Abstract
A Streptomyces albus forearm actinomycetoma that could not be identified by culture was properly identified at the species level by study of an internal fragment of the heat shock protein gene, comparative sequence analysis of 16S ribosomal DNA (rDNA), and the hypervariable γ-region of the 16S rDNA.
CASE REPORT
A 28-year-old male from Oviedo, Asturias, Spain, who was an unemployed bricklayer who had carried out gardening work in the previous few months, was seen in the Casualty Unit of the University Hospital. He had two very painful, swollen, red, nodular lesions of 3 by 3 cm, which had developed over a period of 1 month, located on the internal face of the left forearm.
The patient had been diagnosed and treated for lung tuberculosis 14 years before and for Helicobacter pylori gastritis 4 months before the onset of the arm lesions. The Helicobacter pylori gastritis was successfully treated with clarithromycin and amoxicillin plus omeprazole. One month before the onset of this infection, he had developed a reactive erythema nodosus on the extension face of both legs, which was treated with anti-inflammatory nonesteroid drugs followed by corticosteroids at low dosage. The patient had recent kitten scratches on both ankles.
In the Casualty Unit, the patient's temperature was normal, although it rose slightly in the evening (37.8°C). Except for the lesions on the left forearm, the remainder of the physical examination was normal.
The laboratory tests showed leukocytosis with a count of 13 × 103 per μl and 80% polymorphonuclear cells. An increased erythrocyte sedimentation rate (30 mm for the first hour), increased alkaline phosphatase value (271 U/100 ml; upper limit, 240 U/100 ml), and a slight increase in alpha-2 immunoglobulin (10.4 g/liter; upper limit, 9 g/liter) were found.
A computerized axial tomography scan of the thorax revealed minimal residual inflammation of the right lung base. In an abdominal computerized axial tomography with iodine contrast, a nodular subcapsular lesion in the right hepatic lobe was observed and the patient had a seroconversion to Bartonella henselae. Both findings were monitored fortnightly and improved to resolution without further intervention.
The echographic study of the arm lesion showed heterogeneous echogenicity (suggesting blood versus pus) with poorly defined borders in the distal third of the left forearm, affectation of the subcutaneous cellular tissue, brachial biceps, and adjacent muscles, in communication with deeper areas, all compatible with cellulitis and myositis, without bone involvement.
Whitish pus was drained from the lesion. Pus was drained by puncture aspiration with fine needle, and a biopsy of the granuloma was obtained through an incision made in the upper skin. The biopsy showed abundant polymorphonuclear leukocytes and lymphoplasmocytic infiltrate, with central granulomas, areas of necrosis, and negative Ziehl-Neelsen and Grocott stains, indicating nonspecific tuberculoid granulomas.
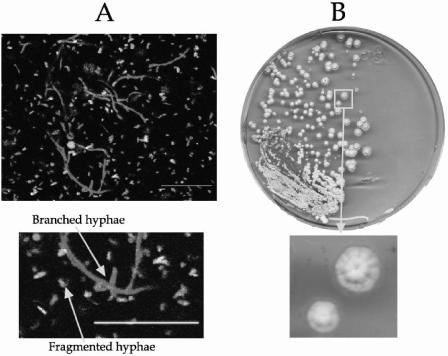
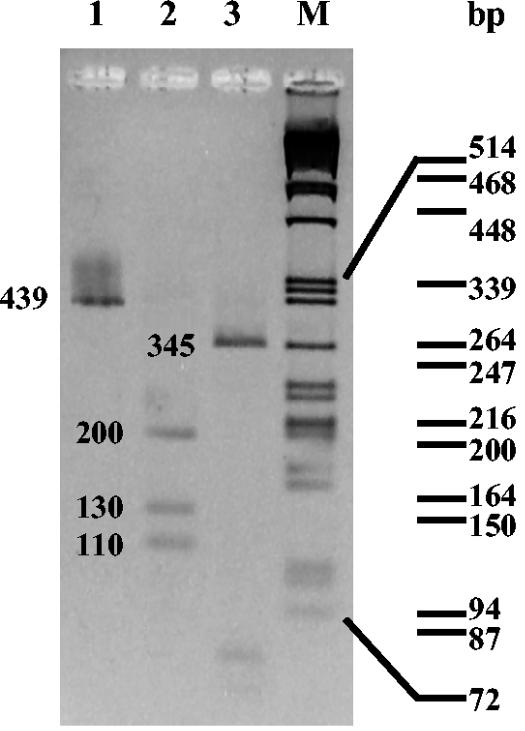
Examination of smears of exudate stained with Gram or Ziehl-Neelsen stain and the bacteria viability assay (LIVE/DEAD Bac-Light Bacterial Viability kit; Molecular Probes; L-13152) previously adapted for Streptomyces (6) showed branched thin hyphae (0.5 to 2 μm in diameter with the typical appearance of actinomycete structures) (Fig. 1A). Remarkably, these hyphae were very fragmented, and some of them were dead. The pus swabbed onto blood agar and chocolate agar plates showed a uniform growth in 48 h of grayish-white, pitting colonies around 3 mm in diameter. Fresh microscope preparations from the plate culture showed branched mycelia with chains of spores and identical morphology to those observed in unstained fresh direct smears of the exudate. A subculture in glucose-asparagine-yeast extract medium (7) at 30°C was positive in 48 h and showed a rich grayish-white growth (Fig. 1B). The microorganism gave a negative result with Kinyoun stain: it was catalase positive and grew on blood agar, chocolate agar, and Lowenstein-Jensen medium. It reduced nitrates to nitrites and hydrolyzed casein, esculin, gelatin, hypoxanthine, starch, and l-tyrosine. All of these findings were consistent with the Streptomyces genus (2). Endopeptidases, exopeptidases, l-glutamine, sugars, lipase, and lecithinase activities and an extensive array of physiological tests located this strain in cluster 2 (16), but this only gives <85% in the subcluster, which is insufficient for an accurate identification at the species level. To further characterize the strain (isolated from pus swab culture and directly from drained exudate), three techniques, PCR-restriction fragment length polymorphism (RFLP) of an internal fragment from heat shock protein (HSP) gene (15), sequencing of 16S ribosomal DNA (rDNA), and analysis of the γ-region of the 16S rDNA (positions 158 to 276) (8), were carried out. These three techniques were found to be identical in the pus and the exudate samples. A rapid identification by a PCR-RFLP scheme that used an amplified 439-bp segment of the 65-kDA HSP gene and restriction endonuclease analysis with MspI and HinfI (15) were employed. An internal fragment of the gene was amplified with TB11 (5′-ACCAACGATGGTGTGTCCAT-3′) and TB12 (5′-CTTGTCGAACCGCATACCCT-3′) oligonucleotides under the following conditions. A loopful of bacteria from agar culture or 1 μl of pus was resuspended in a PCR mixture that consisted of 1× PCR buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.2 mM each oligonucleotide primer, 2 U of DyNAzyme II DNA polymerase (Finnzymes Oy, Espoo, Finland), and nanopure water to make up a total final volume of 50 μl. PCR amplification using a DNA thermal cycler (MJ Research, Inc.) included denaturation at 95°C for 5 min followed by 30 cycles at 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min and a final incubation at 72°C for 10 min. The amplicon was digested with MspI and HinfI according to the manufacturer's instructions (Amersham Bioscience Europe). Fragments were electrophoresed on 3% agarose gel, stained with ethidium bromide, and then visualized on a UV transilluminator (312 nm). Figure 2 shows the results of PCR-RFLP analysis with MspI and HinfI of the strain under study. The RFLP band patterns that were produced coincided with the fragment expectation for the S. albus group: three fragments of 200, 130, and 110 bp with MsfI enzyme (lane 2) and a 345-bp fragment with HinfI enzyme (lane 3). Identical results were found for both samples. To clarify the identification, analysis of 16S rDNA was used. Primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and pH* (5′-AAGGAGGTGATCCAGCCGCA-3′), as described in reference 5, were used for the amplification of 16S rDNA sequences under the same conditions described above, with annealing at 58°C and extension at 72°C for 2 min. The amplicon was automatically sequenced (CSIC, Madrid, Spain). The partial sequence (1,468 bp; EMBL accession no. AJ696941) of the 16S rRNA of this isolate was aligned and compared with all eubacterial 16S rRNA gene sequences available in the GenBank and EMBL databases by multisequence analysis as described in reference 1, giving the maximum identity (99%) with 16S rDNA sequences from Streptomyces albus subsp. albus type strain DSM (AJ621602).
FIG. 1.
(A) Laser confocal observations of exudate stained with the viability assay LIVE/DEAD BacLight Bacterial Viability kit (Molecular Probes). Long branched hyphae and fragmented mycelium are observed. (B) Colonies of the Streptomyces isolate developed on glucose-asparagine-yeast extract medium, obtained from swabbed pus (3 days of culture at 30°C). Bars, 20 μm.
FIG. 2.
PCR-RFLP analysis from the 439-bp segment of the HSP gene of the Streptomyces strain. Lane 1, 439-bp fragment amplified with TB11 and TB12 primers; lane 2, amplicon digested with MspI; lane 3, amplicon digested with HinfI; lane M, lambda DNA cleaved with PstI. Molecular sizes are shown on the right.
To confirm the identification, comparison of the 16S rRNA from nucleotides 158 to 276, including the γ-region, was analyzed (8). Sequences were compared by the program PILEUP (3): the isolate was 100% identical to S. albus and showed lower identity values to Streptomyces cebimarensis (98.41%), Streptomyces albofaciens (97.62%), Streptomyces somaliensis (97.62%), Streptomyces lydicus (96.03%), Streptomyces rimosus (95.24%), and Streptomyces hygroscopicus (92.56%).
Bacterial susceptibility tests were simultaneously performed by a classical procedure (11) and E-test (AD Biodisk, Solna, Sweden) in Mueller-Hinton medium (Biomedics, Madrid, Spain) following the suggestions of the manufacturer. The results were as follows: vancomycin, 0.50 μg/ml; penicillin G, >256 μg/ml; ampicillin, >256 μg/ml; imipenem, 0.125 μg/ml; cefotaxime, 0.250 μg/ml; erythromycin, >256 μg/ml; chloramphenicol, >256 μg/ml; levofloxacin, 0.380 μg/ml; and trimethoprim-sulfamethoxazole (TS), 0.125/4.75 μg/ml. A good concordance between both methods was found.
The patient received a course of 8 weeks of TS at one tablet twice a day and made good progress, with a fair diminution of local pain and nodule size to a complete cure. Six months later, the patient remained asymptomatic.
Streptomyces strains are infrequently encountered in clinical practice but are important potential causes of serious human and animal infections (10). As in this case, the failure to identify isolates by morphological, physiological, and biochemical approaches is a common problem in this genus, which is characterized by a broad biochemical and genetic diversity and the existence of overspeciation (9). In our case, a first approach was carried out by analysis of amplification restriction of the 65-kDA fragment coding for the HSP (15). The confirmation was based on the comparison of the genomic sequence analysis of the γ-region of 16S rDNA, considered critical in differentiation of Streptomyces species because of their high variability (14). This analysis in the BLAST program gave the maximum coincidence (100%) for S. albus. The coincidence of the results obtained by these two genetic approaches confirmed unequivocally the identity of this strain as belonging to S. albus.
As in the case we report, the diagnosis of these infections requires invasive diagnostic biopsy procedures (10). The typical histological appearance of the disease is a granulomatous inflammatory reaction (16), which is coincident with our findings.
In the last 20 years, at least 10 species of the genus Streptomyces have been isolated from mycetomic and nonmycetomic infections (4, 16). S. somaliensis is a well-documented agent of actinomycetoma (10). S. albus is scarcely cited as a human pathogen; in recent literature, cases of extrinsic alveolitis and skin hypersensitivity type II in farmer workers have been reported (13). To our knowledge, no cases of S. albus actinomycetoma have been published before in the Spanish literature.
Clinical manifestations of this infection, severity of disease and the prognosis are extremely variable and may be determined by factors such the route of infection and the presence or absence of a properly functioning immune system (4). In the case we report, the patient could have been slightly immunodepressed, owing to the coinciding illnesses that he was suffering from (Helicobacter pylori gastritis and cat scratch fever). This is especially true in the case of the Bartonella infection, whose control requires an important effort on the part of the host immune system (12).
The route of entry of the bacteria has not been determined, but it could have been a minimal excoriation in the arm area where the lesion developed later. Handling of earth immediately previous to the appearance of the clinical symptoms could also be the source of contamination.
The treatment of mycetoma is still a problem; there are no approved breakpoint antibiotics for Streptomyces species, and the correlation between the data obtained in vitro and the clinical evolution remains unclear (11). Many drugs are being used with variable results. The drugs used most frequently include TS, dapsone, amikacin alone or combined with TS, and amoxicillin-clavulanic acid (17). All treatments have in common the need for long courses of therapy to obtain satisfactory results. In our case, the high susceptibility of the strain to TS obtained in vitro and the good response to this drug reported in the literature (11) made it the first choice.
In summary, the infections caused by this bacterial species are rare and in general not very well documented. The identification of the bacterial isolate causing the mycetoma as S. albus was convincingly demonstrated by using molecular rDNA techniques applied to the bacterial colonies and directly to the exudate obtained in the intraoperatory biopsy.
Acknowledgments
We are grateful to Miguel A. Alvarez (Instituto de Productos Lácteos de Asturias) for helpful discussion. We thank the medical staff and technicians of the Microbiology Service II of the University Hospital for isolation of Streptomyces.
This work was in part financed by the project FICYT PB-MED-09-04. A.M.F. is an FP2000-6023 grant of the Spanish Ministry de Ciencia y Tecnología.
REFERENCES
- 1.Altschul, X., F. Stephen, L. Thomas, X. Madden, A. Alejandro, X. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, A. S., and E. M. Wellington. 2001. The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol. 51:797-814. [DOI] [PubMed] [Google Scholar]
- 3.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunne, E. F., W. J. Burman, and M. L. Wilson. 1998. Streptomyces pneumonia in a patient with human immunodeficiency virus infection: case report and review of the literature on invasive streptomyces infections. Clin. Infect. Dis. 27:93-96. [DOI] [PubMed] [Google Scholar]
- 5.Edwards, U., T. Rogall, H. Böcker, M. Emde, and E. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal DNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez, M., and J. Sanchez. 2002. Nuclease activities and cell death processes associated with the development on surface cultures of Streptomyces antibioticus ETH7451. Microbiology 148:405-412. [DOI] [PubMed] [Google Scholar]
- 7.Hardisson, C., M. B. Manzanal, J. A. Salas, and J. E. Suarez. 1978. Fine structure, physiology and biochemistry of arthrospore germination in Streptomyces antibioticus. J. Gen. Microbiol. 105:203-214. [DOI] [PubMed] [Google Scholar]
- 8.Kataoka, M., K. Ueda, T. Kudo, T. Seki, and T. Yoshida. 1997. Application of the variable region in 16S rDNA to create an index for rapid species identification in the genus Streptomyces. FEMS Microbiol. Lett. 151:249-255. [DOI] [PubMed] [Google Scholar]
- 9.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical streptomyces genetics, p. 2-5. The John Innes Foundation, Norwich, England.
- 10.McNeil, M. M., and J. M. Brown. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasher, M. A., R. J. Hay, E. S. Mahgoub, and S. A. Gumaa. 1989. In vitro studies of antibiotic sensitivities of Streptomyces somaliensis, a cause of human actinomycetoma. Trans. R. Trop. Med. Hyg. 83:265-268. [DOI] [PubMed] [Google Scholar]
- 12.Neves, P. E., M. L. Cintra, A. M. Uthida-Tanaka, A. M. de Moraes, and A. Mariotto. 2003. What do we (not) know about the human bartollenoses? Infect. Dis. 7:1-6. [DOI] [PubMed] [Google Scholar]
- 13.Śpiewak, R., A. Góra, and J. Dutkiewicz. 2001. Work-related skin symptoms and type I allergy among Eastern-Polish farmers growing hops and other crops. Ann. Agric. Environ. Med. 8:51-56. [PubMed] [Google Scholar]
- 14.Stackebrandt, E., D. Witt, C. Kemmerling, R. Kroppenstedt, and W. Liesack. 1991. Designation of streptomycete 16S and 23S rRNA-based target regions for oligonucleotide probes. Appl. Environ. Microbiol. 57:1468-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steingrube, V. A., R. W. Wilson, B. A. Brown, K. C. Jost, Jr., Z. Blacklock, J. L. Gibson, and R. J. Wallace, Jr. 1997. Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J. Clin. Microbiol. 35:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trujillo, M. E., and M. Goodfellow. 2003. Numerical phenetic classification of clinically significant aerobic sporoactinomycetes and related organisms. Antonie Leeuwenhoek 84:39-68. [DOI] [PubMed] [Google Scholar]
- 17.Welsh, O., E. Sauceda, and J. González. 1987. Amikacin alone and in combination with sulphamethoxazole plus trimethoprim. J. Am. Acad. Dermatol. 17:443-448. [DOI] [PubMed] [Google Scholar]