered via detection of multiple RUNX1 copies on ETV6-RUNX1 FISH testing. iAMP21 ALL occurs in 2% of childhood B-ALL and is more prevalent in older children and was previously associated with poor outcomes. Appropriate high-risk therapy intensification has improved survival substantially in this group of patients without the need for HSCT (Harrison, et al 2014). The very rare constitutional Robertsonian translocation rob(15;21)(q10;q10)c is associated with a dramatically increased risk of developing iAMP21 ALL (Li, et al 2014).
DUX4 Rearrangement and ERG Dysregulation
Recurrent IGH-DUX4 (double homeobox 4) fusions, resulting in DUX4 overexpression and a unique gene expression signature, were very recently reported in up to 7% of childhood B-ALL cases. DUX4 rearrangement results in loss of function of ERG (ETS-related gene) via intragenic deletion or via induction of a dominant-negative isoform that inhibits wild-type ERG (Zhang, et al 2016). ERG-DUX4 fusions have also been reported (Lilljebjorn, et al 2016). Despite frequent concomitant IKZF1 deletions, patients with DUX4 rearrangements appear to have excellent clinical prognoses with standard chemotherapy treatment (Clappier, et al 2014, Lilljebjorn, et al 2016, Yasuda, et al 2016, Zhang, et al 2016).
MEF2D and ZN384 Rearrangements
Rearrangements involving MEFD2 (myocyte enhancer factor 2D) or ZNF384 (zinc finger protein 384) were recently described in subsets of childhood B-ALL and are associated with distinct gene expression signatures (Liu, et al 2016). MEF2D-rearranged ALL occurs in 3-6% of childhood B-ALL, more commonly in older children and adolescents, and may be associated with poor outcomes. Preclinical data suggest a possible role for histone deacetylase inhibitors in MEF2D-rearranged ALL. Rearrangements involving ZNF384 were also recently described in approximately 3% of childhood B-ALL and appear associated with an intermediate prognosis. Preclinical data demonstrate upregulation of JAK/STAT signalling in ZNF384-rearranged ALL, also suggesting therapeutic potential with JAK inhibitors (Liu, et al 2016).
RAS pathway-mutant ALL
Activating mutations in various Ras pathway-associated genes, including the GTPases KRAS, NRAS, HRAS and PTPN11 (protein tyrosine phosphatase, non-receptor type 11), CBL (Casitas B-lineage lymphoma) and FLT3, have been reported in various subtypes of childhood ALL, including high hyperdiploid ALL, hypodiploid ALL, infant ALL, and a subset of Ph-like ALL (Holmfeldt, et al 2013, Paulsson, et al 2015). In some studies, RAS mutations are associated with early risk of relapse and poor clinical outcomes (Irving, et al 2014), while other groups have not identified inferior clinical outcomes. Additional studies are required to clarify the driver versus passenger nature of these mutations in ALL.
Targeting of mutant RAS remains of great theoretical interest in human cancer, but has proven quite challenging to accomplish. Recent efforts have instead focused upon targeting aberrant MAPK or PI3K signalling networks. Preclinical studies have demonstrated enhanced sensitivity of RAS-mutant ALL cells to MEK inhibitors or PI3K/mTOR inhibitors (Dail, et al 2010, Irving, et al 2014). Clinical trials of MEK inhibitors in children with relapsed/refractory RAS-mutant leukaemias are now in development.
T-CELL ACUTE LYMPHOBLASTIC LEUKAEMIA (T-ALL)
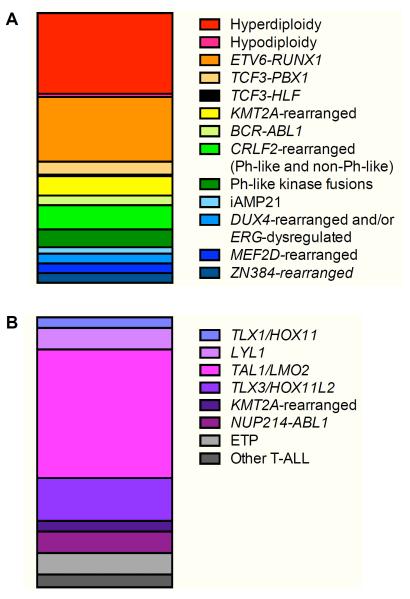
T-ALL comprises 10-15% of childhood ALL and is more common in males, older children and adolescents, and patients of African-American ethnicity (Hunger and Mullighan 2015). While the prognosis for children with T-ALL was historically inferior to that of B-ALL, children treated with contemporary intensive therapies now achieve >85% 5-year EFS (Hunger and Mullighan 2015, Wood, et al 2014). Nonetheless, T-cell immunophenotype remains an adverse risk factor in multivariate analyses of data from several childhood leukaemia clinical trials (Hunger, et al 2012, Vora, et al 2013). Unlike in B-ALL, age at diagnosis and presenting WBC count have very little prognostic impact in T-ALL and are not used for risk stratification of patients (Pui, et al 2015a). Genomic alterations in T-ALL are frequently cytogenetically cryptic, and the prognostic significance of recurrent T-ALL-associated mutations remains incompletely understood (Figure 2, Table I). Risk stratification of patients with T-ALL instead is largely determined by CNS status and early response to therapy, as measured by MRD testing (Schrappe, et al 2011, Wood, et al 2014).
Figure 2. Recurrent genomic alterations in childhood ALL.
Relative incidence of major translocations and other alterations are delineated for (A) B cell acute lymphoblastic leukaemia (B-ALL) and (B) T cell acute lymphoblastic leukaemia (T-ALL).
While exciting advances have occurred in genomic characterization of T-ALL, development of precision medicine treatment approaches for T-ALL has proven more challenging. The nucleoside analogue nelarabine demonstrated impressive early activity in patients with relapsed/refractory T-ALL, but was associated with neurotoxicity that precluded further clinical testing for some time. Subsequently, the safety of adding nelarabine to multi-agent chemotherapy for children and AYAs with newly diagnosed T-ALL was demonstrated. Ongoing analyses of the recently completed Children’s Oncology Group (COG) AALL0434 clinical trial will determine whether addition of nelarabine to combination therapy improves outcome (Winter, et al 2015). A current COG phase 3 trial is studying the potential for enhanced activity of chemotherapy combined with the proteasome inhibitor bortezomib in children and adolescents with de novo T-ALL (NCT02112916).
Recurrent Chromosomal Translocations
T Cell Receptor Gene Rearrangement
Approximately 50% of childhood T-ALL cases have chromosomal translocations involving fusion of T-cell receptor genes to oncogenes or interstitial deletions resulting in juxtaposition of two genes. Comprehensive expression profiling studies have also more fully characterized the genomic and epigenomic landscape of these leukaemias. These data have facilitated “binning” of T-ALL into four major subtypes: (1) TLX1 (previously termed HOX11), (2) LYL1, (3) TAL1/LMO2, and (4) TLX3 (previously termed HOX11L2) (Van Vlierberghe and Ferrando 2012). While the prognostic and therapeutic significance of these T-ALL subsets has not been well-elucidated, patients with TLX1 alterations appear to have more favourable responses to standard therapy. Other rare alterations in T-ALL include t(8;14)(q24;q11) resulting in TRA-MYC or TRD-MYC fusion (Van Vlierberghe and Ferrando 2012).
KMT2A Rearrangement
As in B-ALL, 11q23 translocations resulting in KMT2A rearrangement have been reported in 10-15% of T-ALL. It is not known whether KMT2A-R T-ALL cases overexpress wild-type FLT3, as in infant B-ALL, or if FLT3 inhibitors could improve outcomes for such patients.
PICALM-MLLT10 Rearrangement
Occurring also in acute myeloid leukaemias (Savage, et al 2010), the t(10;11)(p13;q21) translocation resulting in PICALM (phosphatidylinositol binding clathrin assembly protein)-MLLT10 (mixed-lineage leukaemia; translocated to 10) (previously termed CALM-AF10) fusion has been associated with particularly poor survival in children with T-ALL. More recent data demonstrate intermediate outcomes with intensive therapy (Lo Nigro, et al 2013).
ABL1 Rearrangement
NUP214-ABL1 fusion resulting from t(5;14) occurs in 5-10% of T-ALL. Preclinical studies demonstrate activated ABL1 kinase signalling in NUP214-ABL1 T-ALL analogous to that of Ph+ and subsets of Ph-like B-ALL, as well as in vitro inhibition of the ABL1 target phosphoproteins CrkL and STAT5 and increased cell death with dasatinib or nilotinib treatment (De Keersmaecker, et al 2014, Quintas-Cardama, et al 2008). As predicted, anecdotal clinical efficacy of imatinib or dasatinib treatment of patients with refractory NUP214-ABL1 ALL has also been reported (Deenik, et al 2009), but no well-controlled clinical trials have yet been performed.
JAK inhibition (e.g., momelotinib, ruxolitinib, tofacitinib) may also have therapeutic relevance in ABL1-rearranged and other subsets of T-ALL. Point mutations and indels in IL7R (interleukin-7 receptor), JAK1, JAK3, and SH2B3 (SH2B adaptor protein 3) genes are common in T-ALL, particularly in the early thymic precursor (ETP) subtype discussed below (Vicente, et al 2015). Preclinical activity of JAK inhibition was recently reported in models of childhood T-ALL (Maude, et al 2015a).
Alterations in X Chromosome Genes
Inactivating mutations and deletions in PHF6 (plant homeodomain finger 6) occur frequently in the TLX1, TAL1/LMO2, and TLX3 subsets. PHF6 is involved in nucleosome remodelling and deacetylation and may function as a tumour suppressor (Van Vlierberghe, et al 2010). The location of PHF6 on the X chromosome may partly explain the greater observed occurrence of T-ALL in males (Van Vlierberghe, et al 2010). Similarly, somatic loss-of-function mutations in the X-linked histone H3K27me3 demethylase ubiquitously transcribed X chromosome also occur frequently in T-ALL, are enriched in males, and appear sensitive in vitro to inhibitors of the histone methyltransferase EZH2 (Van der Meulen, et al 2015).
NOTCH1 Mutations
Somatic mutations in NOTCH1, which encodes a transmembrane receptor responsible for T-cell lineage commitment and survival, occur in >50% of T-ALL cases. Constitutive activation of PI3K/mTOR signalling has been reported in T-ALL, particularly in leukaemias with NOTCH1 mutations or deletions of PTEN (phosphatase and tensin homolog), a negative regulator of PI3K signalling. Common cooperating deletions of tumour suppressor CDKN2A or mutations in ubiquitin protein ligase FBXW7 may further enhance aberrant PI3K signalling via attenuation of NOTCH1 protein degradation (Weng, et al 2004).
In general, patients with NOTCH1-mutant T-ALL have favourable outcomes with standard chemotherapy treatment. Nonetheless, the high frequency of NOTCH1 mutations in T-ALL has inspired significant efforts to develop new treatment approaches that may further improve outcomes. In addition to potential therapeutic relevance of PI3K inhibition, anti-NOTCH1 antibody immunotherapies and gamma secretase inhibitors (GSIs) that block NOTCH1 degradation have been investigated. While preclinical studies of GSIs demonstrated remarkable leukaemia cytotoxicity (Real, et al 2009), the significant on target/off tumour gastrointestinal toxicity of first-generation GSIs has limited their clinical efficacy to date. Clinical testing of newer GSIs with more favourable toxicity profiles is currently in progress in adults with relapsed T-ALL (Papayannidis, et al 2015).
Early Thymic Precursor ALL
The early thymic precursor or early T-cell precursor (ETP) subtype comprises 10-15% of childhood T-ALL and is diagnosed by its characteristic immature flow cytometric immunophenotype (CD1a−, CD8−, CD5− or CD5dim with co-expression of myeloid or stem cell markers) (Coustan-Smith, et al 2009). Association of ETP-ALL with high rates of chemoresistance and relapse and dismal clinical outcomes was initially reported (Coustan-Smith, et al 2009). Data from more recent clinical trials showed that children and AYAs with ETP-ALL responded more slowly to induction chemotherapy and had frequent end-induction MRD positivity when compared to those with non-ETP-ALL. However, most patients with ETP-ALL achieved molecular remission after three months of chemotherapy, and recent cooperative group data demonstrate comparable outcomes of children with ETP-ALL and non-ETP ALL when stratified by MRD responses with an overall 80-85% 5-year EFS (Patrick, et al 2014, Wood, et al 2014).
As in other ALL subtypes, frequent activating mutations in RAS pathway and cytokine receptor signalling genes, IL7R pathway genes and haematopoietic development and histone modification genes have been reported in ETP-ALL (Zenatti, et al 2011, Zhang, et al 2012). The mutational landscape of ETP-ALL has striking similarity to that of myeloid leukaemias, suggesting that ETP-ALL may be part of a spectrum of leukaemias arising from very early haematopoietic progenitor cells and/or stem cells. Preclinical studies in childhood T-ALL (including ETP) models demonstrate potential therapeutic efficacy of targeting activated cytokine receptor signalling networks with FLT3, JAK, or PI3K/mTOR inhibitors (Maude, et al 2015a, Neumann, et al 2013).
RELAPSED ALL
Modern risk stratification and appropriate therapeutic intensification have resulted in markedly improved outcomes for children with ALL. While relapse risk is now known to be greatest in genomically-defined high-risk subsets of childhood ALL, leukaemia recurrence occurs across the genetic spectrum. Relapsed ALL is frequently associated with treatment resistance, possibly arising from enrichment of pre-existing resistant subclone(s) and/or from mutation acquisition during chemotherapy exposure (Hunger and Mullighan 2015). The roles of leukaemia oligoclonality in initial chemotherapy responses and at time of leukaemia recurrence remain incompletely understood, but are probably critical determinants of relapse risk and of durable remission (Gaynon and Sun 2016).
Detailed genomic profiling of matched diagnosis, remission and relapsed ALL specimens has furthered our understanding of clonal evolution, chemoresistance mechanisms, and evolution of new genetic mutations (Irving, et al 2016, Ma, et al 2015, Mullighan, et al 2008). Most relapsed ALL specimens preserve key genetic features present at initial diagnosis, particularly sentinel chromosome translocations that are probably very early (possibly initiating) events in leukaemogenesis. However, new genetic alterations are detectable in nearly 100% of relapsed ALL cases, which imply further dynamic evolution of leukaemogenesis at recurrence (Ma, et al 2015). These studies further demonstrate the eradication of initial diagnostic bulk leukaemia clone and persistence of rare, pre-existing mutation-prone subclones. Genome-wide association studies have also identified enrichment of several germline single nucleotide polymorphisms in children with relapsed ALL (Yang, et al 2009).
Approximately 20% of relapsed ALL specimens harbour mutations in CREBBP (CREB-binding protein) (Malinowska-Ozdowy, et al 2015, Mullighan, et al 2011). The CREBBP (also known as CBP) protein functions in glucocorticoid-mediated transcription and histone deacetylation, and preclinical activity of histone deacetylase inhibitors in chemoresistant ALL has been reported (Gang, et al 2014, Mullighan, et al 2011). Co-occurrence of CREBBP and KRAS mutations has also been observed frequently in relapsed ALL, which suggests potential therapeutic efficacy of MEK or PI3K/mTOR pathway inhibition (Malinowska-Ozdowy, et al 2015). To date, clinical investigation of kinase inhibitors in children with relapsed/refractory ALL has largely focused upon mTOR inhibition in combination with chemotherapy (NCT01523977, NCT01614197, NCT01403415).
Gain-of-function somatic mutations in NT5C2 (5'-nucleotidase, cytosolic II) have also been identified in nearly 20% of children with relapsed B- or T-ALL, particularly those who relapse while receiving maintenance chemotherapy (Meyer, et al 2013, Tzoneva, et al 2013). The NT5C2 enzyme is involved in nucleoside analogue metabolism and inactivation. Similarly, relapse-specific mutations in PRPS1 (phosphoribosyl pyrophosphate synthetase 1) were recently reported in up to 7% of relapsed B-ALL cases, which appeared to occur independently of NT5C2 mutations (Li, et al 2015). In mechanistic studies, PRPS1-mutant leukaemia cells were associated with impaired thiopurine metabolism and decreased conversion of 6-mercaptopurine to 6-thioguanine and its metabolites (Li, et al 2015). Taken together, the frequent acquisition of NT5C2 and PRPS1 mutations in children with relapsed ALL highlights their probable role in conferring decreased sensitivity to anti-metabolite chemotherapies and facilitating chemoresistant relapse.
Activating mutations in RAS pathway-associated genes (e.g., KRAS, NRAS, FLT3 and PTPN11) have also been identified in up to 50% of relapsed ALL cases, and potent anti-leukaemic activity of MEK inhibition in preclinical models of RAS-mutant childhood ALL has been reported (Irving, et al 2014). Deletions in the DNA mismatch repair gene MSH6 and the glucocorticoid receptor NR3C1 and mutations in the H3K36 trimethyltransferase SETD2, the lysine-specific demethylase KDM6A, and the epigenetic regulator KMT2D (previously termed MLL2) have also been reported, further highlighting the genomic complexity of relapsed ALL and potential for alternative therapies (Mar, et al 2014, Yang, et al 2008).
SUMMARY: GENOMICS INFORM THERAPEUTICS
The complete genomic, epigenomic and transcriptomic landscape of de novo childhood ALL remains incompletely defined, as highlighted by recent discovery of IGH-DUX4 fusions in 5-10% of B-ALL (Lilljebjorn, et al 2016, Yasuda, et al 2016, Zhang, et al 2016). Recent observations of frequent chemoresistance-associated mutations and leukaemia-predisposing gene polymorphisms further highlight the dynamic biological complexity of ALL. Continued elucidation of the biology of this remarkably genetically heterogeneous group of diseases and its associated biochemical, immunological and transcriptional sequelae will continue to inform risk stratification of patients.
Clinical development of “more tailored, less toxic” precision medicine therapies for children with ALL will continue to rely upon sensitive diagnostic testing to identify known (and currently unknown) prognostic leukaemia-associated alterations and to assign patients to appropriately intensive therapy. The rapidity and depth of leukaemia remission after induction chemotherapy remain critical determinants of long-term outcomes (Conter, et al 2014, Paganin, et al 2014). However, the years of multi-agent chemotherapy required to achieve cure are well-associated with deleterious short- and long-term sequelae (Robison 2011). Therapeutic reduction in carefully selected lowest-risk children (e.g., rapid early responders with ETV6-RUNX1 or hyperdiploid ALL) has been investigated or is under current evaluation by several childhood cancer cooperative groups. Several studies have demonstrated excellent feasibility of this approach without compromise in cure rates (Moricke, et al 2008, Vora, et al 2013, Zenatti, et al 2011).
For highest risk patients with targetable genetic or epigenetic lesions, access to biologically relevant inhibitors with established safe paediatric dosing is critical. Incorporation of TKIs into chemotherapy for children with Ph+ ALL has provided a great paradigm for precision medicine in paediatric oncology, and similar therapeutic strategies for children with other potentially targetable ALL-associated mutations are of great interest. However, such approaches are early in development.
A major obstacle in successful clinical realization of precision medicine approaches in childhood ALL is that new agents are usually first studied as monotherapy in patients with multiply relapsed disease. While phase 1 trials are critical to assess drug safety and to define a tolerable dose, these studies do not always include patients with defined genetic mutations who are predicted to respond to a novel agent. Furthermore, lack of therapeutic efficacy in the phase 1 setting does not necessarily predict clinical responses in children with de novo ALL or when the agent is administered on a chemotherapy backbone. It is becoming increasingly apparent that combination therapies are probably required to achieve long-term cure and to minimize development of resistance mutations and escape pathways. Finally, clinical trials to evaluate new drugs in increasingly smaller genetically- or epigenetically-defined “boutique” subsets of patients will surely require (a) comparison to accurate historical control data for standard chemotherapy treatment and (b) development of robust biomarkers to most accurately define biological activity and to prioritize further drug development. Innovative trial designs and international collaboration among childhood cancer consortia are probably necessary to maximize efficient patient accrual and study completion.
While significant advances in the genomic characterization of childhood ALL have identified new potential therapeutic vulnerabilities, additional studies are necessary to decipher more fully the molecular dependencies of childhood ALL and to bring targeted therapies to fullest fruition. Ultimately, it is expected that the paradigm of imatinib for Ph+ ALL will continue to inspire successful precision medicine treatment approaches for other genomically-defined subsets of childhood ALL that will improve outcomes and perhaps also minimize toxicities. In addition, tremendous efficacy of new CD19- and CD22-targeted antibody- and T cell-based immunotherapies has been recently reported with high rates of remission achieved in patients with multiply relapsed or refractory B-ALL (Gore, et al 2014, Kantarjian, et al 2016, Lee, et al 2015, Maude, et al 2014). These exciting new treatment modalities have potential to define new therapeutic paradigms for childhood ALL and are discussed in detail elsewhere (Jabbour, et al 2015, Maude, et al 2015b). In 2016 and beyond, the major current challenges in childhood ALL are to improve cure rates through more precise risk stratification and increased use of molecularly-targeted therapies, as well as to minimize late effects by identifying the least toxic regimens that can cure low-risk patients.
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute (K08CA184418 award to SKT). SPH is the Jeffrey E. Perelman Distinguished Chair in Pediatrics at the Children’s Hospital of Philadelphia. We apologize that space and reference limitations made it impossible to reference many important publications.
Footnotes
FINANCIAL DISCLOSURES AND CONFLICTS OF INTEREST
SPH has received honoraria from Jazz Pharmaceuticals, Sigma Tau Pharmaceuticals and Erytech. SKT receives research funding from Gilead Sciences. SPH is co-inventor of US patent on Gene Expression Signatures for Detection of Underlying Philadelphia Chromosome-Like (Ph-like) Events and Therapeutic Targeting in Leukaemia.
AUTHORSHIP CONTRIBUTIONS
SKT and SPH wrote and edited the manuscript.
REFERENCES
- Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, Nakitandwe J, Holmfeldt L, Parker M, Easton J, Huether R, Kriwacki R, Rusch M, Wu G, Li Y, Mulder H, Raimondi S, Pounds S, Kang G, Shi L, Becksfort J, Gupta P, Payne-Turner D, Vadodaria B, Boggs K, Yergeau D, Manne J, Song G, Edmonson M, Nagahawatte P, Wei L, Cheng C, Pei D, Sutton R, Venn NC, Chetcuti A, Rush A, Catchpoole D, Heldrup J, Fioretos T, Lu C, Ding L, Pui CH, Shurtleff S, Mullighan CG, Mardis ER, Wilson RK, Gruber TA, Zhang J, Downing JR, for The St. Jude Children's Research Hospital-Washington University Pediatric Cancer Genome Project The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–337. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico M, Schrappe M, Hunger SP, Carroll WL, Conter V, Galimberti S, Manabe A, Saha V, Baruchel A, Vettenranta K, Horibe K, Benoit Y, Pieters R, Escherich G, Silverman LB, Pui CH, Valsecchi MG. Clinical outcome of children with newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia treated between 1995 and 2005. J Clin Oncol. 2010;28:4755–4761. doi: 10.1200/JCO.2010.30.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attarbaschi A, Morak M, Cario G, Cazzaniga G, Ensor HM, te Kronnie T, Bradtke J, Mann G, Vendramini E, Palmi C, Schwab C, Russell LJ, Schrappe M, Conter V, Mitchell CD, Strehl S, Zimmermann M, Potschger U, Harrison CJ, Stanulla M, Panzer-Grumayer R, Haas OA, Moorman AV, for the Associazione Italiana di Ematologia ed Oncologia Pediatrica -Berlin-Frankfurt-Munster Study Group. National Cancer Research Institute -Children's Cancer & Leukaemia Study Group Treatment outcome of CRLF2-rearranged childhood acute lymphoblastic leukaemia: a comparative analysis of the AIEOP-BFM and UK NCRI-CCLG study groups. Br J Haematol. 2012;158:772–777. doi: 10.1111/j.1365-2141.2012.09221.x. [DOI] [PubMed] [Google Scholar]
- Bercovich D, Ganmore I, Scott LM, Wainreb G, Birger Y, Elimelech A, Shochat C, Cazzaniga G, Biondi A, Basso G, Cario G, Schrappe M, Stanulla M, Strehl S, Haas OA, Mann G, Binder V, Borkhardt A, Kempski H, Trka J, Bielorei B, Avigad S, Stark B, Smith O, Dastugue N, Bourquin JP, Tal NB, Green AR, Izraeli S. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet. 2008;372:1484–1492. doi: 10.1016/S0140-6736(08)61341-0. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Armstrong SA. Targeting epigenetic programs in MLL-rearranged leukemias. Hematology Am Soc Hematol Educ Program. 2011;2011:354–360. doi: 10.1182/asheducation-2011.1.354. [DOI] [PubMed] [Google Scholar]
- Biondi A, Schrappe M, De Lorenzo P, Castor A, Lucchini G, Gandemer V, Pieters R, Stary J, Escherich G, Campbell M, Li CK, Vora A, Arico M, Rottgers S, Saha V, Valsecchi MG. Imatinib after induction for treatment of children and adolescents with Philadelphia-chromosome-positive acute lymphoblastic leukaemia (EsPhALL): a randomised, open-label, intergroup study. Lancet Oncol. 2012;13:936–945. doi: 10.1016/S1470-2045(12)70377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowitz MJ, Wood BL, Devidas M, Loh ML, Raetz EA, Salzer WL, Nachman JB, Carroll AJ, Heerema NA, Gastier-Foster JM, Willman CL, Dai Y, Winick NJ, Hunger SP, Carroll WL, Larsen E. Prognostic significance of minimal residual disease in high risk B-ALL: a report from Children's Oncology Group study AALL0232. Blood. 2015;126:964–971. doi: 10.1182/blood-2015-03-633685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P, Kairalla J, Wang C, Dreyer Z, Salzer W, Sorenson M, Borowitz M, Carroll A, Heerema N, Gore L, Devidas M, Carroll W, Winick N, Raetz E, Loh M, Hunger S, Hilden J. Addition of FLT3 Inhibitor Lestaurtinib to Post-Induction Chemotherapy Does Not Improve Outcomes in MLL-Rearranged Infant Acute Lymphoblastic Leukemia (All): AALL0631, A Children’s Oncology Group Study. Pediatric blood & cancer (SIOP 2016 Scientific Programme+Index) 2016;63(Suppl S3, S5) abstract O-001. [Google Scholar]
- Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–820. doi: 10.1182/blood-2004-06-2498. [DOI] [PubMed] [Google Scholar]
- Buitenkamp TD, Izraeli S, Zimmermann M, Forestier E, Heerema NA, van den Heuvel-Eibrink MM, Pieters R, Korbijn CM, Silverman LB, Schmiegelow K, Liang DC, Horibe K, Arico M, Biondi A, Basso G, Rabin KR, Schrappe M, Cario G, Mann G, Morak M, Panzer-Grumayer R, Mondelaers V, Lammens T, Cave H, Stark B, Ganmore I, Moorman AV, Vora A, Hunger SP, Pui CH, Mullighan CG, Manabe A, Escherich G, Kowalczyk JR, Whitlock JA, Zwaan CM. Acute lymphoblastic leukemia in children with Down syndrome: a retrospective analysis from the Ponte di Legno study group. Blood. 2014;123:70–77. doi: 10.1182/blood-2013-06-509463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzaniga V, De Lorenzo P, Mottadelli F, GFazio G, Villa T, Rossi V, Pigazzi M, Mansur MB, Zecca M, Barisone E, Locatelli F, Basso G, Valsecchi MG, Greaves M, Biondi A, Ford AM, Cazzaniga G. Clonal Evolution and Lack of BCR-ABL1 Mutations in Pediatric Ph+ ALL Patients Resistant/Refractory to Imatinib Treatment. Blood. 2015;126 abstract 2622. [Google Scholar]
- Chang BH, Willis SG, Stork L, Hunger SP, Carroll WL, Camitta BM, Winick NJ, Druker BJ, Schultz KR. Imatinib resistant BCR-ABL1 mutations at relapse in children with Ph+ ALL: a Children's Oncology Group (COG) study. Br J Haematol. 2012;157:507–510. doi: 10.1111/j.1365-2141.2012.09039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clappier E, Auclerc MF, Rapion J, Bakkus M, Caye A, Khemiri A, Giroux C, Hernandez L, Kabongo E, Savola S, Leblanc T, Yakouben K, Plat G, Costa V, Ferster A, Girard S, Fenneteau O, Cayuela JM, Sigaux F, Dastugue N, Suciu S, Benoit Y, Bertrand Y, Soulier J, Cave H. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28:70–77. doi: 10.1038/leu.2013.277. [DOI] [PubMed] [Google Scholar]
- Conter V, Valsecchi MG, Parasole R, Putti MC, Locatelli F, Barisone E, Lo Nigro L, Santoro N, Arico M, Ziino O, Pession A, Testi AM, Micalizzi C, Casale F, Zecca M, Casazza G, Tamaro P, La Barba G, Notarangelo LD, Silvestri D, Colombini A, Rizzari C, Biondi A, Masera G, Basso G. Childhood high-risk acute lymphoblastic leukemia in first remission: results after chemotherapy or transplant from the AIEOP ALL 2000 study. Blood. 2014;123:1470–1478. doi: 10.1182/blood-2013-10-532598. [DOI] [PubMed] [Google Scholar]
- Cooper TM, Cassar J, Eckroth E, Malvar J, Sposto R, Gaynon P, Chang BH, Gore L, August K, Pollard JA, DuBois SG, Silverman LB, Oesterheld J, Gammon G, Magoon D, Annesley C, Brown PA. A Phase I Study of Quizartinib Combined with Chemotherapy in Relapsed Childhood Leukemia: A Therapeutic Advances in Childhood Leukemia & Lymphoma (TACL) Study. Clin Cancer Res. 2016;22:4014–4022. doi: 10.1158/1078-0432.CCR-15-1998. [DOI] [PubMed] [Google Scholar]
- Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, Biondi A, Pui CH, Downing JR, Campana D. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dail M, Li Q, McDaniel A, Wong J, Akagi K, Huang B, Kang HC, Kogan SC, Shokat K, Wolff L, Braun BS, Shannon K. Mutant Ikzf1, KrasG12D, and Notch1 cooperate in T lineage leukemogenesis and modulate responses to targeted agents. Proc Natl Acad Sci U S A. 2010;107:5106–5111. doi: 10.1073/pnas.1001064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Porcu M, Cox L, Girardi T, Vandepoel R, de Beeck JO, Gielen O, Mentens N, Bennett KL, Hantschel O. NUP214-ABL1-mediated cell proliferation in T-cell acute lymphoblastic leukemia is dependent on the LCK kinase and various interacting proteins. Haematologica. 2014;99:85–93. doi: 10.3324/haematol.2013.088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deenik W, Beverloo HB, van der Poel-van de Luytgaarde SC, Wattel MM, van Esser JW, Valk PJ, Cornelissen JJ. Rapid complete cytogenetic remission after upfront dasatinib monotherapy in a patient with a NUP214-ABL1-positive T-cell acute lymphoblastic leukemia. Leukemia. 2009;23:627–629. doi: 10.1038/leu.2008.318. [DOI] [PubMed] [Google Scholar]
- Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, Horstmann MA, Janka-Schaub GE, Kamps WA, Evans WE, Pieters R. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice MS, Gallego MS, Alonso CN, Alfaro EM, Guitter MR, Bernasconi AR, Rubio PL, Zubizarreta PA, Rossi JG. Prognostic impact of t(1;19)/ TCF3-PBX1 in childhood acute lymphoblastic leukemia in the context of Berlin-Frankfurt-Munster-based protocols. Leuk Lymphoma. 2011;52:1215–1221. doi: 10.3109/10428194.2011.565436. [DOI] [PubMed] [Google Scholar]
- Foa R, Vitale A, Vignetti M, Meloni G, Guarini A, De Propris MS, Elia L, Paoloni F, Fazi P, Cimino G, Nobile F, Ferrara F, Castagnola C, Sica S, Leoni P, Zuffa E, Fozza C, Luppi M, Candoni A, Iacobucci I, Soverini S, Mandelli F, Martinelli G, Baccarani M, Party GALW. Dasatinib as first-line treatment for adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;118:6521–6528. doi: 10.1182/blood-2011-05-351403. [DOI] [PubMed] [Google Scholar]
- Gang EJ, Hsieh YT, Pham J, Zhao Y, Nguyen C, Huantes S, Park E, Naing K, Klemm L, Swaminathan S, Conway EM, Pelus LM, Crispino J, Mullighan CG, McMillan M, Muschen M, Kahn M, Kim YM. Small-molecule inhibition of CBP/catenin interactions eliminates drug-resistant clones in acute lymphoblastic leukemia. Oncogene. 2014;33:2169–2178. doi: 10.1038/onc.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynon PS, Sun W. Oligoclonality and new agent evaluation in acute lymphoblastic leukaemia. Br J Haematol. 2016;173:950–957. doi: 10.1111/bjh.14143. [DOI] [PubMed] [Google Scholar]
- Gore L, Locatelli F, Zugmaier G, Zwaan CM, Bhojwani D, Handgretinger R, Bader P, O'Brien MM, Trippett TM, Brethon B, Rizzari C, DuBois SG, Schlegel PG, Barnette P, Messina C, Hu K, Mergen N, Fischer A, Whitlock J, von Stackelberg A. Initial Results from a Phase 2 Study of Blinatumomab in Pediatric Patients with Relapsed/Refractory B-Cell Precursor Acute Lymphoblastic Leukemia. Blood. 2014;124:3703–3703. [Google Scholar]
- Greaves M. Darwin and evolutionary tales in leukemia. The Ham-Wasserman Lecture. Hematology Am Soc Hematol Educ Program. 2009;2009:3–12. doi: 10.1182/asheducation-2009.1.3. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Moorman AV, Schwab C, Carroll AJ, Raetz EA, Devidas M, Strehl S, Nebral K, Harbott J, Teigler-Schlegel A, Zimmerman M, Dastuge N, Baruchel A, Soulier J, Auclerc MF, Attarbaschi A, Mann G, Stark B, Cazzaniga G, Chilton L, Vandenberghe P, Forestier E, Haltrich I, Raimondi SC, Parihar M, Bourquin JP, Tchinda J, Haferlach C, Vora A, Hunger SP, Heerema NA, Haas OA, Ponte di Legno International Workshop in Childhood Acute Lymphoblastic Leukemia An international study of intrachromosomal amplification of chromosome 21 (iAMP21): cytogenetic characterization and outcome. Leukemia. 2014;28:1015–1021. doi: 10.1038/leu.2013.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen SC, McCastlain K, Becksfort J, Ma J, Wu G, Patel SN, Heatley SL, Phillips LA, Song G, Easton J, Parker M, Chen X, Rusch M, Boggs K, Vadodaria B, Hedlund E, Drenberg C, Baker S, Pei D, Cheng C, Huether R, Lu C, Fulton RS, Fulton LL, Tabib Y, Dooling DJ, Ochoa K, Minden M, Lewis ID, To LB, Marlton P, Roberts AW, Raca G, Stock W, Neale G, Drexler HG, Dickins RA, Ellison DW, Shurtleff SA, Pui CH, Ribeiro RC, Devidas M, Carroll AJ, Heerema NA, Wood B, Borowitz MJ, Gastier-Foster JM, Raimondi SC, Mardis ER, Wilson RK, Downing JR, Hunger SP, Loh ML, Mullighan CG. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger SP, Mullighan CG. Acute Lymphoblastic Leukemia in Children. N Engl J Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ, Reaman GH, Carroll WL. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children's oncology group. J Clin Oncol. 2012;30:1663–1669. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci I, Li Y, Roberts KG, Dobson SM, Kim JC, Payne-Turner D, Harvey RC, Valentine M, McCastlain K, Easton J, Yergeau D, Janke LJ, Shao Y, Chen IM, Rusch M, Zandi S, Kornblau SM, Konopleva M, Jabbour E, Paietta EM, Rowe JM, Pui CH, Gastier-Foster J, Gu Z, Reshmi S, Loh ML, Racevskis J, Tallman MS, Wiernik PH, Litzow MR, Willman CL, McPherson JD, Downing JR, Zhang J, Dick JE, Hunger SP, Mullighan CG. Truncating Erythropoietin Receptor Rearrangements in Acute Lymphoblastic Leukemia. Cancer Cell. 2016;29:186–200. doi: 10.1016/j.ccell.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving J, Matheson E, Minto L, Blair H, Case M, Halsey C, Swidenbank I, Ponthan F, Kirschner-Schwabe R, Groeneveld-Krentz S, Hof J, Allan J, Harrison C, Vormoor J, von Stackelberg A, Eckert C. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood. 2014;124:3420–3430. doi: 10.1182/blood-2014-04-531871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving JA, Enshaei A, Parker CA, Sutton R, Kuiper RP, Erhorn A, Minto L, Venn NC, Law T, Yu J, Schwab C, Davies R, Matheson E, Davies A, Sonneveld E, den Boer ML, Love SB, Harrison CJ, Hoogerbrugge PM, Revesz T, Saha V, Moorman AV. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2016;128:911–922. doi: 10.1182/blood-2016-03-704973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour E, O'Brien S, Ravandi F, Kantarjian H. Monoclonal antibodies in acute lymphoblastic leukemia. Blood. 2015;125:4010–4016. doi: 10.1182/blood-2014-08-596403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian HM, DeAngelo DJ, Stelljes M, Martinelli G, Liedtke M, Stock W, Gokbuget N, O'Brien S, Wang K, Wang T, Paccagnella ML, Sleight B, Vandendries E, Advani AS. Inotuzumab Ozogamicin versus Standard Therapy for Acute Lymphoblastic Leukemia. N Engl J Med. 2016;375:740–753. doi: 10.1056/NEJMoa1509277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, Steinberg SM, Stroncek D, Tschernia N, Yuan C, Zhang H, Zhang L, Rosenberg SA, Wayne AS, Mackall CL. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz K, Thompson S, Goloviznina NA, Huan J, LaTocha DH, Druker BJ, Tyner JW, Kurre P, Chang BH. Dasatinib Shows Therapeutic Potential in the Murine Xenograft Model for TCF3 rearranged Acute Lymphoblastic Leukemia. Blood. 2014;124:3718–3718. [Google Scholar]
- Leoni V, Biondi A. Tyrosine kinase inhibitors in BCR-ABL positive acute lymphoblastic leukemia. Haematologica. 2015;100:295–299. doi: 10.3324/haematol.2015.124016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li H, Bai Y, Kirschner-Schwabe R, Yang JJ, Chen Y, Lu G, Tzoneva G, Ma X, Wu T, Li W, Lu H, Ding L, Liang H, Huang X, Yang M, Jin L, Kang H, Chen S, Du A, Shen S, Ding J, Chen H, Chen J, von Stackelberg A, Gu L, Zhang J, Ferrando A, Tang J, Wang S, Zhou BB. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med. 2015;21:563–571. doi: 10.1038/nm.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Schwab C, Ryan SL, Papaemmanuil E, Robinson HM, Jacobs P, Moorman AV, Dyer S, Borrow J, Griffiths M, Heerema NA, Carroll AJ, Talley P, Bown N, Telford N, Ross FM, Gaunt L, McNally RJ, Young BD, Sinclair P, Rand V, Teixeira MR, Joseph O, Robinson B, Maddison M, Dastugue N, Vandenberghe P, Haferlach C, Stephens PJ, Cheng J, Van Loo P, Stratton MR, Campbell PJ, Harrison CJ. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:98–102. doi: 10.1038/nature13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, Askmyr M, Rissler M, Schrappe M, Cario G, Castor A, Pronk CJ, Behrendtz M, Mitelman F, Johansson B, Paulsson K, Andersson AK, Fontes M, Fioretos T. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. doi: 10.1038/ncomms11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, Jiang L, Li JF, Wang MJ, Dai YJ, Zhang ZG, Wang Q, Kong J, Chen B, Zhu YM, Weng XQ, Shen ZX, Li JM, Wang J, Yan XJ, Li Y, Liang YM, Liu L, Chen XQ, Zhang WG, Yan JS, Hu JD, Shen SH, Chen J, Gu LJ, Pei D, Li Y, Wu G, Zhou X, Ren RB, Cheng C, Yang JJ, Wang KK, Wang SY, Zhang J, Mi JQ, Pui CH, Tang JY, Chen Z, Chen SJ. Genomic Profiling of Adult and Pediatric B-cell Acute Lymphoblastic Leukemia. EBioMedicine. 2016;8:173–183. doi: 10.1016/j.ebiom.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Nigro L, Mirabile E, Tumino M, Caserta C, Cazzaniga G, Rizzari C, Silvestri D, Buldini B, Barisone E, Casale F, Luciani M, Locatelli F, Messina C, Micalizzi C, Pession A, Parasole R, Santoro N, Masera G, Basso G, Arico M, Valsecchi M, Biondi A, Conter V. Detection of PICALM-MLLT10 (CALM-AF10) and outcome in children with T-lineage acute lymphoblastic leukemia. Leukemia. 2013;27:2419–2421. doi: 10.1038/leu.2013.149. [DOI] [PubMed] [Google Scholar]
- Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, Song G, Easton J, Harvey RC, Wheeler DA, Ma J, Doddapaneni H, Vadodaria B, Wu G, Nagahawatte P, Carroll WL, Chen IM, Gastier-Foster JM, Relling MV, Smith MA, Devidas M, Guidry Auvil JM, Downing JR, Loh ML, Willman CL, Gerhard DS, Mullighan CG, Hunger SP, Zhang J. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. doi: 10.1038/ncomms7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowska-Ozdowy K, Frech C, Schonegger A, Eckert C, Cazzaniga G, Stanulla M, Zur Stadt U, Mecklenbrauker A, Schuster M, Kneidinger D, von Stackelberg A, Locatelli F, Schrappe M, Horstmann MA, Attarbaschi A, Bock C, Mann G, Haas OA, Panzer-Grumayer R. KRAS and CREBBP mutations: a relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia. 2015;29:1656–1667. doi: 10.1038/leu.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann G, Attarbaschi A, Schrappe M, De Lorenzo P, Peters C, Hann I, De Rossi G, Felice M, Lausen B, Leblanc T, Szczepanski T, Ferster A, Janka-Schaub G, Rubnitz J, Silverman LB, Stary J, Campbell M, Li CK, Suppiah R, Biondi A, Vora A, Valsecchi MG, Pieters R, for the Interfant-99 Study Group Improved outcome with hematopoietic stem cell transplantation in a poor prognostic subgroup of infants with mixed-lineage-leukemia (MLL)-rearranged acute lymphoblastic leukemia: results from the Interfant-99 Study. Blood. 2010;116:2644–2650. doi: 10.1182/blood-2010-03-273532. [DOI] [PubMed] [Google Scholar]
- Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, Neuberg DS, Sinha AU, Sallan SE, Silverman LB, Kung AL, Lo Nigro L, Ebert BL, Armstrong SA. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun. 2014;5:3469. doi: 10.1038/ncomms4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Tasian SK, Vincent T, Hall JW, Sheen C, Roberts KG, Seif AE, Barrett DM, Chen IM, Collins JR, Mullighan CG, Hunger SP, Harvey RC, Willman CL, Fridman JS, Loh ML, Grupp SA, Teachey DT. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, Mahnke YD, Melenhorst JJ, Rheingold SR, Shen A, Teachey DT, Levine BL, June CH, Porter DL, Grupp SA. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Dolai S, Delgado-Martin C, Vincent T, Robbins A, Selvanathan A, Ryan T, Hall J, Wood AC, Tasian SK, Hunger SP, Loh ML, Mullighan CG, Wood BL, Hermiston ML, Grupp SA, Lock RB, Teachey DT. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood. 2015a;125:1759–1767. doi: 10.1182/blood-2014-06-580480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Teachey DT, Porter DL, Grupp SA. CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Blood. 2015b;125:4017–4023. doi: 10.1182/blood-2014-12-580068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta PA, Zhang MJ, Eapen M, He W, Seber A, Gibson B, Camitta BM, Kitko CL, Dvorak CC, Nemecek ER, Frangoul HA, Abdel-Azim H, Kasow KA, Lehmann L, Gonzalez Vicent M, Diaz Perez MA, Ayas M, Qayed M, Carpenter PA, Jodele S, Lund TC, Leung WH, Davies SM. Transplantation Outcomes for Children with Hypodiploid Acute Lymphoblastic Leukemia. Biol Blood Marrow Transplant. 2015;21:1273–1277. doi: 10.1016/j.bbmt.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, Tang Z, Zumbo P, Li S, Zavadil J, Levine RL, Cardozo T, Hunger SP, Raetz EA, Evans WE, Morrison DJ, Mason CE, Carroll WL. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45:290–294. doi: 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moricke A, Reiter A, Zimmermann M, Gadner H, Stanulla M, Dordelmann M, Loning L, Beier R, Ludwig WD, Ratei R, Harbott J, Boos J, Mann G, Niggli F, Feldges A, Henze G, Welte K, Beck JD, Klingebiel T, Niemeyer C, Zintl F, Bode U, Urban C, Wehinger H, Niethammer D, Riehm H, Schrappe M. Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood. 2008;111:4477–4489. doi: 10.1182/blood-2007-09-112920. [DOI] [PubMed] [Google Scholar]
- Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122:3407–3415. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, Harvey RC, Chen IM, Clifford RJ, Carroll WL, Reaman G, Bowman WP, Devidas M, Gerhard DS, Yang W, Relling MV, Shurtleff SA, Campana D, Borowitz MJ, Pui CH, Smith M, Hunger SP, Willman CL, Downing JR. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009a;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, Ma J, Coustan-Smith E, Harvey RC, Willman CL, Mikhail FM, Meyer J, Carroll AJ, Williams RT, Cheng J, Heerema NA, Basso G, Pession A, Pui CH, Raimondi SC, Hunger SP, Downing JR, Carroll WL, Rabin KR. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009b;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, Buetow KH, Pui CH, Baker SD, Brindle PK, Downing JR. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Jeha S, Pei D, Payne-Turner D, Coustan-Smith E, Roberts KG, Waanders E, Choi JK, Ma X, Raimondi SC, Fan Y, Yang W, Song G, Yang JJ, Inaba H, Downing JR, Leung WH, Bowman WP, Relling MV, Evans WE, Zhang J, Campana D, Pui C-H. Outcome of children with hypodiploid ALL treated with risk-directed therapy based on MRD levels. Blood. 2015;126:2896–2899. doi: 10.1182/blood-2015-09-671131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Coskun E, Fransecky L, Mochmann LH, Bartram I, Sartangi NF, Heesch S, Gokbuget N, Schwartz S, Brandts C, Schlee C, Haas R, Duhrsen U, Griesshammer M, Dohner H, Ehninger G, Burmeister T, Blau O, Thiel E, Hoelzer D, Hofmann WK, Baldus CD. FLT3 mutations in early T-cell precursor ALL characterize a stem cell like leukemia and imply the clinical use of tyrosine kinase inhibitors. PLoS One. 2013;8:e53190. doi: 10.1371/journal.pone.0053190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganin M, Fabbri G, Conter V, Barisone E, Polato K, Cazzaniga G, Giraldi E, Fagioli F, Arico M, Valsecchi MG, Basso G. Postinduction minimal residual disease monitoring by polymerase chain reaction in children with acute lymphoblastic leukemia. J Clin Oncol. 2014;32:3553–3558. doi: 10.1200/JCO.2014.56.0698. [DOI] [PubMed] [Google Scholar]
- Papayannidis C, DeAngelo DJ, Stock W, Huang B, Shaik MN, Cesari R, Zheng X, Reynolds JM, English PA, Ozeck M, Aster JC, Kuo F, Huang D, Lira PD, McLachlan KR, Kern KA, Garcia-Manero G, Martinelli G. A Phase 1 study of the novel gamma-secretase inhibitor PF-03084014 in patients with T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Blood Cancer J. 2015;5:e350. doi: 10.1038/bcj.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick K, Wade R, Goulden N, Mitchell C, Moorman AV, Rowntree C, Jenkinson S, Hough R, Vora A. Outcome for children and young people with Early T-cell precursor acute lymphoblastic leukaemia treated on a contemporary protocol, UKALL 2003. Br J Haematol. 2014;166:421–424. doi: 10.1111/bjh.12882. [DOI] [PubMed] [Google Scholar]
- Paulsson K, Lilljebjorn H, Biloglav A, Olsson L, Rissler M, Castor A, Barbany G, Fogelstrand L, Nordgren A, Sjogren H, Fioretos T, Johansson B. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet. 2015;47:672–676. doi: 10.1038/ng.3301. [DOI] [PubMed] [Google Scholar]
- Perez-Andreu V, Roberts KG, Xu H, Smith C, Zhang H, Yang W, Harvey RC, Payne-Turner D, Devidas M, Cheng IM, Carroll WL, Heerema NA, Carroll AJ, Raetz EA, Gastier-Foster JM, Marcucci G, Bloomfield CD, Mrozek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Rowe JM, Luger SM, Tallman MS, Dean M, Burchard EG, Torgerson DG, Yue F, Wang Y, Pui CH, Jeha S, Relling MV, Evans WE, Gerhard DS, Loh ML, Willman CL, Hunger SP, Mullighan CG, Yang JJ. A genome-wide association study of susceptibility to acute lymphoblastic leukemia in adolescents and young adults. Blood. 2015;125:680–686. doi: 10.1182/blood-2014-09-595744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Pei D, Coustan-Smith E, Jeha S, Cheng C, Bowman WP, Sandlund JT, Ribeiro RC, Rubnitz JE, Inaba H, Bhojwani D, Gruber TA, Leung WH, Downing JR, Evans WE, Relling MV, Campana D. Clinical utility of sequential minimal residual disease measurements in the context of risk-based therapy in childhood acute lymphoblastic leukaemia: a prospective study. Lancet Oncol. 2015b;16:465–474. doi: 10.1016/S1470-2045(15)70082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Yang JJ, Hunger SP, Pieters R, Schrappe M, Biondi A, Vora A, Baruchel A, Silverman LB, Schmiegelow K, Escherich G, Horibe K, Benoit YC, Izraeli S, Yeoh AE, Liang DC, Downing JR, Evans WE, Relling MV, Mullighan CG. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J Clin Oncol. 2015a;33:2938–2948. doi: 10.1200/JCO.2014.59.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintas-Cardama A, Tong W, Manshouri T, Vega F, Lennon PA, Cools J, Gilliland DG, Lee F, Cortes J, Kantarjian H, Garcia-Manero G. Activity of tyrosine kinase inhibitors against human NUP214-ABL1-positive T cell malignancies. Leukemia. 2008;22:1117–1124. doi: 10.1038/leu.2008.80. [DOI] [PubMed] [Google Scholar]
- Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C, Homminga I, Meijerink J, Aifantis I, Basso G, Cordon-Cardo C, Ai W, Ferrando A. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, Ma J, Becksfort J, Rusch M, Chen SC, Easton J, Cheng J, Boggs K, Santiago-Morales N, Iacobucci I, Fulton RS, Wen J, Valentine M, Cheng C, Paugh SW, Devidas M, Chen IM, Reshmi S, Smith A, Hedlund E, Gupta P, Nagahawatte P, Wu G, Chen X, Yergeau D, Vadodaria B, Mulder H, Winick NJ, Larsen EC, Carroll WL, Heerema NA, Carroll AJ, Grayson G, Tasian SK, Moore AS, Keller F, Frei-Jones M, Whitlock JA, Raetz EA, White DL, Hughes TP, Guidry Auvil JM, Smith MA, Marcucci G, Bloomfield CD, Mrozek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Pui CH, Jeha S, Relling MV, Evans WE, Gerhard DS, Gastier-Foster JM, Mardis E, Wilson RK, Loh ML, Downing JR, Hunger SP, Willman CL, Zhang J, Mullighan CG. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison LL. Late Effects of Acute Lymphoblastic Leukemia Therapy in Patients Diagnosed at 0-20 Years of Age. ASH Education Program Book. 2011;2011:238–242. doi: 10.1182/asheducation-2011.1.238. [DOI] [PubMed] [Google Scholar]
- Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, Chandrasekaran T, Chapiro E, Gesk S, Griffiths M, Guttery DS, Haferlach C, Harder L, Heidenreich O, Irving J, Kearney L, Nguyen-Khac F, Machado L, Minto L, Majid A, Moorman AV, Morrison H, Rand V, Strefford JC, Schwab C, Tonnies H, Dyer MJ, Siebert R, Harrison CJ. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114:2688–2698. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- Safavi S, Olsson L, Biloglav A, Veerla S, Blendberg M, Tayebwa J, Behrendtz M, Castor A, Hansson M, Johansson B, Paulsson K. Genetic and epigenetic characterization of hypodiploid acute lymphoblastic leukemia. Oncotarget. 2015;6:42793–42802. doi: 10.18632/oncotarget.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage NM, Kota V, Manaloor EJ, Kulharya AS, Pierini V, Mecucci C, Ustun C. Acute leukemia with PICALM-MLLT10 fusion gene: diagnostic and treatment struggle. Cancer Genet Cytogenet. 2010;202:129–132. doi: 10.1016/j.cancergencyto.2010.07.126. [DOI] [PubMed] [Google Scholar]
- Schrappe M, Valsecchi MG, Bartram CR, Schrauder A, Panzer-Grumayer R, Moricke A, Parasole R, Zimmermann M, Dworzak M, Buldini B, Reiter A, Basso G, Klingebiel T, Messina C, Ratei R, Cazzaniga G, Koehler R, Locatelli F, Schafer BW, Arico M, Welte K, van Dongen JJ, Gadner H, Biondi A, Conter V. Late MRD response determines relapse risk overall and in subsets of childhood T-cell ALL: results of the AIEOP-BFM-ALL 2000 study. Blood. 2011;118:2077–2084. doi: 10.1182/blood-2011-03-338707. [DOI] [PubMed] [Google Scholar]
- Schultz KR, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, Wang C, Davies SM, Gaynon PS, Trigg M, Rutledge R, Burden L, Jorstad D, Carroll A, Heerema NA, Winick N, Borowitz MJ, Hunger SP, Carroll WL, Camitta B. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children's oncology group study. J Clin Oncol. 2009;27:5175–5181. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, Zheng HW, Davies SM, Gaynon PS, Trigg M, Rutledge R, Jorstad D, Winick N, Borowitz MJ, Hunger SP, Carroll WL, Camitta B, Children's Oncology, G. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children's Oncology Group study AALL0031. Leukemia. 2014;28:1467–1471. doi: 10.1038/leu.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C, Ryan SL, Chilton L, Elliott A, Murray J, Richardson S, Wragg C, Moppett J, Cummins M, Tunstall O, Parker CA, Saha V, Goulden N, Vora A, Moorman AV, Harrison CJ. EBF1-PDGFRB fusion in pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL): genetic profile and clinical implications. Blood. 2016;127:2214–2218. doi: 10.1182/blood-2015-09-670166. [DOI] [PubMed] [Google Scholar]
- Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, Wechsler J, Yang J, Hayes J, Klein RJ, Zhang J, Wei L, Wu G, Rusch M, Nagahawatte P, Ma J, Chen SC, Song G, Cheng J, Meyers P, Bhojwani D, Jhanwar S, Maslak P, Fleisher M, Littman J, Offit L, Rau-Murthy R, Fleischut MH, Corines M, Murali R, Gao X, Manschreck C, Kitzing T, Murty VV, Raimondi SC, Kuiper RP, Simons A, Schiffman JD, Onel K, Plon SE, Wheeler DA, Ritter D, Ziegler DS, Tucker K, Sutton R, Chenevix-Trench G, Li J, Huntsman DG, Hansford S, Senz J, Walsh T, Lee M, Hahn CN, Roberts KG, King MC, Lo SM, Levine RL, Viale A, Socci ND, Nathanson KL, Scott HS, Daly M, Lipkin SM, Lowe SW, Downing JR, Altshuler D, Sandlund JT, Horwitz MS, Mullighan CG, Offit K. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayton WB, Kairalla JA, Schultz KR, Devidas M, Helian S, Pulsipher M, Chang BH, Carroll WL, Borowitz MJ, Brown VI, Winick NJ, Carroll AJ, Heerema NA, Gastier-Foster JM, Wood BL, Mizrahy SL, Hunger S. Outcomes of dasatinib plus intensive chemotherapy or stem cell transplant (SCT) for Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL) on Children’s Oncology Group AALL0622. J Clin Oncol. 2015;33(suppl.) Abstract 10006. [Google Scholar]
- Smith M, Arthur D, Camitta B, Carroll AJ, Crist W, Gaynon P, Gelber R, Heerema N, Korn EL, Link M, Murphy S, Pui CH, Pullen J, Reamon G, Sallan SE, Sather H, Shuster J, Simon R, Trigg M, Tubergen D, Uckun F, Ungerleider R. Uniform approach to risk classification and treatment assignment for children with acute lymphoblastic leukemia. J Clin Oncol. 1996;14:18–24. doi: 10.1200/JCO.1996.14.1.18. [DOI] [PubMed] [Google Scholar]
- Stieglitz E, Loh ML. Genetic predispositions to childhood leukemia. Ther Adv Hematol. 2013;4:270–290. doi: 10.1177/2040620713498161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasian SK, Loh ML, Hunger SP. Childhood acute lymphoblastic leukemia: Integrating genomics into therapy. Cancer. 2015;121:3577–3590. doi: 10.1002/cncr.29573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, Paietta E, Racevskis J, Rowe JM, Tallman MS, Paganin M, Basso G, Hof J, Kirschner-Schwabe R, Palomero T, Rabadan R, Ferrando A. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19:368–371. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, Rondou P, Rosen M, Pieters T, Vandenberghe P, Delabesse E, Lammens T, De Moerloose B, Menten B, Van Roy N, Verhasselt B, Poppe B, Benoit Y, Taghon T, Melnick AM, Speleman F, Wendel HG, Van Vlierberghe P. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, De Moerloose B, Philippe J, Gonzalez-Garcia S, Toribio ML, Taghon T, Zuurbier L, Cauwelier B, Harrison CJ, Schwab C, Pisecker M, Strehl S, Langerak AW, Gecz J, Sonneveld E, Pieters R, Paietta E, Rowe JM, Wiernik PH, Benoit Y, Soulier J, Poppe B, Yao X, Cordon-Cardo C, Meijerink J, Rabadan R, Speleman F, Ferrando A. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente C, Schwab C, Broux M, Geerdens E, Degryse S, Demeyer S, Lahortiga I, Elliott A, Chilton L, La Starza R, Mecucci C, Vandenberghe P, Goulden N, Vora A, Moorman AV, Soulier J, Harrison CJ, Clappier E, Cools J. Targeted sequencing identifies associations between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica. 2015;100:1301–1310. doi: 10.3324/haematol.2015.130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora A, Goulden N, Wade R, Mitchell C, Hancock J, Hough R, Rowntree C, Richards S. Treatment reduction for children and young adults with low-risk acute lymphoblastic leukaemia defined by minimal residual disease (UKALL 2003): a randomised controlled trial. Lancet Oncol. 2013;14:199–209. doi: 10.1016/S1470-2045(12)70600-9. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris J.P.t., Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Wiemels J. Perspectives on the causes of childhood leukemia. Chem Biol Interact. 2012;196:59–67. doi: 10.1016/j.cbi.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SS, Dunsmore KP, Devidas M, Eisenberg N, Asselin BL, Wood BL, Leonard Rn MS, Murphy J, Gastier-Foster JM, Carroll AJ, Heerema NA, Loh ML, Raetz EA, Winick NJ, Carroll WL, Hunger SP. Safe integration of nelarabine into intensive chemotherapy in newly diagnosed T-cell acute lymphoblastic leukemia: Children's Oncology Group Study AALL0434. Pediatr Blood Cancer. 2015;62:1176–1183. doi: 10.1002/pbc.25470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood BL, Winter SS, Dunsmore KP, Devidas M, Chen S, Asselin B, Esiashvili N, Loh ML, Winick NJ, Carroll WL, Raetz EA, Hunger SP. T-Lymphoblastic Leukemia (T-ALL) Shows Excellent Outcome, Lack of Significance of the Early Thymic Precursor (ETP) Immunophenotype, and Validation of the Prognostic Value of End-Induction Minimal Residual Disease (MRD) in Children’s Oncology Group (COG) Study AALL0434. Blood. 2014;123 abstract 1. [Google Scholar]
- Yang JJ, Bhojwani D, Yang W, Cai X, Stocco G, Crews K, Wang J, Morrison D, Devidas M, Hunger SP, Willman CL, Raetz EA, Pui CH, Evans WE, Relling MV, Carroll WL. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112:4178–4183. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Cheng C, Yang W, Pei D, Cao X, Fan Y, Pounds SB, Neale G, Trevino LR, French D, Campana D, Downing JR, Evans WE, Pui CH, Devidas M, Bowman WP, Camitta BM, Willman CL, Davies SM, Borowitz MJ, Carroll WL, Hunger SP, Relling MV. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, Imoto N, Kohsaka S, Kunita A, Doi K, Sakura T, Yujiri T, Kondo E, Fujimaki K, Ueda Y, Aoyama Y, Ohtake S, Takita J, Sai E, Taniwaki M, Kurokawa M, Morishita S, Fukayama M, Kiyoi H, Miyazaki Y, Naoe T, Mano H. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet. 2016;48:569–574. doi: 10.1038/ng.3535. [DOI] [PubMed] [Google Scholar]
- Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, Tritapoe J, Hixon JA, Silveira AB, Cardoso BA, Sarmento LM, Correia N, Toribio ML, Kobarg J, Horstmann M, Pieters R, Brandalise SR, Ferrando AA, Meijerink JP, Durum SK, Yunes JA, Barata JT. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43:932–939. doi: 10.1038/ng.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, Wu G, Li Y, Wei L, Iacobucci I, Liu Y, Qu C, Mardis E, Wilson RK, Dick JE, Hunger SP, Loh ML, Downing JR, Mullighan CG. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016 doi: 10.1038/ng.3691. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Churpek JE, Keel SB, Walsh T, Lee MK, Loeb KR, Gulsuner S, Pritchard CC, Sanchez-Bonilla M, Delrow JJ, Basom RS, Forouhar M, Gyurkocza B, Schwartz BS, Neistadt B, Marquez R, Mariani CJ, Coats SA, Hofmann I, Lindsley RC, Williams DA, Abkowitz JL, Horwitz MS, King MC, Godley LA, Shimamura A. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47:180–185. doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]