Abstract
Real-time PCR has become an important method for the rapid identification of Bacillus anthracis since the 2001 anthrax mailings. Most real-time PCR assays for B. anthracis have been developed to detect virulence genes located on the pXO1 and pXO2 plasmids. In contrast, only two published chromosomal targets exist, the rpoB gene and the gyrA gene. In the present study, subtraction-hybridization with a plasmid-cured B. anthracis tester strain and a Bacillus cereus driver was used to find a unique chromosomal sequence. By targeting this region, a real-time assay was developed with the Ruggedized Advanced Pathogen Identification Device. Further testing has revealed that the assay has 100% sensitivity and 100% specificity, with a limit of detection of 50 fg of DNA. The results of a search for sequences with homology with the BLAST program demonstrated significant alignment to the recently published B. anthracis Ames strain, while an inquiry for protein sequence similarities indicated homology with an abhydrolase from B. anthracis strain A2012. The importance of this chromosomal assay will be to verify the presence of B. anthracis independently of plasmid occurrence.
Bacillus anthracis is a spore-forming gram-positive bacterium well known for its recent use as a bioterrorist agent. Identification of B. anthracis can be done clinically by Gram stain, colony morphology, and various biochemical tests (19). However, these methods are time-consuming, and more rapid tests, such as PCR, have been used to detect B. anthracis in clinical samples (20). Real-time PCR is preferred over conventional PCR methods for the identification of organisms because it is fast, is less labor-intensive, and adds the specificity of a probe. While real-time PCR assays have been used to identify B. anthracis on the basis of the virulence genes associated with the toxin-encoding plasmid (pXO1) and the capsule-encoding plasmid (pXO2) (11, 20, 22), a reliable chromosomal assay has not been developed. Chromosomal assays can be valuable tools when they are used in conjunction with virulence gene assays because they provide information on the genetic contexts of the pXO1 and pXO2 plasmids. While the chromosomal assays may not prove useful as initial screening assays, they can certainly have a significant role in confirmatory testing as part of an integrated diagnostic approach.
Past attempts to develop a chromosomal real-time PCR assay have failed due to the close genetic relationship of Bacillus species. B. anthracis, Bacillus cereus, and Bacillus thuringiensis have very little variability and are genetically indistinguishable by multilocus enzyme electrophoresis (10). Recent work by repetitive PCR has shown that the previously listed species of Bacillus, as well as Bacillus mycoides, Bacillus pseudomycoides, and Bacillus weihenstephanensis, do have some genetic differences (3). Real-time PCR assays based on the chromosomal rpoB and gyrA genes of B. anthracis have been developed (5, 13, 25). However, these assays are based on single-nucleotide differences between B. anthracis and other Bacillus species. In our hands, the performance of assays based on point mutations has been notoriously dependent on assay conditions and cycling parameters, and small alterations in these conditions can result in the loss of specificity, especially with TaqMan-based probes.
Thus, a real-time PCR assay based on a unique chromosomal nucleotide sequence in B. anthracis is needed to complement other available assays for identification of the organism. Plasmids can be transmitted among bacterial species; therefore, a chromosomal marker to identify B. anthracis as the organism harboring the plasmids is important in a time of bioengineering and terrorist threats. While the presence of both pXO1 and pXO2 is needed to give B. anthracis its virulence, it is conceivable that these plasmids could be passed to its genetic neighbors, with unknown implications.
Bacterial subtraction has previously been used to find unique sequences in bacterial species (6, 8, 24, 26). In this study, nucleic acid isolated from B. cereus ATCC 21769 and genomic DNA from plasmid-cured B. anthracis strain Delta ANR-SWS (dANR) was used in a genomic subtraction-hybridization to identify possible chromosomal sequences unique to B. anthracis that could be used to develop a real-time PCR assay.
MATERIALS AND METHODS
Nucleic acid isolation.
DNA from B. anthracis dANR and B. cereus ATCC 21769 (BACI177) were extracted by the use of QIAamp DNA mini kits (Qiagen, Valencia, Calif.), as follows. An overnight culture of cells was pelleted by centrifugation at 5,000 × g for 10 min, and the pellet was resuspended in 180 μl of Dulbecco's phosphate-buffered saline (Gibco BRL, Rockville, Md.). Next, 20 μl of proteinase K (Qiagen) and 100 μl of RNase A (4 mg/ml; Promega, Madison, Wis.) were added to the cell suspension and the mixture was incubated at room temperature for 15 min. A 200-μl volume of AL buffer was added to the tubes containing the cell suspension, and the contents were mixed and then incubated at 55°C for 1 h. Then, 210 μl of 100% ethanol was added to the lysed cells and the mixture was vortexed. This mixture was transferred to a QIAamp spin column in a collection tube and centrifuged at 6,000 × g for 2 min. The spin column was transferred to a new collection tube, 500 μl of AW1 buffer was added to the spin column, and the tubes were centrifuged at 6,000 × g for 2 min. The spin column was transferred to another new collection tube, 500 μl of AW2 buffer was added, and the tubes were centrifuged at 6,000 × g for 2 min. The spin column was then transferred to a 1.5-ml Eppendorf tube, 50 μl of preheated (70°C) AE buffer was added to the spin column, and the tubes were incubated at 70°C for 5 min. The spin column and Eppendorf tube were spun at 6,000 × g for 1 min to elute the DNA. To assess the quality of the DNA prior to the subtraction, 2 μl of each DNA sample was analyzed by gel electrophoresis on a 1% agarose gel run at 90 V for 65 min.
Bacterial subtraction.
A PCR-Select Bacterial Genome Subtraction kit (Clontech, Palo Alto, Calif.) was used to identify sequences unique to B. anthracis. Strain BACI177 was chosen for use as the driver DNA, and strain dANR was chosen for use as the tester. The genomic Escherichia coli provided in the kit was used as a control. The majority of the subtraction-hybridization was performed according to the manufacturer's specifications, but the modifications described below were used. Restriction enzyme digestion was performed according to the instructions provided with the kit, and the digest was analyzed for efficiency by gel electrophoresis, as described above. The digested nucleic acid was purified, and adaptor ligations were performed with the tester and the control DNA. Ligation efficiency analysis was performed by PCR with the following components: 35.5 μl of molecular biology-grade (MBG) H2O, 2 μl of a 10 mM deoxynucleoside triphosphate (dNTP) mix (Roche, Indianapolis, Ind.), 5 μl of 10× PCR buffer, 4 μl of 25 mM MgCl2, 0.5 μl of AmpliTaq Gold (Applied Biosystems, Foster City, Calif.), 1 μl of each primer, and 1 μl of ligated DNA per 50-μl reaction mixture. The primers used with the tester DNA were 704F (5′-AGATTTTCCGACGGCAGGTT-3′) at a concentration of 50 pmol/μl and 829R (5′-TTTCAATCAATCGCGCCTTATT-3′) at a concentration of 50 pmol/μl; these primer sequences span a region of the gyrA gene in B. anthracis. One tester DNA reaction mixture incorporated primer 704F with PCR primer 1 at a concentration of 0.2 μM/μl, while primer 704F and primer 829R were used in the other tester DNA reactions. The PCR conditions were 72°C for 2 min and 95°C for 10 min, followed by 35 cycles of 95°C for 2 min, 60°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min. The PCRs were performed with an MJ Research (Waltham, Mass.) PTC-100 thermal cycler. The DNA products were analyzed by gel electrophoresis on a 2% gel run at 150 V for 30 min. Following the ligation efficiency analysis, the two hybridization steps were performed at an incubation temperature of 62°C for both the tester DNA and the driver DNA due to their lower G+C contents. The PCRs used to amplify tester nucleic acid followed the manufacturer's instructions, with the exception that the nested PCR was run for 17 cycles, and were performed on an MJ Research PTC-100 thermal cycler. The DNA products were analyzed by gel electrophoresis, as described directly above.
Bacterial cloning.
The nested PCR products subtracted from strain dANR were cloned into the pCR2.1-TOPO vector by using the TOPO TA Cloning kit (Invitrogen, Carlsbad, Calif.), according to the manufacturer's instructions for chemical transformation. White colonies were selected and boiled in 50 μl of H2O for 10 min to break up the cells and release the nucleic acid. PCR was performed with the following components per 100-μl reaction mixture: 71 μl of MBG H2O, 4 μl of a 10 mM dNTP mixture, 10 μl of 10× PCR buffer, 8 μl of 25 mM MgCl2, 1 μl of AmpliTaq Gold, 2 μl of M13 forward primer (50 pmol/μl; Invitrogen), 2 μl of M13 reverse primer (50 pmol/μl; Invitrogen), and 2 μl of supernatant from the boiled preparation. PCR conditions were 72°C for 2 min and 95°C for 10 min, followed by 30 cycles of 95°C for 2 min, 55°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min. The PCRs were performed with an MJ Research PTC-100 thermal cycler. PCR products were analyzed by gel electrophoresis, as described above. Clones with positive inserts were selected for sequencing and further analysis.
Bacterial sequencing.
PCR products from the selected clones were purified with the Montage PCR96 cleanup kit (Millipore, Billerica, Mass.). Sequencing reactions were carried out with 8 μl of Big Dye (Applied Biosystems), 1 μl of M13 forward primer (50 pmol/μl) or 1 μl of M13 reverse primer (50 pmol/μl; Invitrogen), 1 μl of purified PCR product, and 10 μl of MBG H2O. The sequencing reactions were cycled on an MJ Research PTC-100 thermal cycler at 85°C for 30 s, followed by 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min, with a final extension at 60°C for 10 min. Sequencing was performed on an ABI 3100 sequencer (Applied Biosystems), and the sequences were aligned by using DNASTAR software (DNASTAR, Inc., Madison, Wis.). Searches with the BLAST program were conducted for all sequences to eliminate regions not unique to B. anthracis and those with possible cross-reactivities with other organisms. A single clone (clone B26) was picked for further analysis.
Primer and probe design and testing.
Prospective primers and probes were generated with Primer Express software (version 2.0; Applied Biosystems) and the imported sequence from clone B26. Primers were further analyzed for palindromes, hairpins, dimer formation, and annealing temperature with NetPrimer software (BioSoft International, Palo Alto, Calif.).
Primer testing by standard PCR.
Two forward primers and two reverse primers were assessed by standard PCR on an MJ Research PTC-100 thermal cycler with the following components per 20-μl reaction mixture: 10.44 μl of MBG H2O, 2 μl of 10× 2 mM dNTPs (Idaho Technology, Salt Lake City, Utah), 2 μl of 10× PCR buffer containing 50 mM MgCl2 (Idaho Technology), 0.2 μl of forward primer (50 pmol/μl), 0.2 μl of reverse primer (50 pmol/μl), 0.16 μl of of Platinum Taq DNA polymerase (5 U/μl; Invitrogen), and 5 μl of Bacillus species DNA at either 1 ng/μl or 100 pg/μl. The reaction mixtures were cycled at 94°C for 2 min, followed by 34 cycles of 94°C for 30 s, 60°C for 30 min, and 72°C for 1 min, with a final extension at 72°C for 10 min, on an MJ Research PTC-100 thermal cycler. Reactions were analyzed by gel electrophoresis as described above. Primer F41 (5′-TGGCGGAAAAGCTAATATAGTAAAGTA-3′) and primer R146 (5′-CCACATATCGAATCTCCTGTCTAAAA-3′) were selected for further testing. Forty-two B. anthracis strains and 53 Bacillus species were tested by PCR under the conditions defined above. The reaction products were analyzed by gel electrophoresis, as described above.
Probe testing by real-time PCR.
Two Taqman minor groove binder (MGB) probes were tested by real-time PCR on the Ruggedized Advanced Pathogen Identification Device (RAPID; Idaho Technology) with primers F41 and R146. The background fluorescence of the probes was obtained by diluting the 100 μM MGB Taqman probes to 1 μM so that the fluorescence was between 10 and 20 with a gain setting of 16. The probes were tested against 16 B. anthracis isolates and 13 B. cereus isolates. The master mixture contained 8.44 μl of MBG H2O, 2 μl of 10× 2 mM dNTPs, 2 μl of 10× PCR buffer containing 50 mM MgCl2, 0.2 μl of primer F41 (50 pmol/μl), 0.2 μl of primer R146 (50 pmol/μl), 2 μl of 1 μM probe, 0.16 μl of Platinum Taq (5 U/μl), and 5 μl of DNA (1 ng/μl) per 20-μl reaction mixture. The PCR cycling conditions for all runs on the RAPID were 94°C for 2 min, followed by 45 cycles of 94°C for 20 s and 60°C for 20 s. Probe BALB26588-MGB (P88; 5′-ACTTCTAAAAAGCAGATAGAAAT-3′) was chosen for optimization of the RAPID. Five concentrations of MgCl2 (3, 4, 5, 6, and 7 mM) and 10 concentrations of primers F41 and R146 (0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, and 1.0 μM) were tested in triplicate with 1 pg of B. anthracis Ames DNA. The combination that exhibited the earliest crossing threshold (CT) and highest end fluorescence (Ef) was chosen as the optimal condition, which consisted of 5 mM MgCl2 and 0.7 μM each primer. The optimized master mixture contained 8.28 μl of H2O, 2 μl of 10× 2 mM dNTPs, 2 μl of 10× PCR buffer containing 50 mM MgCl2, 0.28 μl of primer F41 (50 pmol/μl), 0.28 μl of primer R146 (50 pmol/μl), 2 μl of probe P88 (1 mM), and 0.16 μl of Platinum Taq (5 U/μl) per 20-μl reaction mixture. Tenfold dilutions of Ames strain DNA from 4 pg/μl to 0.4 fg/μl were made so that 2.5 μl of each dilution along with 2.5 μl of human DNA (5 ng) or 2.5 μl of H2O could be added to 15 μl of the optimized master mixture.
Cross-reactivity and other tests under real-time optimized conditions.
A panel of 77 organisms was run by real-time PCR with the optimized conditions to test for cross-reactivity. The master mixture contained 5 μl of DNA (20 pg/μl) isolated from each organism. Sensitivity and specificity testing was also accomplished as described above by using 45 B. anthracis strains and 62 additional Bacillus species.
Limit of detection and efficacy of the assay.
The limit of detection of the assay was established by running 60 samples of strain Ames DNA at a single concentration with two negative controls. The lowest concentration that was positive for at least 57 of the 60 samples (95%) was determined to be the limit of detection. The efficacy, or efficiency, of the PCR assay was determined by testing 10-fold serial dilutions from 100 pg to 10 fg in triplicate. The crossing point of each dilution was determined by the second derivative method in the LightCycler software (version 3.3; Roche). Amplification efficacy (E) was calculated on the basis of the slope of the crossing point versus the input DNA concentration by the equation E = 10(−1/slope) (23).
RESULTS
Bacterial subtraction, cloning, and sequencing to obtain a unique chromosomal sequence.
Bacterial subtraction was used to find chromosomal DNA sequences specific to B. anthracis. Figure 1 shows the results of subtraction after nested PCR and shows the bands present in unsubtracted dANR that do not occur in subtracted dANR. A prominent band of approximately 1,000 bp existed in the subtracted dANR lane. The entire PCR product from subtracted dANR was cloned and analyzed by PCR with M13 primers. These primer sequences flank the inserted region of the PCR product on the pCR2.1-TOPO vector. Four clones generated a PCR product with an insert of about 1,000 bp, and the product was sequenced and was determined to be unique. All searches with the BLAST program exhibited low bit scores and E values, but clone B26 presented the least amount of homologous hits; therefore, it was selected for continued testing.
FIG. 1.
DNA products following nested PCR. Lanes 1 and 8, DNA PCR ladder (Gene Choice, Frederick, Md.); lane 2, subtracted B. anthracis dANR; lane 3, unsubtracted B. anthracis dANR; lane 4, subtracted E. coli; lane 5, unsubtracted E. coli; lane 6, subtracted control E. coli; lane 7, empty. The values at the sides of the gel are in base pairs.
Primer testing by standard PCR.
Primer sequences were designed from a portion of the B26 sequence. Initial testing was performed with two forward primers and two reverse primers in four different combinations. All combinations produced amplification products, but the primer combination F41 and R146 generated the largest amount of DNA and was selected to test 42 B. anthracis isolates and 53 Bacillus species. All 42 B. anthracis isolates produced an amplicon of about 100 bp. In contrast, among the other Bacillus species tested, only B. cereus ATCC 21771 (BACI179) generated any product, which was a very faint band at 1,000 bp (data not shown).
Probe testing by real-time PCR.
After the primers were tested and selected by standard PCR, two probes previously designed with the primers were tested against 16 B. anthracis and 13 B. cereus strains with the RAPID. The specificity and sensitivity were each 100% for each probe tested. Both probes generated similar CT values for the samples tested, but P88 generated a higher Ef for the samples tested, so it was chosen for optimization. After optimization of the MgCl2 and primer concentrations, serial 10-fold dilutions made with strain Ames DNA gave positive results to 1 fg in triplicate assays (data not shown). When the reactions were spiked with 5 ng of human DNA, positive results to 1 fg in triplicate assays were still obtained, with no changes in CT or Ef observed.
Cross-reactivity panel.
The optimized assay was tested with a cross-reactivity panel of 76 organisms (Table 1), including 11 B. anthracis isolates. All 11 isolates were detected, regardless of the plasmid profile, by the chromosomal marker test, while the remaining 66 organisms failed to produce a signal. In addition, sensitivity and specificity were evaluated by running 45 B. anthracis isolates and 62 Bacillus species by the optimized assay (these isolates included the original 16 B. anthracis isolates and 13 B. cereus isolates tested), and the results are shown in Table 2. All 45 B. anthracis strains were found to be positive, and all of the other 62 Bacillus species were negative, including BACI179, providing 100% sensitivity and specificity.
TABLE 1.
Organisms used to test cross-reactivity by the real-time chromosomal PCR assay
| Organism | ATCC accession no.a |
|---|---|
| Acinetobacter baumannii | 19606 |
| Alcaligenes xylosoxidans | 27061 |
| Bacillus anthracis Delta Sterne | NA |
| Bacillus anthracis 4728 | |
| Bacillus anthracis Delta-NH-1 | NA |
| Bacillus anthracis ST1 | NA |
| Bacillus anthracis NH | NA |
| Bacillus anthracis Vollum | NA |
| Bacillus anthracis SPS 97.13.079 | NA |
| Bacillus anthracis Ames | NA |
| Bacillus anthracis V770-NP-1R | NA |
| Bacillus anthracis SPS 97.13.213 | NA |
| Bacillus anthracis Sterne | NA |
| Bacillus cereus | 10876 |
| Bacillus cereus | 13061 |
| Bacillus coagulans | 7050 |
| Bacillus licheniformis | 12759 |
| Bacillus macerans (Paenibacillus macerans) | 8244 |
| Bacillus megaterium | NA |
| Bacillus polymyxa | 842 |
| Bacillus popilliae | 14706 |
| Bacillus sphaericus | 4525 |
| Bacillus stearothermophilus | 7953 |
| Bacillus subtilis var. niger | NA |
| Bacillus subtilis var. niger | 6633 |
| Bacillus subtilis var. niger | 9372 |
| Bacillus thuringiensis | 39152 |
| Bacillus thuringiensis | 35646 |
| Bacillus thuringiensis | NA |
| Bacteroides distasonis | 8503 |
| Bordetella bronchiseptica | 10580 |
| Budvicia aquatica | 35567 |
| Burkholderia cepacia | 25416 |
| Burkholderia pseudomallei | NA |
| Clostridium perfringens | 13124 |
| Clostridium sporogenes | 3584 |
| Comanonas acidovorans | 15668 |
| Enterococcus durans | 6056 |
| Enterococcus faecalis | 29212 |
| Enterococcus faecalis | 700802D |
| Escherichia coli | 25922 |
| Francisella tularensis | NA |
| Francisella tularensis | NA |
| Francisella tularensis | NA |
| Francisella tularensis | NA |
| Francisella tularensis | NA |
| Francisella tularensis | NA |
| Haemophilus influenzae | 10211 |
| Haemophilus influenzae | 51907D |
| Klebsiella pneumoniae | 13883 |
| Klebsiella pneumoniae subsp. pneumoniae | 700721D |
| Listeria monocytogenes | 15313 |
| Moraxella cattaharalis | 25240 |
| Neisseria lactamica | 23970 |
| Proteus mirabilis | 7002 |
| Proteus vulgaris | 49132 |
| Providencia stuartii | 33672 |
| Pseudomonas aeruginosa | 17933D |
| Ralstonia pickettii | 27511 |
| Salmonella choleraesuis subsp. choleraesuis serotype Paratyphi | 9150D |
| Serratia odorifera | 33077 |
| Shigella flexneri | 12022 |
| Shigella sonnei | 9290 |
| Staphylococcus aureus | 29247 |
| Staphylococcus aureus subsp. aureus | 35556D |
| Staphylococcus hominis | 27844 |
| Stenotrophomonas maltophilia | 13637 |
| Streptococcus pneumoniae | 33400 |
| Streptococcus pyogenes | 19615 |
| Streptococcus pyogenes | 12344D |
| Yersinia enterocolitica | NA |
| Yersinia frederiksenii | 33641 |
| Yersinia kristensenii | 33639 |
| Yersinia pestis (CO92; PW) | NA |
| Yersinia pseudotuberculosis | 6904 |
| Yersinia ruckeri | 29473 |
ATCC, American Type Culture Collection; NA, not applicable.
TABLE 2.
Bacillus species used for sensitivity and specificity testing of the real-time chromosomal PCR assay
| Organism | Strain | ATCC accession no. | PA | Cap | |
|---|---|---|---|---|---|
| Bacillus anthracis | CDC 471 | NEG | NEG | ||
| Bacillus anthracis | Delta-UM23-1-1 | NEG | NEG | ||
| Bacillus anthracis | 183 | NEG | NEG | ||
| Bacillus anthracis | Delta-ANR-SWS | NEG | NEG | ||
| Bacillus anthracis | Delta Sterne | NEG | NEG | ||
| Bacillus anthracis | Delta-NH-1 | NEG | POS | ||
| Bacillus anthracis | 4229 | NEG | POS | ||
| Bacillus anthracis | 1928 | NEG | POS | ||
| Bacillus anthracis | 4728 | NEG | POS | ||
| Bacillus anthracis | 107 | NEG | POS | ||
| Bacillus anthracis | V770-NP-1R | POS | NEG | ||
| Bacillus anthracis | ST1 | POS | NEG | ||
| Bacillus anthracis | Arkansas | POS | NEG | ||
| Bacillus anthracis | SK-465 | POS | NEG | ||
| Bacillus anthracis | SK-102 | POS | NEG | ||
| Bacillus anthracis | SK-61 | POS | NEG | ||
| Bacillus anthracis | ST-15 | POS | NEG | ||
| Bacillus anthracis | Ger. LVS | POS | NEG | ||
| Bacillus anthracis | M | POS | NEG | ||
| Bacillus anthracis | Sterne (British) | POS | NEG | ||
| Bacillus anthracis | V770-NP1-R | POS | NEG | ||
| Bacillus anthracis | SPS 97.13.213 | POS | NEG | ||
| Bacillus anthracis | CDC 607 | POS | POS | ||
| Bacillus anthracis | Delta-Ames-1 | POS | POS | ||
| Bacillus anthracis | Vollum 1 | POS | POS | ||
| Bacillus anthracis | Vollum 1B | POS | POS | ||
| Bacillus anthracis | English Vollum | POS | POS | ||
| Bacillus anthracis | Ames | POS | POS | ||
| Bacillus anthracis | Buffalo | POS | POS | ||
| Bacillus anthracis | NH | POS | POS | ||
| Bacillus anthracis | V770-2P | POS | POS | ||
| Bacillus anthracis | FLA-V770 | POS | POS | ||
| Bacillus anthracis | V770 | POS | POS | ||
| Bacillus anthracis | 108 | POS | POS | ||
| Bacillus anthracis | 205 | POS | POS | ||
| Bacillus anthracis | 57 | POS | POS | ||
| Bacillus anthracis | N994 | POS | POS | ||
| Bacillus anthracis | SK-162 | POS | POS | ||
| Bacillus anthracis | SK-128 | POS | POS | ||
| Bacillus anthracis | N-99 | POS | POS | ||
| Bacillus anthracis | SK-31 | POS | POS | ||
| Bacillus anthracis | G-28 | POS | POS | ||
| Bacillus anthracis | SPS 97.13.079 | POS | POS | ||
| Bacillus anthracis | NH | POS | POS | ||
| Bacillus anthracis | Vollum | POS | POS | ||
| Bacillus cereus | 7039 | NEG | NEG | ||
| Bacillus cereus | 12480 | NEG | NEG | ||
| Bacillus cereus | 13472 | NEG | NEG | ||
| Bacillus cereus | 13824 | NEG | NEG | ||
| Bacillus cereus | 14603 | NEG | NEG | ||
| Bacillus cereus | 14893 | NEG | NEG | ||
| Bacillus cereus | 15816 | NEG | NEG | ||
| Bacillus cereus | 19625 | NEG | NEG | ||
| Bacillus cereus | 19637 | NEG | NEG | ||
| Bacillus cereus | 21182 | NEG | NEG | ||
| Bacillus cereus | 21366 | NEG | NEG | ||
| Bacillus cereus | 21634 | NEG | NEG | ||
| Bacillus cereus | 21768 | NEG | NEG | ||
| Bacillus cereus | 21769 | NEG | NEG | ||
| Bacillus cereus | 21770 | NEG | NEG | ||
| Bacillus cereus | 21771 | NEG | NEG | ||
| Bacillus cereus | 21772 | NEG | NEG | ||
| Bacillus cereus | 21928 | NEG | NEG | ||
| Bacillus cereus | 25621 | NEG | NEG | ||
| Bacillus cereus | 27348 | NEG | NEG | ||
| Bacillus cereus | 27522 | NEG | NEG | ||
| Bacillus cereus | 27877 | NEG | NEG | ||
| Bacillus cereus | 31293 | NEG | NEG | ||
| Bacillus cereus | 31429 | NEG | NEG | ||
| Bacillus cereus | 31430 | NEG | NEG | ||
| Bacillus cereus | 33018 | NEG | NEG | ||
| Bacillus cereus | 33019 | NEG | NEG | ||
| Bacillus cereus | 43881 | NEG | NEG | ||
| Bacillus cereus | 53522 | NEG | NEG | ||
| Bacillus cereus | 55055 | NEG | NEG | ||
| Bacillus cereus | 700282 | NEG | NEG | ||
| Bacillus cereus | 9139 | NEG | NEG | ||
| Bacillus cereus | 9818 | NEG | NEG | ||
| Bacillus cereus | 10876 | NEG | NEG | ||
| Bacillus cereus | 13061 | NEG | NEG | ||
| Bacillus cereus | 10987 | NEG | NEG | ||
| Bacillus thuringiensis | 10792 | NEG | NEG | ||
| Bacillus thuringiensis | 13366 | NEG | NEG | ||
| Bacillus thuringiensis | 13367 | NEG | NEG | ||
| Bacillus thuringiensis | 19266 | NEG | NEG | ||
| Bacillus thuringiensis | 19267 | NEG | NEG | ||
| Bacillus thuringiensis | 19268 | NEG | NEG | ||
| Bacillus thuringiensis | 19269 | NEG | NEG | ||
| Bacillus thuringiensis | 19270 | NEG | NEG | ||
| Bacillus thuringiensis | 29730 | NEG | NEG | ||
| Bacillus thuringiensis | 33679 | NEG | NEG | ||
| Bacillus thuringiensis | 39152 | NEG | NEG | ||
| Bacillus thuringiensis | 35646 | NEG | NEG | ||
| Bacillus thuringiensis | NA | NEG | NEG | ||
| Bacillus subtilis | 6633 | NEG | NEG | ||
| Bacillus subtilis var. niger | NA | NEG | NEG | ||
| Bacillus subtilis var. niger | 9372 | NEG | NEG | ||
| Bacillus macerans | 8244 | NEG | NEG | ||
| Bacillus stearothermophilus | 7953 | NEG | NEG | ||
| Bacillus popilliae | 14706 | NEG | NEG | ||
| Bacillus coagulane | 7050 | NEG | NEG | ||
| Bacillus licheniforms | 12759 | NEG | NEG | ||
| Bacillus halodurans | 21591D | NEG | NEG | ||
| Bacillus brevis | 8246 | NEG | NEG | ||
| Bacillus mycoides | 31101 | NEG | NEG | ||
| Bacillus mycoides | 21929 | NEG | NEG | ||
| Bacillus species | 813+ #11 | NEG | NEG |
Abbreviations: ATCC, American Type Culture Collection; PA, protective antigen protein encoded by the pXO1 plasmid; Cap, capsule proteins encoded by the pXO2 plasmid; NA, not available; POS, positive; NEG, negative.
Limit of detection and efficacy.
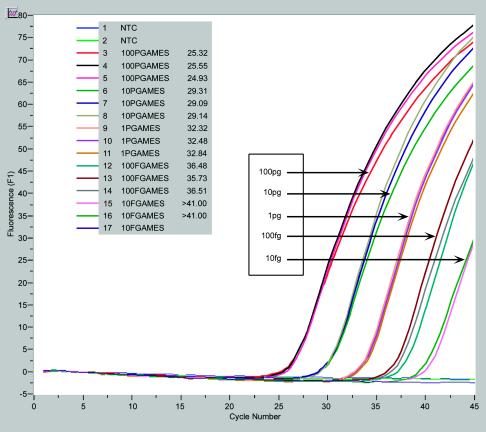
The chromosomal assay developed resulted in positive results when as little as 1 fg of DNA was incorporated into the reaction mixture. However, the limit of detection was determined to be 50 fg because all 60 samples tested simultaneously at this concentration produced a signal. At 40 fg only 47 of 60 (78.3%) samples produced a signal. Furthermore, an amplification efficacy value of 1.896 was determined, and the assay used to determine this value is depicted in Fig. 2. For an assay to produce unambiguous and identical results, its amplification efficacy should be as close as possible to 2. This value corresponds to a doubling of the template after each PCR cycle.
FIG. 2.
Real-time PCR results for determination of efficacy. NTC, no template control; PGAMES, picograms of strain Ames; FGAMES, femtograms of strain Ames.
DISCUSSION
Differentiation of B. anthracis from its close relatives has traditionally relied upon phenotypic characterization performed in a clinical setting (12). However, these methods can often take from 24 to 48 h to complete (19). Recently, increased awareness about biological weapons and the use of anthrax as a bioterrorist agent have led to the necessity for improved and more rapid methods of identification (12, 14, 30). Real-time PCR assays may offer an improved means of identification of B. anthracis due to their speed, specificity, sensitivity, and throughput. After the 2001 anthrax spore mailings, many of these rapid PCR assays developed for the detection of B. anthracis were used to confirm the presence or absence of the organism (11).
The importance of designing a chromosomal assay for B. anthracis has arisen due to the discovery of isolates cured of plasmids pX01 and pX02 (27, 31). Identification of avirulent B. anthracis strains could be a potential indicator of virulent strains in the environment (31). Real-time PCR based on chromosomal markers could provide a powerful tool for identifying B. anthracis strains either containing or lacking virulence plasmids. To date, the majority of B. anthracis targets for real-time PCR assays have been developed from genes located on the pXO1 and pXO2 plasmids (2, 7, 20). These assays are specific because the targets code for the toxins and capsule proteins essential for virulence. However, complete identification of B. anthracis should involve the detection of both plasmids and a chromosomal marker due to the possibility of the transmission, transfer, or loss of one or both plasmids between Bacillus species seen both in vivo and in vitro (18, 27, 29, 31). Bioengineering may also allow the transfer of virulence plasmids to other species, including bioengineered organisms (12), exemplifying the need to detect these agents made by humans.
The development of chromosomal real-time PCR assays has been slow due to the genetic similarities between B. anthracis and its neighbors (10). To date there are very few chromosomal markers to distinguish B. anthracis from its close relatives. Analysis of the 16S rRNA genes from B. anthracis, B. cereus, B. mycoides, and B. thuringiensis revealed that they have almost identical sequences (9); and some argue that due to the similarity of the B. anthracis, B. thuringiensis, and B. cereus genomes, they should be considered to belong to the same species (10). Some chromosomal markers are available, such as the Ba813 fragment (21) and the vrrA variable number tandem repeat (1, 15, 17); however, these markers have failed because they are not specific to B. anthracis (4, 12), and assays incorporating these targets require extra steps after the PCR, making them impractical for the rapid identification of B. anthracis (30). Recently, the rpoB and gyrA genes, located on the B. anthracis chromosome, have been targeted for the development of a real-time PCR assay (13, 25). These assays are based on theoretical differences in melting temperatures due to single-nucleotide differences, but in practice, false-positive results with other Bacillus species have occurred (7, 25). In fact, Bacillus sp. strain Ba813+ #11, which was found to be positive by the published rpoB assay (25), was negative by the chromosomal assay developed here. The present assay has the potential for increased specificity because it is not dependent on probable point mutations or theoretical differences in melting temperatures, and all primers and probes are based on a sequence unique to B. anthracis.
The assay developed in this study is based on a chromosomal sequence unique to B. anthracis. This region was found by subtractive hybridization incorporating a plasmid-cured B. anthracis (tester) strain and a B. cereus (driver) isolate. Previous reports have shown success with the use of this method to find sequences in the tester strain's genome that are absent in the driver (8, 24), and recently, it has been used to find unique targets in B. anthracis (26). The forward and reverse primers as well as the reverse complements from the previous report (26) were checked for alignment with our sequence, without success. By the use of subtractive hybridization, a sequence unique to B. anthracis was found and was used to develop a chromosomal real-time PCR assay that had 100% sensitivity in tests with all B. anthracis strains studied and 100% specificity when it was tested with other Bacillus species. In addition, a cross-reactivity panel of 76 organisms that included 27 Bacillus strains was used to test the sensitivity and the specificity of the assay. The organisms in this panel were chosen because of their genetic proximity to B. anthracis and their possible similar environmental and clinical niches with B. anthracis.
The recent release of the B. anthracis genome allowed the comparison of the B26 clone sequence with this new sequence information by use of the BLAST program. Forward primer F41 and probe P88 produced significant alignments with B. anthracis strain Ames and strain Sterne, having the highest scores compared to any other alignment and E values of 7 × 10−4. Reverse primer R146 also produced the highest scores with B. anthracis strain Ames and strain Sterne, with E values of 2 × 10−5; however, an alignment to B. cereus ATCC 10987 with an E value of 0.005 also occurred. This B. cereus strain has been tested and was not detected by the assay. A search of the assay sequence and surrounding nucleotides (527 nucleotides in total) against all protein sequences with the BLAST program produced a high score of 191, with an E value of 5 × 10−48 for the homology of B. anthracis (GenBank accession number NP_656566.1) with an abhydrolase (alpha/beta-hydrolase) fold. The 106-bp product produced in this investigation is just outside the open reading frame of this protein. Other significant alignments included a prolyl aminopeptidase of an Oceanobacillus iheyensis strain with a score of 115 and an E value of 2 × 10−29 and a proline iminopeptidase of B. cereus ATCC 14579 with a score of 94 and an E value of 4 × 10−21.
Further testing with other B. anthracis strains from other sources must be performed to ensure that this sequence is conserved between additional B. anthracis strains. In addition, the phenotypic necessity of this genetic region has yet to be determined. While amplified fragment length polymorphism analysis (16) has shown little variability among B. anthracis isolates, despite the diversity that exists, previous work with multiple-locus variable number tandem repeat analysis has classified B. anthracis into 89 genotypes (17). In our study, genotype information was available for 17 of the 45 B. anthracis strains studied. These 17 B. anthracis strains represented 8 of the 89 genotypes. Recent sequencing of a B. anthracis isolate from a victim of the 2001 anthrax attack has revealed single-nucleotide polymorphisms, insertions, deletions, and changes in tandem repeats when the strain's sequence is compared to those of reference B. anthracis strains (28). It is conceivable that the region targeted in this study may not be conserved. Genetic variation and molecular diversity in sequences could potentially decrease the sensitivity and the specificity of this particular assay. However, molecular identification of B. anthracis is a polyphasic process, requiring assays for pXO1, pXO2, and the chromosome. Furthermore, an integrated diagnostic approach that uses multiple genetic markers decreases the risk of technological surprise. The assay developed in this study could be an important component of an integrated diagnostic system for the detection of B. anthracis.
The present findings demonstrate that the assay developed is sensitive and specific for the identification of the B. anthracis chromosome. The assay did not cross-react with any genetic neighbors available for the study, and all strains of B. anthracis were identified. The assay was independent of the plasmid profile of the strains tested and had a DNA detection limit of 50 fg. In summary, the assay developed from the sequence identified in this study is a rapid, sensitive, and specific tool for the identification of B. anthracis. The assay can be used to complement assays for virulence markers by providing the genetic background of the organism containing plasmid pXO1 or pXO2, or both.
Acknowledgments
We thank Gerry Howe, Brian Walker, David Kulesh, and Bruce Ivins for technical assistance; Melissa Frye and Melanie Ulrich for critical suggestions on the manuscript; Guy Patra and Vito DelVecchio for providing us with Bacillus sp. strain Ba813+ #11; and everyone in the Diagnostic System Division for their significant support with this project.
The assay described in this report has a provisional patent pending as of the date of publication.
REFERENCES
- 1.Andersen, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, C. A., J. R. Uhl, T. L. Hadfield, J. C. David, R. F. Meyer, T. F. Smith, and F. R. Cockerill III. 2002. Detection of Bacillus anthracis DNA by LightCycler PCR. J. Clin. Microbiol. 40:2897-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherif, A., L. Brusetti, S. Borin, A. Rizzi, A. Boudabous, H. Khyami-Horani, and D. Daffonchio. 2003. Genetic relationship in the ‘Bacillus cereus group’ by rep-PCR fingerprinting and sequencing of a Bacillus anthracis-specific rep-PCR fragment. J. Appl. Microbiol. 94:1108-1119. [DOI] [PubMed] [Google Scholar]
- 4.Daffonchio, D., S. Borin, G. Frova, R. Gallo, E. Mori, R. Fani, and C. Sorlini. 1999. A randomly amplified polymorphic DNA marker specific for the Bacillus cereus group is diagnostic for Bacillus anthracis. Appl. Environ. Microbiol. 65:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drago, L., A. Lombardi, E. D. Vecchi, and M. R. Gismondo. 2002. Real-time PCR assay for rapid detection of Bacillus anthracis spores in clinical samples. J. Clin. Microbiol. 40:4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dwyer, K. G., J. M. Lamonica, J. A. Schumacher, L. E. Williams, J. Bishara, A. Lewandowski, R. Redkar, G. Patra, and V. G. DelVecchio. 2004. Identification of Bacillus anthracis specific chromosomal sequences by suppressive subtractive hybridization. BMC Genomics 5:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellerbrok, H., H. Nattermann, M. Ozel, L. Beutin, B. Appel, and G. Pauli. 2002. Rapid and sensitive identification of pathogenic and apathogenic Bacillus anthracis by real-time PCR. FEMS Microbiol. Lett. 214:51-59. [DOI] [PubMed] [Google Scholar]
- 8.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrell, L. J., G. L. Andersen, and K. H. Wilson. 1995. Genetic variability of Bacillus anthracis and related species. J. Clin. Microbiol. 33:1847-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins, J. A., M. Cooper, L. Schroeder-Tucker, S. Black, D. Miller, J. S. Karns, E. Manthey, R. Breeze, and M. L. Perdue. 2003. A field investigation of Bacillus anthracis contamination of U.S. Department of Agriculture and other Washington, D.C., buildings during the anthrax attack of October 2001. Appl. Environ. Microbiol. 69:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmaster, A. R., R. F. Meyer, M. D. Bowen, C. K. Marston, R. S. Weyant, K. Thurman, S. L. Messenger, E. E. Minor, J. M. Winchell, M. V. Rassmussen, B. R. Newton, J. T. Parker, W. E. Morrill, N. McKinney, G. A. Barnett, J. J. Sejvar, J. A. Jernigan, B. A. Perkins, and T. Popovic. 2002. Evaluation and validation of a real-time polymerase chain reaction assay for rapid identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurtle, W., E. Bode, D. A. Kulesh, R. S. Kaplan, J. Garrison, D. Bridge, M. House, M. S. Frye, B. Loveless, and D. Norwood. 2004. Detection of the Bacillus anthracis gyrA gene by using a minor groove binder probe. J. Clin. Microbiol. 42:179-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inglesby, T. V., D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Hauer, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, K. Tonat, et al. 1999. Anthrax as a biological weapon: medical and public health management. JAMA 281:1735-1745. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Andersen, K. H. Wilson, M. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keim, P., A. Kalif, J. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koehler, T. M. 2002. Bacillus anthracis genetics and virulence gene regulation. Curr. Top. Microbiol. Immunol. 271:143-164. [DOI] [PubMed] [Google Scholar]
- 19.Logan, N. A., and P. C. B. Turnbull. 2004. Bacillus and other aerobic endospore-forming bacteria, p. 445-460. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 20.Oggioni, M. R., F. Meacci, A. Carattoli, A. Ciervo, G. Orru, A. Cassone, and G. Pozzi. 2002. Protocol for real-time PCR identification of anthrax spores from nasal swabs after broth enrichment. J. Clin. Microbiol. 40:3956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patra, G., P. Sylvestre, V. Ramisse, J. Therasse, and J. L. Guesdon. 1996. Isolation of a specific chromosomic DNA sequence of Bacillus anthracis and its possible use in diagnosis. FEMS Immunol. Med. Microbiol. 15:223-231. [DOI] [PubMed] [Google Scholar]
- 22.Patra, G., L. E. Williams, Y. Qi, S. Rose, R. Redkar, and V. G. DelVecchio. 2002. Rapid genotyping of Bacillus anthracis strains by real-time polymerase chain reaction. Ann. N. Y. Acad. Sci. 969:106-111. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pradel, N., S. Leroy-Setrin, B. Joly, and V. Livrelli. 2002. Genomic subtraction to identify and characterize sequences of Shiga toxin-producing Escherichia coli O91:H21. Appl. Environ. Microbiol. 68:2316-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi, Y., G. Patra, X. Liang, L. E. Williams, S. Rose, R. J. Redkar, and V. G. DelVecchio. 2001. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl. Environ. Microbiol. 67:3720-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radnedge, L., P. G. Agron, K. K. Hill, P. J. Jackson, L. O. Ticknor, P. Keim, and G. L. Andersen. 2003. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 69:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramisse, V., G. Patra, H. Garrigue, J. L. Guesdon, and M. Mock. 1996. Identification and characterization of Bacillus anthracis by multiplex PCR analysis of sequences on plasmids pXO1 and pXO2 and chromosomal DNA. FEMS Microbiol. Lett. 145:9-16. [DOI] [PubMed] [Google Scholar]
- 28.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 29.Reddy, A., L. Battisti, and C. B. Thorne. 1987. Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J. Bacteriol. 169:5263-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards, C. F., J. L. Burstein, J. F. Waeckerle, and H. R. Hutson. 1999. Emergency physicians and biological terrorism. Ann. Emerg. Med. 34:183-190. [DOI] [PubMed] [Google Scholar]
- 31.Turnbull, P. C., R. A. Hutson, M. J. Ward, M. N. Jones, C. P. Quinn, N. J. Finnie, C. J. Duggleby, J. M. Kramer, and J. Melling. 1992. Bacillus anthracis but not always anthrax. J. Appl. Bacteriol. 72:21-28. [DOI] [PubMed] [Google Scholar]