Abstract
The 1H-MRS spectrum contains information about the concentration of tissue metabolites within a predefined region of interest (a voxel). The conventional spectrum in some cases obscures information about less abundant metabolites due to limited separation and complex splitting of the metabolite peaks. One method to detect these metabolites is to reduce the complexity of the spectrum using editing. This review provides an overview of the one-dimensional editing methods available to interrogate these obscured metabolite peaks. These methods include: sequence optimizations, echo-time averaging, J-difference editing methods (single BASING, dual BASING and MEGA-PRESS), constant-time PRESS and quantum filtering. It then overviews the brain metabolites whose detection can benefit from one or more of these editing approaches, including ascorbic acid, γ-aminobutyric acid (GABA), lactate, aspartate, N-acetyl aspartyl glutamate, 2-hydroxyglutarate, glutathione, glutamate, glycine, and serine.
Keywords: J-coupling, echo-time averaging, J-difference editing, constant-time PRESS, quantum filtering, metabolites, magnetic resonance spectroscopy (MRS)
Introduction
Proton Magnetic Resonance Spectroscopy (1H-MRS) is a non-invasive methodology that allows the detection and quantification of endogenous tissue metabolites. Signals arising from spins in different chemical environments are separated along the chemical shift axis, revealing a spectrum with a number of identifiable peaks. In the brain, these peaks include N-acetyl aspartate (NAA), creatine (Cr), myoInositol (mI), and choline (Cho). For many signals, the chemical shift dispersion is limited compared to the in vivo linewidth and splittings due to scalar (J-) couplings, and therefore all the information that is potentially available in an MR spectrum is not easily resolved. As a result, some metabolites are present at potentially detectable levels (of the order of 1 mM), but cannot be associated with any single resolved peak in the in vivo spectrum. Thus, the 1H-MR spectrum often contains too much information spread over too narrow a parameter-space. There are two approaches to resolving this – either extending the space over which signals are spread by adding a second dimension to the MR experiment, or by reducing the information content of the one-dimensional spectrum. The latter strategy, which is the focus of this review article, is referred to as editing the spectrum. The most common editing approaches exploit known J-coupling relationships within molecules-of-interest to separate their signals from stronger, overlying signals of more concentrated molecules. J-coupling (or simply coupling) is a through-bond interaction between adjacent proton spins and results in the splitting of peaks in the spectrum. (For more details, see the Appendix.)
Metabolites that can benefit from editing include ascorbic acid (Asc), γ-aminobutyric acid (GABA), lactate (Lac), aspartate (Asp), N-acetyl aspartyl glutamate (NAAG), 2-hydroxyglutarate (2HG), glutathione (GSH), glutamate (Glu), glycine (Gly), serine (Ser).
This review aims to describe methods to edit the 1H-MRS spectrum in human experiments, the metabolites that are measured and to review some of the main findings of applying these measurements. Many of these applications are in the brain, reflecting the bias of the MRS literature. The review is aimed at MR-familiar readers without an extensive technical training in the physics of (N)MR spectroscopy, and aims to use the least technical level of language that sufficiently describes the methods to maintain accessibility for a wide audience. Additional detail on common terms of reference, such as scalar (J-) couplings and coherences, is included in Appendix 1.
Methods to Edit the MRS Spectrum
Using the broad definition for editing as “a method that simplifies the 1H-MR spectrum”, Figure 1 presents the pulse sequences that will be discussed here. All these sequences have two features in common: the localization of signal (usually with PRESS (1)); and a mechanism for reducing the information content of the spectrum (the core principle of editing). These two features are largely independent, and thus most editing approaches can, in principle, be incorporated within a number of spin-echo-based localization schemes, such as PRESS (1), SPECIAL (2), or semi-LASER (3).
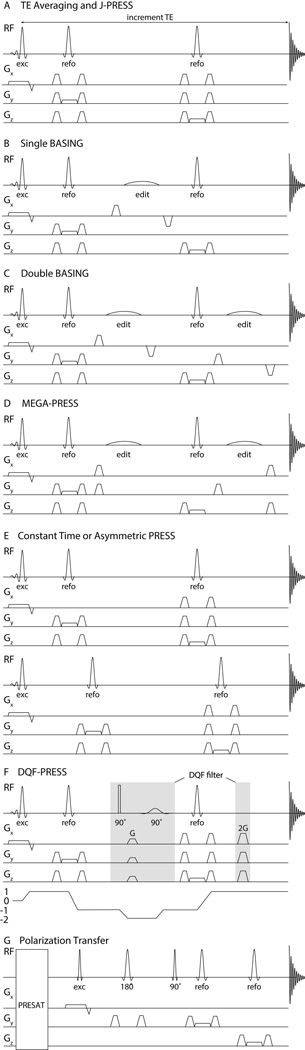
Figure 1.
Pulse sequences that are used to edit the spectrum. A. TE-averaging: the echo time is varied during the acquisition. The typical PRESS localization scheme is used with gradients applied during the 90 excitation pulse and the two 180 refocussing pulses. B. Single BASING: a single frequency-selective editing pulse is placed between the two refocusing pulses. C. Dual Basing: in half of the transients, two frequency-selective editing pulses are applied, one after each refocusing pulse. These editing pulses refocus the evolution of selected couplings. In the remaining half of the transients, these pulses are not applied (pulse sequence not shown) such that in the subtraction spectrum, overlapping larger resonances are removed, revealing only the spins impacted by the editing pulses. D. MEGA (MEscher-GArwood): Similar to the dual BASING scheme, a pair of frequency-selective editing pulses refocus the evolution of the coupling in half of the transients, the ‘On’ condition. The difference between the subspectra with and without the refocusing pulses subtracts the overlapping metabolites to reveal the metabolite of interest. E. Asymmetric PRESS: Two spectra with same TE but different interpulse delays are acquired. Timings are optimized to maximize differences in the modulation of strongly coupled spins, so their signals are enhanced in the difference spectrum, and resonances from singlets are removed. F. Example of a double quantum filter experiment and the associated coherence transfer pathway. The double-quantum coherence is formed by the excitation pulse, first refocusing pulse and an additional 90° pulse. Subsequently, the 90° frequency-selective pulses convert the desired double quantum signals into observable coherence. G. Polarization transfer: First, signals in the spectral range of interest are pre-saturated (PRESAT). Signal is then excited on a spin outside the saturated range and transfered to a coupled partner. Within the saturated range, only signal that arises from such coherence transfer give detectable signals in the acquired spectrum. Coherence transfer is achieved by the pulse marked 90°.
The most widely used localization scheme for editing is PRESS (1). Readers are directed to the review by Yahya (4) for a more specific review of the PRESS sequence and its modifications. The dual spin echo of the PRESS experiment refocuses evolution of the chemical shift offset during TE, and allows scalar couplings (referred to hence forward simply as couplings) to evolve. For uncoupled and weakly coupled spin systems, it does not greatly matter how TE is broken down into its two constituent spin echoes, referred to as TE1 and TE2. Typically, TE1 is kept as short as possible to minimize unwanted coherences or signal evolution. Unwanted coherences may occur due to imperfect pulse calibration, at the edges of the voxel, or in the case of strong coupling (when refocusing pulses act to some extent 90° pulses). The extent to which these factors cause undesirable formation of multiple quantum and/or coherence transfer between coupled spins is minimized by keeping TE1 short (i.e. TE1 ≪ 1/2J), or harnessed by parameter optimization to simplify the spectrum. Beyond this, the selection of TE will be influenced by the limitations of the pulse sequence and characteristics of the metabolites of interest. Alternatives to PRESS localization for editing applications are STEAM (5), SPECIAL (2) and semi-LASER (3). In the STEAM experiment, a stimulated echo is detected resulting from three slice-selective 90° pulses. In the SPECIAL experiment, the voxel is localized through the subtraction of two spin-echo acquisitions. In the first acquisition, a column is selected by applying a 90° slice selective pulse and a perpendicular 180° refocusing pulse. In the second acquisition, an inversion pulse perpendicular to the refocused column is applied prior to the 90° pulse such that the difference between these acquisitions results in a spectrum located at the intersection of the three pulses (2). In the semi-LASER experiment, two pairs of adiabatic 180° pulses are used to select the second and third dimensions of the voxel after the initial slice-selective 90° pulse (3).
Foundations of Editing
The in vivo human methods reviewed here are all built on a rich history of NMR spectroscopy, and in many cases, on animal experiments performed in vivo. Selective excitation methods (e.g. (6)) were shown to simplify the 13C-coupled proton spectrum, and selective heteronuclear polarization transfer methods (e.g. (7, 8)), edited the 13C-decoupled spectrum. A number of selective one-dimensional (1D) analogues to two-dimensional homonuclear methods were developed, including 1D correlation spectroscopy and 1D nuclear Overhauser effect spectroscopy (9). Such methods are useful to simplify more complex small-molecule NMR spectra, as well as to investigate scalar couplings and cross-relaxation between resonances. In vivo, editing faces the additional hurdle of localization to a region/tissue of interest; some early animal (10) and human (11) edited experiments relied on depth-selective excitation and surface-coil detection. Once established, single-voxel localization for in vivo applications (1, 5) was very rapidly modified to include editing (12).
Spin Echo TE optimization
When sequence parameters are explicitly optimized so as to improve the resolution of a desired metabolite signal, sequence parameters approaches fall within our broad definition of editing. In particular the TE in a PRESS sequence and the mixing time (TM) and TE in a STEAM sequence have been altered reduce signal overlap. Sequence optimizations have been applied to improve separation of glutamate and glutamine, and optimize the detection of glycine, GABA and 2HG (4, 13–16). For example when using the PRESS sequence at 3T, it has been suggested that setting the TE equal to 80 ms optimizes the detection of glutamate due to the decay of the background signals, improved lineshape and SNR of the 2.35 ppm peak. With improved glutamate detection, the separation of glutamate from glutamine may likewise be improved (17). Similarly, in a STEAM acquisition optimizing the TE and TM can improve the detection of glutamate/glutamine (14). In a different implementation, a STEAM sequence was optimized to simultaneously detect glutamate, glutamine and GABA (18) and STEAM optimization has been preformed to measure GABA alone (15). By contrast, Near et al. (19) measured GABA using short-TE (8.5 ms) with SPECIAL localization. Using a PRESS sequence, measurement of 2HG was illustrated by optimizing the total TE to 97 ms and TE1/TE2 to 32/65 ms (16).
In general, such approaches can be shown to work well in simulations and phantoms. In vivo, performance is often tightly linked to linewidth and subject compliance, with excellent performance in the best-case scenario and rapidly diminishing performance in sub-optimal conditions (19). Since the simplification of the spectrum that most such optimizations offer is relatively small, signal overlap is often rapidly restored with increasing linewidth due to subject motion or scanner instabilities or frequency drift.
TE-averaging
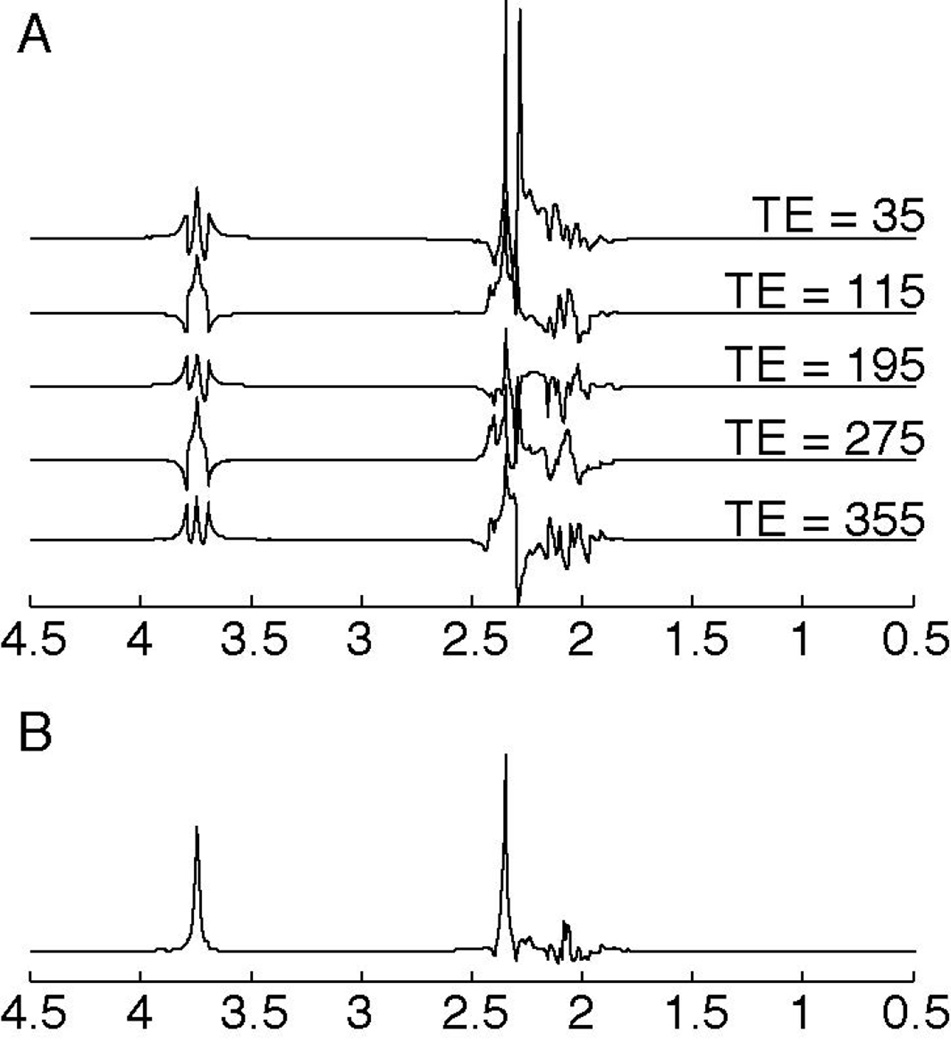
Coupling evolves during a spin echo, so that the appearance of coupled signals in the spectrum is TE-dependent. A doublet signal evolves under the coupling so that the two peaks acquire equal but opposite phases. With a triplet signal, the outer two peaks acquire equal and opposite phases, but the center peak does not evolve during TE. A TE-averaged experiment acquires and averages data at a range of TEs (20, 21). For triplets, the resulting spectrum is substantially simplified, compared to the spectrum at each TE, as the outer peaks tend to be cancelled and the center-peak of the triplet remains. The signal that remains after TE-averaging is at the frequency that is coincident with the chemical shift. This simplification is demonstrated in Figure 2 for glutamate, as this approach is often used to simplify the spectrum to isolate glutamate from glutamine (the combination of glutamate and glutamine are often referred to as Glx). In this example, the complex multiplets from the C3 protons of glutamate and glutamine (at ~2 ppm) are removed and the C4 protons from glutamate at 2.35 are fully resolved as are the Glx C2 protons at 3.75ppm (21). In the brain, TE-averaging substantially simplifies the spectrum, leaving only the methyl singlets (of Cho, Cr, and NAA) and glutatmate triplet center-peaks, and some of the mI signal (20, 21).
Figure 2.
Simulation of TE-averaged data for glutamate. A. Simulated glutamate spec- trum at various TEs ranging from 35 ms to 355 ms. Notice the multiplet structure changes with incrementing TE. B. Simulation of the TE-averaged spectrum from glutamate, using a minimum TE = 35 ms, incrementing in steps of 10 ms up to TE = 355 ms. In the TE-averaged spectrum, the peaks are simplified as the outer wings are effectively cancelled. Spectra were simulated using FID-A (115).
As with long-TE methods in general, TE-averaging suppresses short-T2 macromolecules and lipids, resulting in a flatter baseline and aiding quantification (20, 21). However, with longer effective TE, metabolite quantification becomes more sensitive to T2 uncertainties (22). Several studies (20, 23–26) have compared TE-averaged PRESS to single-TE PRESS with mixed results: TE-averaged PRESS may be more prone to inhomogeneity and frequency drifts, thus is less reliable (23, 25, 26) but may be more accurate (24) and more sensitive to some disease conditions (20).
J-PRESS
Although J-resolved methods lie beyond the scope of this review, it is worth emphasizing their close relationship to TE-averaging. Whereas TE-averaged PRESS simply adds up spectra acquired at a range of echo times, it is possible to acquire the same dataset and then Fourier transform with respect to the echo time (in addition to the usual Fourier transformation of the acquisition dimension). This experiment is referred to as J-PRESS (27), and gives a two-dimensional spectrum in which F2, the acquired dimension, contains coupling and chemical shift information, while F1, the indirect dimension only contains coupling information. Multiplets in the J-PRESS spectrum appear along diagonals centered on (F2=Ω, F1=0). Mathematically, the TE-averaged spectrum is the same as the F1=0 line of the J-PRESS spectrum.
Single BASING
Band-selective inversion with gradient dephasing (BASING) (28) was originally developed as a water and lipid suppression method. Editing developed as a secondary application. Single BASING (Figure 1) uses one frequency-selective inversion pulse to refocus evolution of coupling during TE, which can improve the visibility of coupled signals, such as lactate, without removing other signals. Frequency-selective editing pulses are more commonly applied in pairs, and within a J-difference framework, described next.
J-Difference Methods
J-difference editing requires two sub-experiments that differ in their treatment of a molecule of interest. Subtracting these two experiments removes most signals from the spectrum, while retaining the signal of interest.
The most common schemes, including both MEGA (MEscher-GArwood) (29) and the contemporaneously published dual BASING (30), is to acquire one experiment in which a pair of frequency-selective editing pulses refocus the evolution of a coupling of interest (the On sub-experiment), and one in which the coupling is allowed to evolve without intervention (the Off sub-experiment), both shown in Figure 3. The difference between the On and Off subspectra (the Diff spectrum) only contains those signals that are impacted by the editing pulses – those signals directly affected by editing pulses appear with negative polarity, while signals coupled to spins inverted by the editing pulse usually appear with positive polarity. The editing targets are selected such that the overlapping signals of the detected signal (e.g., the creatine signal that overlaps the GABA peak at 3 ppm) are not impacted by the frequency-selective editing pulses and are removed in the subtraction, shown in Figure 4.
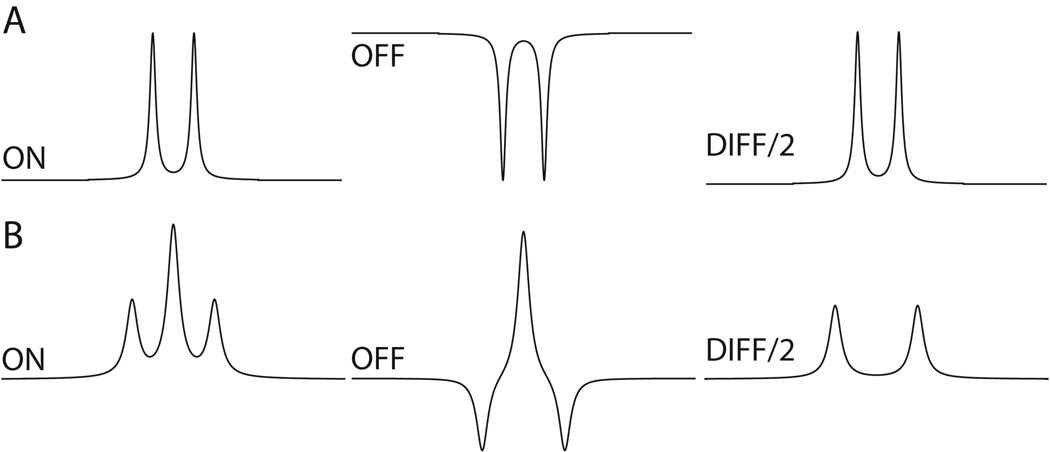
Figure 3.
The appearance of the detected peaks in the On, Off and Diff spectra of (A) a doublet (e.g., lactate) and (B) a triplet (e.g., approximately GABA) for J-difference editing.
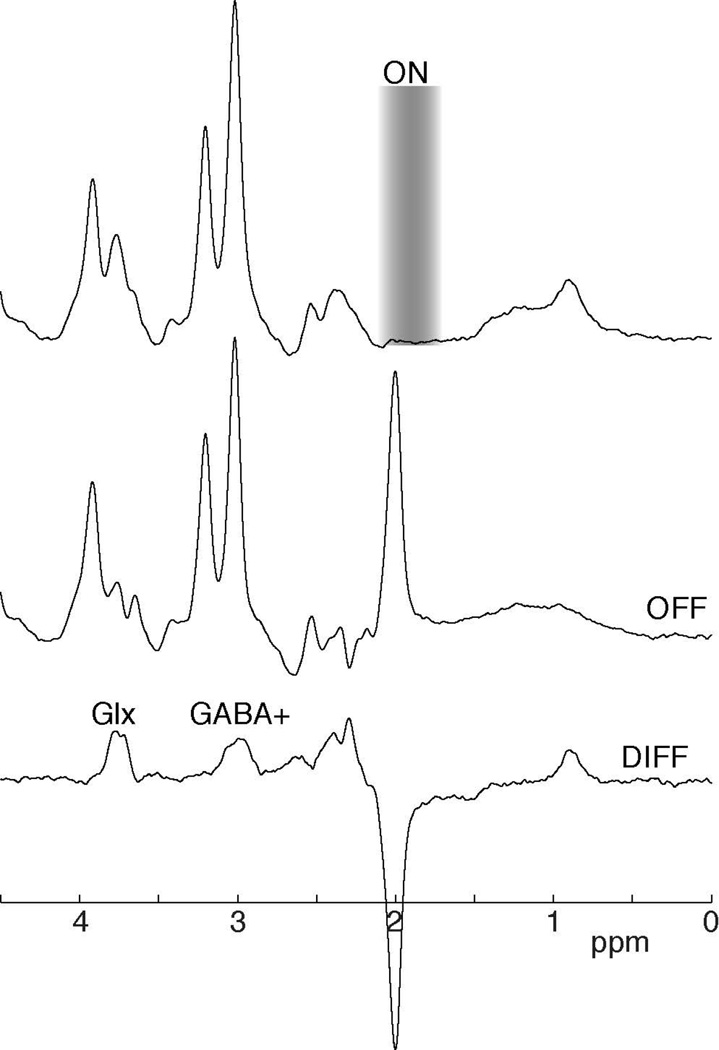
Figure 4.
Example (A) On, (B) Off and (C) difference spectra for a GABA-edited experiment. In the On subspectrum, a frequency-selective editing pulse is applied, in this case a 1.9 ppm. In the Off subspectrum, no editing pulse is applied so in the Diff spectrum the overlapping creatine peak is removed. The frequency-selective editing pulse (On sub-spectrum) co-edits MM and Glx. The co-edited MM peak is also at 3 ppm, hence the term GABA+. The co-edited Glx peak is seen at 3.75 ppm.
MEGA and BASING differ subtly in the placement of gradients about the editing pulses (compare Figure 1D and 1E) but the terms are often used interchangeably. As with single BASING, MEGA was originally proposed as a method of water suppression (28), as well as editing, although the water suppression aspect of both methods is often not applied.
Edited experiments are generally optimized to detect a single metabolite, with acquisition parameters tuned to the spin-system of interest, often determined by extensive density-matrix simulations e.g. (31–34). In particular, the echo time (TE) for J-difference editing is usually selected so that signals are maximally negative in the Off sub-spectrum, so as to maximize the difference with the refocused, positive signals of the On spectrum. Effectively, this means that doublet-like signals (such as lactate) are edited at around TE = 1/J and triplet-like signals (such as GABA) around 1/2J. This simplification ignores T2 relaxation, strong coupling and the complexity of the spin systems of real molecules in vivo, and as a result the ‘optimal’ TE for editing is sometimes controversial (31, 35).
J-difference editing, in particular MEGA-PRESS, is a powerful method to resolve overlapped signals, and has become the most widely used MRS method to detect some metabolites (e.g., GABA (36)). However, there are limitations to the technique. As a difference method, it is particularly susceptible to instability, such as subject motion and frequency drifts (37). An additional issue that may hinder measurements that is specifically relevant for J-difference editing is co-editing.
Co-Editing and Accelerated Difference Editing
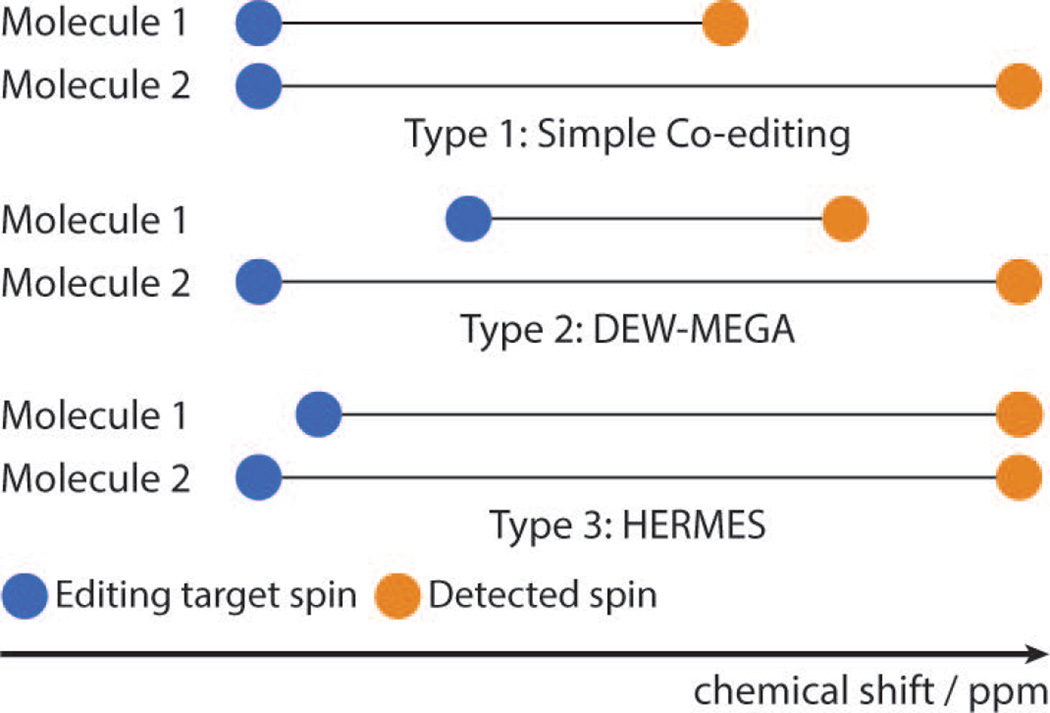
Editing is rarely perfectly selective, and molecules other than the editing target often give signal in the difference spectrum, a process referred to as co-editing. Co-editing that does not result in overlapping signals can allow the quantification of more than one metabolite, whereas overlapping co-edited signals substantially complicate the interpretation of edited spectra. In the case of GABA editing, for example, both occur: glutamine and glutamate (Glx) give an edited signal that does not overlap with the intended GABA signal (see Figures 4 and 5); whereas a macromolecular resonance co-edits at a similar frequency to GABA. This co-editing of macromolecules (MM) can hinder the quantification of GABA, as the resulting signal at 3 ppm includes approximately 50% macromolecules (38–40). These measures are often therefore referred to as GABA+ to indicate GABA+MM. The co-edited Glx signal results in a peak in the edited-spectrum appearing at 3.75 ppm and does not interfere with the GABA measurement.
Figure 5.
Schematic to illustrate the classes of co-editing. A. Non-overlapping co-editing: the editing pulse modulates two metabolites that have coupled spins at different chemical shifts. B. DEW: the On sub-spectrum for metabolite-1 serves as the Off subspectrum for metabolite 2 and vice versa. C. HERMES: the detected signals have similar chemical shift but can still be resolved using Hadamard-encoded editing as the editing targets spins are at different chemical shifts.
Insofar as editing usually proceeds at a rate of “one-metabolite-per-experiment”, co-editing of this sort can be thought of as the simplest case of accelerated editing. In cases where the editing target spins and the detected resonances of two spin systems are resolved, it is possible to edit both in the same double-edited (DEW) experiment (41). In this motif, the ‘On’ experiment for one metabolite is the ‘Off’ for the other, and vice versa, so that two metabolites can be edited at the same time (with opposite polarity in the difference spectrum). This has been developed to edit for ascorbate and GSH in which editing pulses are applied alternately at 4.01 and 4.56 ppm to refocus the coupled spins of ascorbate at 3.73 ppm and GSH at 2.95 ppm, respectively. As such, the On subspectrum for ascorbate acts as the Off subspectrum for GSH and vice versa.
Hadamard-based editing (HERMES) has recently been developed, in which multiple metabolites can be edited orthogonally and simultaneously, even if their detected signals overlap, so long as the editing target spins can be sufficiently resolved. HERMES has been demonstrated for the measurement of NAA and N-acetyl aspartyl glutamate (NAAG) (42) and for simultaneous editing of GABA and glutathione (43), and to be expandable to simultaneously edit more than two metabolites within an acquisition.
Constant time PRESS (Asymmetric PRESS)
As stated above, the chemical shift offset is refocused across the echo time, whereas couplings evolve. For PRESS detection of weakly coupled spin systems, it does not greatly matter how TE is broken down into the two constituent spin echoes. For strongly coupled spin systems, even perfectly calibrated refocusing pulses act to some extent like 90° pulses e.g. by causing coherence transfer between spins. This effect can be harnessed to selectively edit strongly coupled spin systems, by acquiring two PRESS experiments with the same TE but differing TE1 and TE2 as shown in Figure 1. Since uncoupled signals and weakly coupled signals behave the same in these two cases, subtraction will remove such signals, and only strongly coupled signals will remain (33, 44). In the Sum spectrum, the peaks from singlets and non-varying coupled spins remain (34) , as with J-difference methods.
This method of constant time or asymmetric difference editing relies heavily upon simulation-based optimization of TE1 and TE2 to maximize signal differences between the two subspectra (44). The optimal values of TE1 and TE2 are field-strength-dependent because chemical shift offset frequencies are proportional to field strength (and couplings are not); smaller adjustments in TE1 are required at higher field strength (44). This approach has also been applied to resolve weakly coupled systems by deliberately using reduced flip angle refocusing pulses (34).
Multiple-quantum and longitudinal-scalar-order filtering
It is not possible to describe multiple-quantum coherences (MQC, taken to include both double-quantum and zero-quantum coherences) or longitudinal scalar order (LSO) in classical vector terms, nor is it possible to visualize them intuitively. Readers without a strong technical background should know that:
MQC and LSO can only exist in coupled spin systems;
Neither MQC, nor LSO, is directly observable;
MQC and LSO can be converted to observable single-quantum coherence (SQC) by RF pulses (usually 90° pulses); and
MQC and LSO can be separated from SQC by using gradients or phase cycling for coherence transfer pathway selection.
Multiple-quantum-filtered (MQF) and LSO-filtered experiments are challenging to describe without resorting to technical language and/or the product operator description. Simply, MQF can be thought of as a black-box mechanism for separating signals from a coupled spin system of interest from stronger overlying signals from other metabolites.
In more detail, MQC or LSO filtering exploits aforementioned statement 1 that only coupled spin systems can generate multiple-quantum coherences (MQC) and/or LSO. An experiment that only acquires signal derived from multiple-quantum coherence will therefore not contain singlet signals. Furthermore, multiple-quantum coherence from a particular spin system of interest can be isolated by using frequency-selective pulses to form or read out the MQC. A particular advantage of gradient-selected MQF is that unwanted signals are removed from the spectrum experimentally, within each TR, rather than relying upon the subtraction of two TRs, making MQF experiments less sensitive to subject motion and scanner instability than J-difference editing.
In its simplest implementation, a double-quantum filter (DQF) can be added to a localized measurement as a pair of 90° pulses, between which MQC is selected using either gradients or phase cycling. In Figure 1, this implementation is shown, as applied by McLean et al. (45). As can be seen from the coherence transfer pathway diagram, the first 90° pulse converts single-quantum coherence into double-quantum coherence, which is then converted back to observable single-quantum coherence by the second 90° pulse. Only coherence that is antiphase with respect to the coupling (i.e. product operator terms such as 2I1xI2z) can be converted to MQC during the filter – the efficiency of formation and retrieval of MQC will determine the sensitivity of the approach. The advantage of the DQF approach is all uncoupled resonances are reduced in each scan by the static magnetic field gradients regardless of their chemical shift offset (46). This results in 25%-100% of available signal from coupled molecules being maintained and non-coupled spins being suppressed by factor up to 1000.
MQC-edited experiments applied in vivo in humans build upon a foundation of methodological developments in phantoms and animals. An early development by Sotak et al. (12) employed a STEAM-localized MQC experiment for to measure lactate without the overlapping lipid signal. He et al. (47) and Hurd et al (48) performed related selective-MQC experiments in vivo to detect lactate. Melkus et al. (49) combined selective-MQC with short-echo spectroscopic imaging to characterize tumor-specific metabolites, such as choline and lactate.
There are two main disadvantages to the MQF approach – one is that there is no internal reference signal preserved by an MQF experiment, making quantification challenging. One solution is to this challenge is to acquire the unedited spectrum as a separate echo (e.g. (49)). The second disadvantage is the sensitivity of the experiment may be reduced by 50% due to loss of signal to zero-quantum coherence, and by a further 50% as the filter selects either +2 or –2 coherences, but not both (50).
Longitudinal scalar order (i.e. product operator terms such as 2I1xI2z) can similarly only be made for coupled spin systems. Most of the above discussion of MQC applies equally to LSO – however, separation of LSO and zero-quantum coherence is far from trivial. MQC and LSO methods do not rely upon subtraction to remove overlapping signals, therefore are likely to be more robust to subject motion and scanner instability.
Polarization/Coherence Transfer methods
Another approach to separating a coupled signal from overlying signals is to make use of coherence transfer between coupled spins, i.e. to deliberately excite signal on one coupled spin and detect it on the other. In these methods, presaturation (or spectrally selective refocusing) is used to suppress all signals in the region of the spectrum where the coupled signal is to be detected. Coherence can then be transferred from unsaturated spins (outside the presaturation range) to spins that give rise to a signal in the (otherwise) suppressed region of the spectrum. This approach was one of the first applied in vivo to edit lactate (51) and more recently for GABA detection (52, 53). It has been applied using nonselective coherence transfer (54) and expanded using selective spin-locked Hartmann-Hahn coherence transfer (55).
The pulse sequence shown in Figure 1G represents the homonuclear transfer experiment proposed by Shen et al. (52), first suppressing GABA signal (and overlying creatine signal) at 3 ppm, then transferring coherence from the coupled signal at 1.9 ppm to 3 ppm before acquiring. Such polarization transfer methods do not rely upon subtraction to remove overlapping signals, therefore are likely to be more robust to subject motion and scanner instability than difference methods.
Analytical tools for signal quantification and fitting
There are various analytical tools to measure edited signals. The most common tools are LCModel (56), Tarquin (57), AMARES (58) in jMRUI (59) and Gannet (60). Edited signals from some of the aforementioned methods, such as MQF, single basing and asymmetric press, require in-house tools for analysis, limiting the more widespread implementation of these methods.
LCModel is commercially available and the most widely used software for spectroscopy data analysis, including use for MEGA-edited (61–63), TE-averaged (25), parameter optimized sequences (e.g., (19)) and J-resolved spectra (63). LCModel has a ‘black-box’ approach, does not have retrospective frequency correction, and depends on the prior knowledge of the individual metabolite spectra with the acquisition parameters used (a basis set) to fit edited signals. Tarquin, AMARES in jMRUI and Gannet are freely available software. Tarquin incorporates both basis-set simulation and fitting and fits in the time domain. It can be used to analyze conventional spectroscopy and MEGA-edited analyses are currently limited to GABA. AMARES also performs time-domain fitting using user-defined a priori information. AMARES in jMRUI can provide fitting for MEGA-edited, TE-average and short echo-time data. Gannet is MATLAB-based open source software developed specifically for (GABA-) edited spectra. For MEGA-edited GABA, both Tarquin and Gannet use a simple Gaussian for fitting.
In discussing quantification and fitting, it is worth establishing the various kinds of signals that make up a spectrum and the ways in which an algorithm can fail. Each spectrum consists of: S1, signal of interest; S2, signals from other metabolites; N, true noise; A, artifacts, including subtraction artifacts and out-of-voxel artifacts. Most quantification errors involve the misattribution of one kind of signal as another. For example, LCModel might incorrectly fit some glutamate signal as GABA, if the basis functions are not sufficiently independent – this is a misattribution of S2 as S1. Further, subtraction artifacts in J-difference spectroscopy have been shown to bias GABA estimates (37, 64) – this is a misattribution of A and S1. Pursuing an edited strategy amounts to a judgment that reducing S1/S2 attribution errors will improve the quantification of S1, even if it also increases A signals and reduces the size of S1 relative to N.
One useful example in this discussion is (19). In this paper, non-edited strategies for quantifying GABA are developed using simulations and compared to editing. For the unedited acquisition, an exceptionally short TE of 8.5 ms was made possible by SPECIAL localization (compared to typical short-TE values of ~30 ms). At such a short TE, J-coupling evolution is much reduced in addition to the minimization of signal decay from T2 relaxation. This substantially improves the ability of LCModel fitting algorithm to fit GABA in a reliable manner. At this short TE, the decay of macromolecular and lipid signals is also minimized and therefore these signals become highly influential on the accuracy and performance of the fit. This paper shows that an inappropriate baseline results in underestimation of GABA due to a misattribution of GABA signal as macromolecules even with high SNR. The paper presents a correlation of unedited measurements with J-difference edited measurement, and demonstrates a moderate correlation (0.58). Thus, with excellent SNR and linewidth (650, 6 Hz) and very-short-TE (<9 ms) it is possible to quantify GABA, even at 3T, without editing. However, simulations indicate that even moderate reductions of SNR and linewidth (e.g. to 450, 8 Hz) substantially worsen reproducibility errors and Cramer-Rao lower bound fitting estimates, suggesting that shorter measurements of less compliant clinical subjects might not perform as well with this methodology. In the context of that paper, J-difference editing was considered the gold standard, for establishing an unedited alternative, but limitations of both methodologies contribute to the low correlation value.
Metabolites that have been assessed using editing
Metabolites that are candidates for editing share a number of characteristics. They generally have low-to-medium concentration and give signals at the same chemical shift as stronger peaks in the spectrum. They usually have coupled spin systems, providing a mechanism to separate their signals from the rest of the spectrum. This coupling may be weak coupling, as in the case of J-difference editable molecules, or strong coupling as in the case of metabolites targeted with CT-PRESS. In the following sections, metabolites of the brain (whose molecular structures are shown in Figure 6) that can be observed with editing will be reviewed, briefly defining the role and pathophysiological interest in each, and outlining the spin system and successful approaches to editing each.
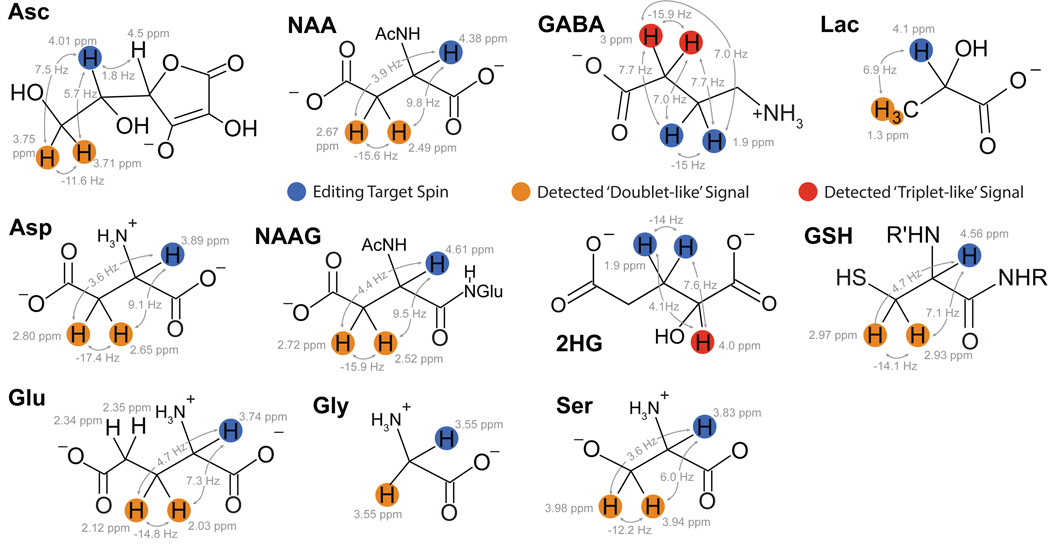
Figure 6.
Chemical structures and coupling constants of metabolites that can be measured using edited MRS methods. Asc: ascorbic acid, NAA: N-acetyl aspartate, GABA: γ-aminobutyric acid, Lac: lactate, Asp: aspartate, NAAG: N-acetyl aspartyl glutamate, 2HG: 2-hydroxyglutarate, GSH: glutathione, Glu: glutamate, Gly: glycine, Ser: serine.
2HG
2HG is an oncometabolite, having low concentration in healthy brain, but elevated concentrations in some tumors. It is formed by a mutation in isocitrate dehydrogenase (IDH1 and 2, in the cytosol and mitochondria respectively (16, 65)). There is a clear association between IDH mutations and overall survival level; therefore, detection of 2HG has prognostic value (16). 2HG has been detected in tumors of patients confirmed to have IDH1 mutations, while it was not detected in patients with wild-type IDH or healthy controls, supporting the sensitivity of measuring 2HG to IDH1 mutations (16, 65). 2HG detection has great potential as a sensitive, specific biomarker.
The structure of 2HG, shown in Figure 6, results in a methine (CH) signal at 1.9 ppm, coupled to a methylene (CH2) signal at 4.02 ppm. J-difference editing can be used to detect 2HG by applying an editing pulse at 1.9 ppm to isolate the 4.02 ppm resonance from the signals of creatine and phosphocreatine (3.92 ppm and 3.94 ppm, respectively), choline (4.05 ppm), myo-inositol (4.06 ppm) and lactate (4.1 ppm) (16, 65).
Ascorbate
Ascorbate (vitamin C) is an antioxidant that appears to have a preventative role in chronic degenerative disorders and cancer (66). The 1H-MR spectrum of ascorbate has multiplets at 3.73 ppm, 4.01 ppm and 4.50 ppm. MEGA-PRESS editing, applying editing pulses to 4.01 ppm spins and detecting edited signal at 3.73 ppm, has been developed (66). However, even editing pulses of 40-ms duration in this 4 T implementation were not sufficiently selective to avoid partially impacting the 3.73 ppm resonance. Therefore, the editing pulse is applied off-resonance to 4.13 ppm, with slightly reduced sensitivity. The DEW-MEGA-PRESS implementation has been applied to simultaneously detect ascorbate and GSH, referred to as the antioxidant profile (41).
GABA
GABA (γ-amino butyric acid) is the primary inhibitory neurotransmitter in the human brain and has been extensively studied using MRS editing methods. GABAergic inhibition is involved in the tuning and control of cortical responses and cerebral plasticity. Approximately 20% of cortical neurons are GABAergic and virtually all cortical neurons receive some form of GABA input. Due to the widespread presence of GABA in the cortex, the role of GABA in healthy function and GABAergic dysfunction implicated in many conditions has been widely studied. GABA has been shown to be correlated with functional imaging with tasks (67–72); however, inability to reproduce this work may hinder interpretations (73, 74). Changes in GABA as measured by MRS have been widely observed, including in healthy aging (75), developmental disorders (ADHD, autism spectrum disorder (76–78)), psychiatric disorders (high risk psychosis (79), schizophrenia (80–82)) and neurological disease (ALS (83), epilepsy (84) and chronic pain (85, 86)).
GABA has three methylene groups that appear on the 1H-MR spectrum at 3 ppm, 2.3 ppm and 1.9 ppm. The most common method to measure GABA is using MEGA-PRESS (36), detecting the GABA signal at 3 ppm (2CH2) that is coupled to the 1.9 ppm peak (3CH2) (Figure 6). Frequency-selective editing pulses are applied at 1.9 ppm to modulate the coupled 3-ppm spins without impacting the overlapping Cr peak. The Cr peak is then removed through subtraction in the difference spectrum, and GABA is retained. GABA is the metabolite most commonly detected using MEGA-PRESS editing, and other reviews have been written that cover the area in further detail (36, 87).
Due to the limited selectivity of the editing pulses (typically 14–16 ms in duration), as mentioned above, a limitation of this method is co-editing of the macromolecular resonance at 1.7 ppm and therefore measurements are often referred to as GABA+ (to indicate GABA+MM). One solution is to increase the selectivity of the editing pulses and place the Off editing pulse at 1.5 ppm, which is symmetric about the 1.7-ppm macromolecular peak (88, 89). This method has increased specificity for measuring GABA and limits the impact of inter-individual differences of the co-edited macromolecular signal, which may greatly impact results (38); however, it will be more susceptible to instabilities, motion and frequency drift (37) due to the more selective editing pulses which results in the rapid breakdown of symmetric suppression of macromolecules (90).
DQF experiments have been designed to detect GABA (45, 50). While these experiments demonstrated robust and reproducible measurements at 1.5 T, and sequence optimization of the STEAM acquisition has also been demonstrated (15) MEGA-PRESS is much more widely used.
Glutamate
Glutamate is the most concentrated metabolite in brain tissue. It is the primary excitatory neurotransmitter in the human brain but also has multiple metabolic roles and is closely associated with the tricarboxylic acid cycle. Glutamate levels, similar to NAA, can report on neuronal health and therefore reductions are often seen with degenerative diseases. However, excessive glutamate release can be neurotoxic, and elevated glutamate is seen in a number of disorders, including ALS (91) and potentially in traumatic brain injury (92).
Glutamate and glutamine have very similar chemical structures and therefore very similar spectra. As a result these metabolites overlap and are often referred to in combination as ‘Glx’. It is this highly overlapped structure that makes editing attractive as a method to suppress glutamine and improve the resolution and differentiation of glutamate (23). TE-averaging allows measurements of glutamate in isolation from glutamine, and has shown that glutamate has higher concentration in GM than WM (22). TE-averaging has also demonstrated reduced glutamate in HIV patients, particularly those with cognitive decline (93). Sequence optimization is an alternative approach to separate glutamate and glutamate (17, 54, 94). While these approaches show it is possible to differentiate glutamate and glutamine with ideal conditions and high-quality data, performance of these methods in sub-optimal conditions, as seen in clinical studies, is less clear. Otherwise, the use of edited methods specifically to measure glutamate is relatively rare, given that excellent reproducibility can be achieved for Glx (total glutamate+glutamine) measurements.
Glutathione (GSH)
Glutathione is the brain’s most abundant antioxidant, protecting cells against reactive oxygen compounds and is therefore considered a marker of oxidative stress. In addition, GSH is necessary for the synthesis and breakdown of proteins and DNA precursors. The concentration of GSH in the brain is on the order of 2–3 mM, although GSH is found throughout the body. GSH appears to decrease with age (95). Preliminary work suggests that GSH may be reduced in stroke lesions (96). Due to the role of GSH in mitigating against oxidative stress, GSH may be involved in the pathophysiology of schizophrenia and lower GSH levels have been observed in the middle temporal lobe of patients with first episode psychosis (97).
GSH is a tripeptide consisting of glutamate, cysteine and glycine. As a result, its spectrum is complex, with peaks from the cysteine moiety at 2.93, 2.98 and 4.56 ppm, peaks from the glutamate-moiety at 2.15, 2.55 and 3.77 ppm and the glycine peak at 3.77 ppm. With MRS, either the cysteinyl moiety, relying on the J-coupling between the 2.95 ppm and the 4.56 ppm peaks (35), or the glycine moiety, relying on the J-coupling between the 3.77 ppm and 2.1 ppm (NH) signal (98) can be targets for editing.
MEGA-PRESS (35), polarization transfer (54) and double quantum filtering (96) methods have been used to measure GSH. For MEGA-PRESS methods, the most common approach is to apply editing pulses at 4.56 ppm and measure the difference peak at 2.95 ppm (35, 98). The spectrum is substantially simplified in this method, however, the NAA-aspartyl peaks at 2.45 and 2.67 ppm are co-edited, making quantification more challenging. The impact of this coediting was minimized after simulations showed maximal GSH signal at echo time of 68 ms (35). Since then, additional experiments have suggested an longer echo times of 120 ms or 131 ms result in signal maximization; however the optimal TE will depend on many factors in addition to the echo time (31, 96). In the double-quantum experiments, the cysteinyl group was also targeted (99). Simulations and phantom work indicate high signal yields with polarization transfer methods, but this has yet to be applied in vivo (54).
Glycine
Glycine is an inhibitory neurotransmitter and coagonist at glutamatergic NMDA receptors. Since glycine is associated NMDA activity and glycine been suggested as a treatment for NMDA dysfunction in schizophrenia (100). Glycine has also been suggested to be a biomarker for tumor malignancy as increased glycine is seen in astrocytomas and glioblastoma but not low-grade tumors or normal tissue (101).
Glycine appears as a singlet at 3.55 ppm, however, its detection is complicated by the over-lapping, more highly concentrated myo-inositol resonances at 3.61ppm and 3.52 ppm. Because these myo-inositol resonances are coupled and phase modulated, TE-averaging can suppress the myo-inositol peaks to permit quantification of glycine (102). Quantification of glycine has not been widely applied, however using the TE-averaging approach, oral glycine supplements over the course of two weeks increased glycine levels in the brain by 260% (100).
Lactate
Lactate is a by-product of anaerobic metabolism. As such, an increase in lactate often indicates altered metabolism, as found in cancer. It is becoming more recognized that lactate is an essential metabolic intermediate in many organs, in particular the brain, lactate is likely shuttled between astrocytes and neurons to meet high-energy demands (82, 103).
Lactate is an AX3 spin system, with a doublet at 1.33 and a quartet at 4.1 ppm. Its low concentration and the macromolecule signals at 1.24 ppm and 1.43 ppm limit the detection of lactate in typical, healthy brain (104). Lactate has been measured using MEGA-PRESS (105–107), single and dual BASING (30), MQF (108) and polarization transfer (54). In these editing methods, the editing pulse is applied at 4.1 ppm and the signal of the 1.33 ppm peak is detected.
Lactate is often observed in tumors and stroke due to increased anaerobic metabolism in these conditions (109, 110) and lactate levels may be useful to indicate response to therapy (107). While some studies have relied on the increased concentration of lactate alone as a biomarker (109) others have shown utility and reliability in editing of lactate in tumor, including J-difference editing and MQF methods (107, 108). In healthy participants, increased lactate has been observed during an inspiratory hypoxic challenge (105, 106).
NAAG
N-acetyl aspartyl glutamate (NAAG) is a peptide neuromodulator in the human brain. It is formed from NAA and glutamate. The functional profile of NAA and NAAG remain incomplete, despite the fact that the most predominant signal in the 1H-MR spectrum of the brain is NAA. Functions of NAAG include inhibiting synaptic release of GABA, glutamate and dopamine, regulating GABA receptor expression and reducing cyclic AMP levels (88, 111).
MEGA-PRESS can be used to isolate NAAG from NAA, based on the aspartyl spin systems in both. To measure NAAG, On editing pulses are applied at 4.61 ppm to refocus the coupled spins at 2.6 ppm. Due to the limited selectivity of editing pulses, symmetric suppression of NAA in NAAG measurements is beneficial, so Off editing pulses are placed symmetrically about the 4.38 ppm NAA peak at 4.15 ppm. Similarly, to isolate the 2.5 ppm NAA peak from NAAG, On editing pulses are placed at 4.38 ppm and in the Off condition, editing pulses are symmetric about the 4.61 ppm NAAG peak at 4.84 ppm. This method has been validated at 3T (88) and 7T (112).
TE-averaging with regularized lineshape deconvolution has also been used to isolate the NAAG signal (113). In this method, strongly coupled multiple resonance lines and macromolecules are suppressed, while the singlets of NAAG and NAA are not affected and therefore the spectral resolution of NAAG and NAA is improved.
Few studies have specifically examined NAAG, using editing to isolate it from the larger, overlapping NAA peak. In healthy participants, the concentration of NAAG in white matter appears to be twice that of grey matter; however, this result was derived from using 2 voxels (one WM rich and one GM rich) and small cohorts of participants (112, 113). In schizophrenia, a correlation between centrum semiovale NAAG levels and symptom severity has been shown and that NAAG lower in an older cohort and is higher in a younger cohort (80). A greater number of publications not using an editing method and showing NAAG results, typically derived from LCModel have been published; however, due to the overlapping nature of NAAG and NAA, the accuracy of this analysis method is highly dependent on spectral quality and difficult to evaluate.
Serine
The endogenous amino acid serine modulates the activity of glutamate at NMDA receptors. In schizophrenia and psychosis, alterations of glutamatergic transmission have been found, which may include alterations of the coagonist serine (114).
The serine spectrum consists of peaks at 3.8, 3.94 and 3.83 ppm that are strongly coupled, and overlapped with a creatine peak at 3.92 ppm, making serine a candidate for asymmetric difference editing (i.e., CT-PRESS). A constant-TE triple-refocusing difference editing strategy for serine has been proposed and tested at 7T (114). This method adds an additional frequency-selective 180° pulse between the two 180° refocusing pulses of the PRESS sequence to refocuses all resonances between 1.8 and 4.0 ppm. As described above, uncoupled, singlet resonances, in this case the 3.9 ppm creatine peak, are not impacted by the sub-echo times, only strongly coupled spins will be impacted. In the difference spectrum, the serine peak remains while the overlapping creatine peak is removed. Even with optimized subecho times, the total TE is relatively long; thus, even at 7T, this method still suffers from low SNR.
Summary
MRS measures the concentration of tissue metabolites in order to interrogate tissue status and therefore can be used to understand disease processes. Many tissue metabolites are not easily resolved with conventional spectroscopy. In this manuscript, we have presented editing methods that reduce the information in the one dimensional 1H-MR spectrum in order to resolve information about other metabolites. We then summarized the metabolites that can then be better resolved by applying these methods. The application of these editing procedures is somewhat technically challenging but can yield useful and applicable information about specific metabolites and the associated understanding of metabolic function and dysfunction in disease.
Acknowledgments
We acknowledge funding from the Alberta Children’s Hospital Research Institute and the Hotchkiss Brain Institute, University of Calgary and NIH grants: R01 EB016089 and P41 EB015090.
Appendix 1: Supplementary Notes on MR Physics
Coupling
Spins are coupled if the spin-state of one impacts the energy levels, or resonant frequency, of the other. In the case of scalar (J-) coupling, this occurs through the bonding network of a molecule and can be conceptualized as a transfer of information within the molecule – ‘one spin knows what is going on with the other’. The size of a scalar coupling is expressed as the coupling constant (J, measured in Hz). The 1H-MR spectrum for most small organic molecules (including metabolites) consists of a number of signals with different chemical shifts (reflecting differing electronic environments within the molecule) and splittings due to J-couplings (reflecting interactions with adjacent, coupled protons). For example, lactate has two peaks: the doublet at 1.31 ppm arises from the three magnetically equivalent protons of the methyl group (3CH3); the quartet at 4.1 ppm corresponds to the methine proton (2CH). Both are split due to a mutual 7-Hz coupling. The methyl signal is split into a doublet, reflecting the two spin-states of the methine protons. Similarly, the methine signal is split into a quartet reflecting permutations of the independent spin states of the three methyl protons. Coupling has two main impacts on signals in the spectrum, reducing their peak amplitude and widening their footprint along the chemical shift axis, both of which make it harder to resolve coupled signals in the in vivo MR spectrum.
Couplings can be classified as weak or strong - a coupling is weak when the coupling constant is much smaller than the chemical shift difference between the coupled spins, and otherwise it is strong. The 7 Hz coupling of lactate is much smaller than the ~350 Hz separation (at 3T) between the two peaks and therefore the lactate spin system is weakly coupled.
Coherence Transfer
Coherence transfer is a process by which transverse magnetization (observable coherence) associated with one spin is converted into transverse magnetization associated with another spin. Usually this transfer occurs between spins that are J-coupled, and is caused by 90° RF pulses. Coherences can be classified by order – single-quantum coherences are observable (only −1, by convention), which multiple-quantum coherences are not. Coherences of different orders can be differentiated by phase cycling or gradient selection. Coherence pathway diagrams are used to illustrate the coherences retained at various points during a pulse sequence, particularly for MQF experiments.
References
- 1.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Proc Natl Acad Sci U S A. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 2.Mlynarik V, Gambarota G, Frenkel H, Gruetter R. Localized short-echo-time proton MR spectroscopy with full signal-intensity acquisition. Magn Reson Med. 2006;56(5):965–970. doi: 10.1002/mrm.21043. [DOI] [PubMed] [Google Scholar]
- 3.Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H–MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59(1):1–6. doi: 10.1002/mrm.21302. [DOI] [PubMed] [Google Scholar]
- 4.Yahya A. Metabolite detection by proton magnetic resonance spectroscopy using PRESS. Progress in Nuclear Magnetic Resonance Spectroscopy. 2009;55(3):183–198. [Google Scholar]
- 5.Frahm J, Merboldt K, Hänicke W, Haase A. Stimulated echo imaging. Journal of Magnetic Resonance. 1985;64(1):81–93. [Google Scholar]
- 6.Morris GA, Freeman R. Selective excitation in Fourier transform nuclear magnetic resonance. Journal of Magnetic Resonance (1969) 1978;29(3):433–462. doi: 10.1016/j.jmr.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 7.Bendall MR, Pegg DT. Complete accurate editing of decoupled 13 C spectra using DEPT and a quaternary-only sequence. Journal of Magnetic Resonance (1969) 1983;53(2):272–296. [Google Scholar]
- 8.Doddrell D, Pegg D, Bendall MR. Distortionless enhancement of NMR signals by polarization transfer. Journal of Magnetic Resonance (1969) 1982;48(2):323–327. [Google Scholar]
- 9.Kessler H, Oschkinat H, Griesinger C, Bermel W. Transformation of homonuclear two-dimensional NMR techniques into one-dimensional techniques using Gaussian pulses. Journal of Magnetic Resonance (1969) 1986;70(1):106–133. [Google Scholar]
- 10.He Q, Shungu DC, Vanzijl PC, Bhujwalla ZM, Glickson JD. Single-scan in vivo lactate editing with complete lipid and water suppression by selective multiple-quantum-coherence transfer (Sel-MQC) with application to tumors. Journal of Magnetic Resonance, Series B. 1995;106(3):203–211. doi: 10.1006/jmrb.1995.1035. [DOI] [PubMed] [Google Scholar]
- 11.Rothman DL, Petroff O, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proceedings of the National Academy of Sciences. 1993;90(12):5662–5666. doi: 10.1073/pnas.90.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sotak CH, Freeman DM. A method for volume-localized lactate editing using zero-quantum coherence created in a stimulated-echo pulse sequence. Journal of Magnetic Resonance (1969) 1988;77(2):382–388. doi: 10.1002/mrm.1910070315. [DOI] [PubMed] [Google Scholar]
- 13.Yahya A, Mädler B, Fallone BG. Exploiting the chemical shift displacement effect in the detection of glutamate and glutamine (Glx) with PRESS. Journal of Magnetic Resonance. 2008;191(1):120–127. doi: 10.1016/j.jmr.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Thompson RB, Allen PS. Response of metabolites with coupled spins to the STEAM sequence. Magnetic resonance in medicine. 2001;45(6):955–965. doi: 10.1002/mrm.1128. [DOI] [PubMed] [Google Scholar]
- 15.Hanstock CC, Coupland NJ, Allen PS. GABA X2 multiplet measured pre- and post-administration of vigabatrin in human brain. Magnetic resonance in medicine. 2002;48(4):617–623. doi: 10.1002/mrm.10265. [DOI] [PubMed] [Google Scholar]
- 16.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schubert F, Gallinat J, Seifert F, Rinneberg H. Glutamate concentrations in human brain using single voxel proton magnetic resonance spectroscopy at 3 Tesla. Neuroimage. 2004;21(4):1762–1771. doi: 10.1016/j.neuroimage.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Hu J, Yang S, Xuan Y, Jiang Q, Yang Y, Haacke EM. Simultaneous detection of resolved glutamate, glutamine, and γ-aminobutyric acid at 4T. Journal of Magnetic Resonance. 2007;185(2):204–213. doi: 10.1016/j.jmr.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Near J, Andersson J, Maron E, Mekle R, Gruetter R, Cowen P, et al. Unedited in vivo detection and quantification of gamma-aminobutyric acid in the occipital cortex using short-TE MRS at 3 T. NMR Biomed. 2013;26(11):1353–1362. doi: 10.1002/nbm.2960. [DOI] [PubMed] [Google Scholar]
- 20.Hancu I, Zimmerman EA, Sailasuta N, Hurd RE. 1H MR spectroscopy using TE averaged PRESS: A more sensitive technique to detect neurodegeneration associated with Alzheimer’s disease. Magnetic Resonance in Medicine. 2005;53(4):777–782. doi: 10.1002/mrm.20419. [DOI] [PubMed] [Google Scholar]
- 21.Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn Reson Med. 2004;51(3):435–440. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Shen J. Regional and tissue-specific differences in brain glutamate concentration measured by in vivo single voxel MRS. J Neurosci Methods. 2015;239:94–99. doi: 10.1016/j.jneumeth.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonenc A, Govind V, Sheriff S, Maudsley AA. Comparison of spectral fitting methods for overlapping J-coupled metabolite resonances. Magn Reson Med. 2010;64(3):623–628. doi: 10.1002/mrm.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancu I. Optimized glutamate detection at 3T. J Magn Reson Imaging. 2009;30(5):1155–1162. doi: 10.1002/jmri.21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullins PG, Chen H, Xu J, Caprihan A, Gasparovic C. Comparative reliability of proton spectroscopy techniques designed to improve detection of J-coupled metabolites. Magn Reson Med. 2008;60(4):964–969. doi: 10.1002/mrm.21696. [DOI] [PubMed] [Google Scholar]
- 26.Yang S, Salmeron BJ, Ross TJ, Xi ZX, Stein EA, Yang Y. Lower glutamate levels in rostral anterior cingulate of chronic cocaine users - A (1)H-MRS study using TE-averaged PRESS at 3 T with an optimized quantification strategy. Psychiatry Res. 2009;174(3):171–176. doi: 10.1016/j.pscychresns.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryner LN, Sorenson JA, thomas MA3D. Localized 2D NMR Spectroscopy on an MRI Scanner. J Magn Reson B. 1995;107:126–137. doi: 10.1006/jmrb.1995.1068. [DOI] [PubMed] [Google Scholar]
- 28.Star-Lack J, Nelson SJ, Kurhanewicz J, Huang LR, Vigneron DB. Improved Water and Lipid Suppression for 3D PRESS CSI Using RF Band Selective Inversion with Gradient Dephasing (BASING) Magn Reson Med. 1997;38(2):311–321. doi: 10.1002/mrm.1910380222. [DOI] [PubMed] [Google Scholar]
- 29.Mescher M, Tannus A, O’Neil Johnson M, Garwood M. Solvent Suppression Using Selective Echo Dephasing. Journal of Magnetic Resonance, Series A. 1996;123(2):226–229. [Google Scholar]
- 30.Star-Lack J, Spielman D, Adalsteinsson E, Kurhanewicz J, Terris DJ, Vigneron DB. In Vivo Lactate Editing with Simultaneous Detection of Choline, Creatine, NAA and Lipid Singlets at 1.5 T Using PRESS Excitation with Applications to the Study of Brain and Head and Neck Tumors. J Magn Reson. 1998;133(2):243–254. doi: 10.1006/jmre.1998.1458. [DOI] [PubMed] [Google Scholar]
- 31.Chan KL, Puts NA, Snoussi K, Harris AD, Barker PB, Edden RA. Echo time optimization for J-difference editing of glutathione at 3T. Magn Reson Med. 2016 doi: 10.1002/mrm.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Near J, Evans CJ, Puts NA, Barker PB, Edden RA. J-difference editing of gamma-aminobutyric acid (GABA): simulated and experimental multiplet patterns. Magn Reson Med. 2013;70(5):1183–1191. doi: 10.1002/mrm.24572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder J, Thompson RB, Wilman AH. Difference spectroscopy using PRESS asymmetry: application to glutamate, glutamine, and myo-inositol. NMR Biomed. 2010;23(1):41–47. doi: 10.1002/nbm.1424. [DOI] [PubMed] [Google Scholar]
- 34.Snyder J, Hanstock CC, Wilman AH. Spectral editing of weakly coupled spins using variable flip angles in PRESS constant echo time difference spectroscopy: application to GABA. J Magn Reson. 2009;200(2):245–250. doi: 10.1016/j.jmr.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Terpstra M, Henry PG, Gruetter R. Measurement of reduced glutathione (GSH) in human brain using LCModel analysis of difference-edited spectra. Magn Reson Med. 2003;50(1):19–23. doi: 10.1002/mrm.10499. [DOI] [PubMed] [Google Scholar]
- 36.Mullins PG, McGonigle DJ, O’Gorman RL, Puts NA, Vidyasagar R, Evans CJ, et al. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage. 2014;86:43–52. doi: 10.1016/j.neuroimage.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris AD, Glaubitz B, Near J, John Evans C, Puts NA, Schmidt-Wilcke T, et al. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med. 2014;72(4):941–948. doi: 10.1002/mrm.25009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris AD, Puts NA, Barker PB, Edden RA. Spectral-editing measurements of GABA in the human brain with and without macromolecule suppression. Magn Reson Med. 2015;74(6):1523–1529. doi: 10.1002/mrm.25549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Near J, Simpson R, Cowen P, Jezzard P. Efficient gamma-aminobutyric acid editing at 3T without macromolecule contamination: MEGA-SPECIAL. NMR Biomed. 2011;24(10):1277–1285. doi: 10.1002/nbm.1688. [DOI] [PubMed] [Google Scholar]
- 40.Mikkelsen M, Singh KD, Sumner P, Evans CJ. Comparison of the repeatability of GABA-edited magnetic resonance spectroscopy with and without macromolecule suppression. Magn Reson Med. 2016;75(3):946–953. doi: 10.1002/mrm.25699. [DOI] [PubMed] [Google Scholar]
- 41.Terpstra M, Marjanska M, Henry PG, Tkac I, Gruetter R. Detection of an antioxidant profile in the human brain in vivo via double editing with MEGA-PRESS. Magn Reson Med. 2006;56(6):1192–1199. doi: 10.1002/mrm.21086. [DOI] [PubMed] [Google Scholar]
- 42.Chan KL, Puts NA, Schar M, Barker PB, Edden RA. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn Reson Med. 2016 doi: 10.1002/mrm.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleh MG, Oeltzschner G, Chan KL, Puts NA, Mikkelsen M, Schär M, et al. Simultaneous edited MRS of GABA and glutathione. Neuroimage. 2016 doi: 10.1016/j.neuroimage.2016.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gambarota G, van der Graaf M, Klomp D, Mulkern RV, Heerschap A. Echo-time independent signal modulations using PRESS sequences: a new approach to spectral editing of strongly coupled AB spin systems. J Magn Reson. 2005;177(2):299–306. doi: 10.1016/j.jmr.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 45.McLean MA, Busza AL, Wald LL, Simister RJ, Barker GJ, Williams SR. In vivo GABA+ measurement at 1.5T using a PRESS-localized double quantum filter. Magn Reson Med. 2002;48(2):233–241. doi: 10.1002/mrm.10208. [DOI] [PubMed] [Google Scholar]
- 46.Wilman AH, Allen PS. Double-Quantum Filtering of Citrate for in Vivo Observation. J Magn Reson Series B. 1994;105:58–60. doi: 10.1006/jmrb.1994.1100. [DOI] [PubMed] [Google Scholar]
- 47.He Q, Bhujwalla ZM, Maxwell RJ, Griffiths JR, Glickson JD. Proton NMR Observation of the Antineoplastic Agent Iproplatin In Vivo by Selective Multiple Quantum Coherence Transfer (Sel-MQC) Magnetic resonance in medicine. 1995;33(3):414–416. doi: 10.1002/mrm.1910330315. [DOI] [PubMed] [Google Scholar]
- 48.Hurd RE, Freeman D. Proton editing and imaging of lactate. NMR in biomedicine. 1991;4:73–80. doi: 10.1002/nbm.1940040208. [DOI] [PubMed] [Google Scholar]
- 49.Melkus G, Mörchel P, Behr VC, Kotas M, Flentje M, Jakob PM. Short-echo spectroscopic imaging combined with lactate editing in a single scan. NMR in biomedicine. 2008;21(10):1076–1086. doi: 10.1002/nbm.1284. [DOI] [PubMed] [Google Scholar]
- 50.Keltner JR, Wald LL, Frederick BD, Renshaw PF. In Vivo Detection of GABA in Human Brain Using a Localized Double-Quantum Filter Technique. Magn Reson Med. 1997;37:366–371. doi: 10.1002/mrm.1910370312. [DOI] [PubMed] [Google Scholar]
- 51.Von Kienlin M, Albrand J, Authier B, Blondet P, Lotito S, Decorps M. Spectral editing in vivo by homonuclear polarization transfer. Journal of Magnetic Resonance. 1987;75(2):371–377. [Google Scholar]
- 52.Shen J, Yang J, Choi I-Y, Li SS, Chen Z. A new strategy for in vivo spectral editing Application to GABA editing using selective homonuclear polarization transfer spectroscopy. Journal of Magnetic Resonance. 2004;170(2):290–298. doi: 10.1016/j.jmr.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Pan J, Duckrow R, Spencer D, Avdievich N, Hetherington H. Selective homonuclear polarization transfer for spectroscopic imaging of GABA at 7T. Magnetic resonance in medicine. 2013;69(2):310–316. doi: 10.1002/mrm.24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yahya A, Gino Fallone B. Incorporating homonuclear polarization transfer into PRESS for proton spectral editing: illustration with lactate and glutathione. J Magn Reson. 2007;188(1):111–121. doi: 10.1016/j.jmr.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Choi IY, Lee SP, Shen J. Selective homonuclear Hartmann-Hahn transfer method for in vivo spectral editing in the human brain. Magn Reson Med. 2005;53(3):503–510. doi: 10.1002/mrm.20381. [DOI] [PubMed] [Google Scholar]
- 56.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR in Biomedicine. 2001;14(4):260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 57.Wilson M, Reynolds G, Kauppinen RA, Arvanitis TN, Peet AC. A constrained least-squares approach to the automated quantitation of in vivo 1H magnetic resonance spectroscopy data. Magnetic resonance in medicine. 2011;65(1):1–12. doi: 10.1002/mrm.22579. [DOI] [PubMed] [Google Scholar]
- 58.Vanhamme L, van den Boogaart A, Van Huffel S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. Journal of Magnetic Resonance. 1997;129(1):35–43. doi: 10.1006/jmre.1997.1244. [DOI] [PubMed] [Google Scholar]
- 59.Naressi A, Couturier C, Devos J, Janssen M, Mangeat C, De Beer R, et al. Java-based graphical user interface for the MRUI quantitation package. Magnetic Resonance Materials in Physics, Biology and Medicine. 2001;12(2–3):141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- 60.Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. Journal of Magnetic Resonance Imaging. 2014;40(6):1445–1452. doi: 10.1002/jmri.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saleh MG, Near J, Alhamud A, Robertson F, van der Kouwe AJ, Meintjes EM. Reproducibility of macromolecule suppressed GABA measurement using motion and shim navigated MEGA-SPECIAL with LCModel, jMRUI and GANNET. Magnetic Resonance Materials in Physics, Biology and Medicine. 2016:1–12. doi: 10.1007/s10334-016-0578-8. [DOI] [PubMed] [Google Scholar]
- 62.O’Gorman RL, Michels L, Edden RA, Murdoch JB, Martin E. In vivo detection of GABA and glutamate with MEGA-PRESS: Reproducibility and gender effects. Journal of magnetic resonance Imaging. 2011;33(5):1262–1267. doi: 10.1002/jmri.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henry ME, Lauriat TL, Shanahan M, Renshaw PF, Jensen JE. Accuracy and stability of measuring GABA, glutamate, and glutamine by proton magnetic resonance spectroscopy: a phantom study at 4Tesla. Journal of Magnetic Resonance. 2011;208(2):210–218. doi: 10.1016/j.jmr.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans CJ, Puts NA, Robson SE, Boy F, McGonigle DJ, Sumner P, et al. Subtraction artifacts and frequency (mis-)alignment in J-difference GABA editing. Journal of magnetic resonance imaging : JMRI. 2013;38(4):970–975. doi: 10.1002/jmri.23923. [DOI] [PubMed] [Google Scholar]
- 65.Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, et al. Detectio of 2-Hydroxyglutarate in IDH-Mutated Glioma Patients by In Vivo Spectral-Editing and 2D Correlation Magnetic Resonance Spectroscopy. Sci Transl Med. 2012;4(116):116ra4. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Terpstra M, Gruetter R. 1H NMR detection of vitamin C in human brain in vivo. Magn Reson Med. 2004;51(2):225–229. doi: 10.1002/mrm.10715. [DOI] [PubMed] [Google Scholar]
- 67.Muthukumaraswamy SD, Edden RA, Jones DK, Swettenham JB, Singh KD. Resting GABA concentration predicts peak gamma frequency and fMRI amplitude in response to visual stimulation in humans. Proc Natl Acad Sci U S A. 2009;106(20):8356–8361. doi: 10.1073/pnas.0900728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Donahue MJ, Rane S, Hussey E, Mason E, Pradhan S, Waddell KW, et al. gamma-Aminobutyric acid (GABA) concentration inversely correlates with basal perfusion in human occipital lobe. J Cereb Blood Flow Metab. 2014;34(3):532–541. doi: 10.1038/jcbfm.2013.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stagg CJ, Bachtiar V, Johansen-Berg H. The role of GABA in human motor learning. Curr Biol. 2011;21(6):480–484. doi: 10.1016/j.cub.2011.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Puts NA, Edden RA, Evans CJ, McGlone F, McGonigle DJ. Regionally specific human GABA concentration correlates with tactile discrimination thresholds. J Neurosci. 2011;31(46):16556–16560. doi: 10.1523/JNEUROSCI.4489-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Northoff G, Walter M, Schulte RF, Beck J, Dydak U, Henning A, et al. GABA concentrations in the human anterior cingulate cortex predict negative BOLD responses in fMRI. Nat Neurosci. 2007;10(12):1515–1517. doi: 10.1038/nn2001. [DOI] [PubMed] [Google Scholar]
- 72.Duncan NW, Wiebking C, Northoff G. Associations of regional GABA and glutamate with intrinsic and extrinsic neural activity in humans-a review of multimodal imaging studies. Neurosci Biobehav Rev. 2014;47:36–52. doi: 10.1016/j.neubiorev.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 73.Harris AD, Puts NA, Anderson BA, Yantis S, Pekar JJ, Barker PB, et al. Multi-regional investigation of the relationship between functional MRI blood oxygenation level dependent (BOLD) activation and GABA concentration. PLoS One. 2015;10(2):e0117531. doi: 10.1371/journal.pone.0117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cousijn H, Haegens S, Wallis G, Near J, Stokes MG, Harrison PJ, et al. Resting GABA and gluatmate concentrations do not predict visual gamma frequency or amplitude. PNAS. 2014;111(25):9301–9306. doi: 10.1073/pnas.1321072111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao F, Edden RA, Li M, Puts NA, Wang G, Liu C, et al. Edited magnetic resonance spectroscopy detects an age-related decline in brain GABA levels. Neuroimage. 2013;78:75–82. doi: 10.1016/j.neuroimage.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bollmann S, Ghisleni C, Poil SS, Martin E, Ball J, Eich-Hochli D, et al. Developmental changes in gamma-aminobutyric acid levels in attention-deficit/hyperactivity disorder. Transl Psychiatry. 2015;5:e589. doi: 10.1038/tp.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Puts NA, Harris AD, Crocetti D, Nettles C, Singer HS, Tommerdahl M, et al. Reduced GABAergic inhibition and abnormal sensory symptoms in children with Tourette syndrome. J Neurophysiol. 2015;114(2):808–817. doi: 10.1152/jn.00060.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edden RAE, Crocetti D, Zhu H, Gilbert DL, Mostofsky SH. Reduced GABA concentration in attention-deficit/hyperactivity disorder. Arch Gen Psychiatry. 2012;69(7):750–753. doi: 10.1001/archgenpsychiatry.2011.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de la Fuente-Sandoval C, Reyes-Madrigal F, Mao X, Leon-Ortiz P, Rodriguez-Mayoral O, Solis-Vivanco R, et al. Cortico-Striatal GABAergic and Glutamatergic Dysregulations in Subjects at Ultra-High Risk for Psychosis Investigated with Proton Magnetic Resonance Spectroscopy. Int J Neuropsychopharmacol. 2015;19(3) doi: 10.1093/ijnp/pyv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rowland LM, Kontson K, West J, Edden RA, Zhu H, Wijtenburg SA, et al. In vivo measurements of glutamate, GABA, and NAAG in schizophrenia. Schizophr Bull. 2013;39(5):1096–1104. doi: 10.1093/schbul/sbs092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rowland LM, Krause BW, Wijtenburg SA, McMahon RP, Chiappelli J, Nugent KL, et al. Medial frontal GABA is lower in older schizophrenia: a MEGA-PRESS with macromolecule suppression study. Mol Psychiatry. 2016;21(2):198–204. doi: 10.1038/mp.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maddock RJ, Buonocore MH. MR spectroscopic studies of the brain in psychiatric disorders. Curr Top Behav Neurosci. 2012;11:199–251. doi: 10.1007/7854_2011_197. [DOI] [PubMed] [Google Scholar]
- 83.Foerster BR, Pomper MG, Callaghan BC, Petrou M, Edden RA, Mohamed MA, et al. An imbalance between excitatory and inhibitory neurotransmitters in amyotrophic lateral sclerosis revealed by use of 3-T proton magnetic resonance spectroscopy. JAMA Neurol. 2013;70(8):1009–1016. doi: 10.1001/jamaneurol.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simister RJ, McLean MA, Barker GJ, Duncan JS. Proton magnetic resonance spectroscopy of malformations of cortical development causing epilepsy. Epilepsy Res. 2007;74(2–3):107–115. doi: 10.1016/j.eplepsyres.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 85.Foerster BR, Petrou M, Edden RA, Sundgren PC, Schmidt-Wilcke T, Lowe SE, et al. Reduced insular gamma-aminobutyric acid in fibromyalgia. Arthritis Rheum. 2012;64(2):579–583. doi: 10.1002/art.33339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Petrou M, Pop-Busui R, Foerster BR, Edden RA, Callaghan BC, Harte SE, et al. Altered excitation-inhibition balance in the brain of patients with diabetic neuropathy. Acad Radiol. 2012;19(5):607–612. doi: 10.1016/j.acra.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Puts NA, Edden RA. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Reson Spectrosc. 2012;60:29–41. doi: 10.1016/j.pnmrs.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edden RA, Pomper MG, Barker PB. In vivo differentiation of N-acetyl aspartyl glutamate from N-acetyl aspartate at 3 Tesla. Magn Reson Med. 2007;57(6):977–982. doi: 10.1002/mrm.21234. [DOI] [PubMed] [Google Scholar]
- 89.Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA Editing Without Macromolecule Contamination. Magn Reson Med. 2001;45:517–520. doi: 10.1002/1522-2594(200103)45:3<517::aid-mrm1068>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 90.Edden RAE, Oeltzschner G, Harris AD, Puts NAJ, Chan K, Boer VO, et al. Symmetrical spectral editing at 3T requires prospective frequency correction. J Magn Reson Imaging. 2016 doi: 10.1002/jmri.25304. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han J, Ma L. Study of the features of proton MR spectroscopy ((1)H-MRS) on amyotrophic lateral sclerosis. J Magn Reson Imaging. 2010;31(2):305–308. doi: 10.1002/jmri.22053. [DOI] [PubMed] [Google Scholar]
- 92.Ashwal S, Holshouser B, Tong K, Serna T, Osterdock R, Gross M, et al. Proton MR spectroscopy detected glutamate/glutamine is increased in children with traumatic brain injury. J Neurotrauma. 2004;21(11):1539–1552. doi: 10.1089/neu.2004.21.1539. [DOI] [PubMed] [Google Scholar]
- 93.Ernst T, Jiang CS, Nakama H, Buchthal S, Chang L. Lower brain glutamate is associated with cognitive deficits in HIV patients: a new mechanism for HIV-associated neurocognitive disorder. J Magn Reson Imaging. 2010;32(5):1045–1053. doi: 10.1002/jmri.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thompson RB, Allen PS. Response of metabolites with coupled spins to the STEAM sequence. Magnetic resonance in medicine. 2001;45:955–965. doi: 10.1002/mrm.1128. [DOI] [PubMed] [Google Scholar]
- 95.Emir UE, Raatz S, McPherson S, Hodges JS, Torkelson C, Tawfik P, et al. Noninvasive quantification of ascorbate and glutathione concentration in the elderly human brain. NMR Biomed. 2011;24(7):888–894. doi: 10.1002/nbm.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.An L, Zhang Y, Thomasson DM, Latour LL, Baker EH, Shen J, et al. Measurement of glutathione in normal volunteers and stroke patients at 3T using J-difference spectroscopy with minimized subtraction errors. J Magn Reson Imaging. 2009;30(2):263–270. doi: 10.1002/jmri.21832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wood SJ, Berger GE, Wellard RM, Proffitt TM, McConchie M, Berk M, et al. Medial temporal lobe glutathione concentration in first episode psychosis: a 1H–MRS investigation. Neurobiol Dis. 2009;33(3):354–357. doi: 10.1016/j.nbd.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 98.Kaiser LG, Marjanska M, Matson GB, Iltis I, Bush SD, Soher BJ, et al. (1)H MRS detection of glycine residue of reduced glutathione in vivo. J Magn Reson. 2010;202(2):259–266. doi: 10.1016/j.jmr.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trabesinger AH, Weber OM, Duc CO, Boesiger P. Detection of glutathione in the human brain in vivo by means of double quantum coherence filtering. Magn Reson Med. 1999;42:283–289. doi: 10.1002/(sici)1522-2594(199908)42:2<283::aid-mrm10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 100.Kaufman MJ, Prescot AP, Ongur D, Evins AE, Barros TL, Medeiros CL, et al. Oral glycine administration increases brain glycine/creatine ratios in men: a proton magnetic resonance spectroscopy study. Psychiatry Res. 2009;173(2):143–149. doi: 10.1016/j.pscychresns.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Choi C, Ganji SK, DeBerardinis RJ, Dimitrov IE, Pascual JM, Bachoo R, et al. Measurement of glycine in the human brain in vivo by 1H–MRS at 3 T: application in brain tumors. Magn Reson Med. 2011;66(3):609–618. doi: 10.1002/mrm.22857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Prescot AP, de BFB, Wang L, Brown J, Jensen JE, Kaufman MJ, et al. In vivo detection of brain glycine with echo-time-averaged (1)H magnetic resonance spectroscopy at 4.0 T. Magn Reson Med. 2006;55(3):681–686. doi: 10.1002/mrm.20807. [DOI] [PubMed] [Google Scholar]
- 103.Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci U S A. 1994;91(22):10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Behar KL, Rothman DL, Spencer DD, Petroff OAC. Analysis of Macromolecule Resonance in 1H NMR Spectra of Human Brain. Magn Reson Med. 1994;32:294–302. doi: 10.1002/mrm.1910320304. [DOI] [PubMed] [Google Scholar]
- 105.Edden RA, Harris AD, Murphy K, Evans CJ, Saxena N, Hall JE, et al. Edited MRS is sensitive to changes in lactate concentration during inspiratory hypoxia. J Magn Reson Imaging. 2010;32(2):320–325. doi: 10.1002/jmri.22233. [DOI] [PubMed] [Google Scholar]
- 106.Harris AD, Roberton VH, Huckle DL, Saxena N, Evans CJ, Murphy K, et al. Temporal dynamics of lactate concentration in the human brain during acute inspiratory hypoxia. J Magn Reson Imaging. 2013;37(3):739–745. doi: 10.1002/jmri.23815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McLean MA, Sun A, Bradstreet TE, Schaeffer AK, Liu H, Iannone R, et al. Repeatability of edited lactate and other metabolites in astrocytoma at 3T. J Magn Reson Imaging. 2012;36(2):468–475. doi: 10.1002/jmri.23673. [DOI] [PubMed] [Google Scholar]
- 108.Harris LM, Tunariu N, Messiou C, Hughes J, Wallace T, DeSouza NM, et al. Evaluation of lactate detection using selective multiple quantum coherence in phantoms and brain tumours. NMR Biomed. 2015;28(3):338–343. doi: 10.1002/nbm.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morana G, Piccardo A, Puntoni M, Nozza P, Cama A, Raso A, et al. Diagnostic and prognostic value of 18F–DOPA PET and 1H–MR spectroscopy in pediatric supratentorial infiltrative gliomas: a comparative study. Neuro Oncol. 2015;17(12):1637–1647. doi: 10.1093/neuonc/nov099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Howe FA, Opstad KS. 1H MR spectroscopy of brain tumours and masses. NMR Biomed. 2003;16(3):123–131. doi: 10.1002/nbm.822. [DOI] [PubMed] [Google Scholar]
- 111.Landim RC, Edden RA, Foerster B, Li LM, Covolan RJ, Castellano G. Investigation of NAA and NAAG dynamics underlying visual stimulation using MEGA-PRESS in a functional MRS experiment. Magn Reson Imaging. 2016;34(3):239–245. doi: 10.1016/j.mri.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choi C, Ghose S, Uh J, Patel A, Dimitrov IE, Lu H, et al. Measurement of N-acetylaspartylglutamate in the human frontal brain by 1H–MRS at 7 T. Magn Reson Med. 2010;64(5):1247–1251. doi: 10.1002/mrm.22536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y, Li S, Marenco S, Shen J. Quantitative measurement of N-acetyl-aspartyl-glutamate at 3 T using TE-averaged PRESS spectroscopy and regularized lineshape deconvolution. Magn Reson Med. 2011;66(2):307–313. doi: 10.1002/mrm.23029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choi C, Dimitrov I, Douglas D, Zhao C, Hawesa H, Ghose S, et al. In vivo detection of serine in the human brain by proton magnetic resonance spectroscopy (1H–MRS) at 7 Tesla. Magn Reson Med. 2009;62(4):1042–1046. doi: 10.1002/mrm.22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Near J, Devenyi GA, Simpson R, editors. FID-A: An open-source, MATLAB-based toolbox for magnetic resonance spectroscopy simulation and data processing. Proc 23 Inter Soc Magn Reson Med; Toronto, Ont, Canada. 2015. p. 4726. [DOI] [PubMed] [Google Scholar]