Abstract
An oligonucleotide biochip that specifically detects point mutations in the gyrA and parC genes of Neisseria gonorrhoeae was designed and subsequently evaluated with 87 untreated clinical specimens. The susceptibilities of the N. gonorrhoeae strains were tested to determine the prevalence of ciprofloxacin-resistant strains in Anhui Province, People's Republic of China. Conventional DNA sequencing was also performed to identify mutations in gyrA and parC and to confirm the biochip data. The study demonstrates that all of the point mutations in the gyrA and parC genes of N. gonorrhoeae were easily discriminated by use of the oligonucleotide biochip. Fifteen different alteration patterns involved in the formation of ciprofloxacin resistance were identified by the biochip assay. Double mutations in both Ser91 and Asp95 of the GyrA protein were seen in all nonsensitive isolates. Double mutations in Ser91 and Asp95 of GyrA plus mutation of Glu91 or Ser87 of the ParC protein lead to significant high-level resistance to ciprofloxacin in N. gonorrhoeae isolates. The results obtained by use of the oligonucleotide biochip were identical to those obtained by use of DNA sequencing. In conclusion, the oligonucleotide biochip technology has potential utility for the rapid and reliable identification of point mutations in the drug resistance genes of N. gonorrhoeae.
The existence of isolates resistant to ciprofloxacin is an emerging theme in the clinical treatment of Neisseria gonorrhoeae infections. In the Far East and Western Pacific region, including the People's Republic of China, resistant strains may account for at least 10% of all N. gonorrhoeae isolates (18, 27). Several mechanisms of ciprofloxacin resistance have been identified and characterized in N. gonorrhoeae strains that have decreased susceptibilities to ciprofloxacin (2, 6, 7, 8, 16, 21, 23, 25). Genotyping studies of genetic mutations resulting in ciprofloxacin resistance in N. gonorrhoeae isolates have shown that amino acid substitutions within the quinolone (i.e., ciprofloxacin) resistance-determining regions (QRDRs) in the gyrA and parC genes play crucial roles in resistance to these agents. Mutations in gyrA that lead to substitutions of tyrosine for serine at position 91 (Ser91/Tyr) and aspartic acid for asparagine at position 95 (Asp95/Asn) were identified as novel single-amino-acid changes in N. gonorrhoeae that contribute to ciprofloxacin resistance. Additionally, alterations in ParC (Asp86/Asn, Ser87/Ile, and Glu91/Gly) have also been characterized (7, 8).
The advantages and limitations of several methods for the detection of mutations, including DNA sequencing, single-strand conformation polymorphism analysis, multiple PCR analyses, and denaturing gradient gel electrophoresis, have been described elsewhere (11). DNA sequencing provides the highest resolution for the molecular biology-based diagnosis of hereditary diseases and mutant microorganisms; however, it is not feasible in most laboratories for the generation of high-throughput mutation information. Oligonucleotide biochips are promising tools for molecular biology-based diagnosis due to their ability to perform a multitude of tests simultaneously (3-5, 9, 11, 12, 22, 26, 28). Biochips enable the rapid identification of mutations in thousands of genes as well as gene products with desired properties, such that much information about biomolecules can be collected on various scales in a short time (13-15). Although biochip technologies present their own challenges, they have been used extensively in surveys of gene expression (4, 20, 28), for the detection of genetic polymorphisms (9), and for the serological detection of pathogens (10) and have been broadly used to screen clinical samples for microorganisms and to characterize microorganisms (3, 4, 12). Many investigators have successfully immobilized dozens of biomolecular elements onto biochips for research and commercial purposes in order to detect changes in the genes or proteins of interest. Low-density biochips certainly have the potential for custom fabrication and the detection of all desired features and could conveniently provide reliable results and decrease production costs considerably.
In the study described here, we used an oligonucleotide biochip to detect N. gonorrhoeae infection and analyze the gyrA and parC genes of N. gonorrhoeae isolates from clinical samples for mutations. A set of known allele-specific probes designed according to the nucleotide sequence data in the GenBank database was immobilized onto a glass surface. The probes specific for a mutation at a given site had the same sequence except for a base substitution in the middle of the probes to allow the discrimination of wild-type and mutant genotypes. The probes on the biochips were hybridized with cyanine 5 (Cy5)-labeled complementary targets amplified from the suspected N. gonorrhoeae clinical isolates. The diagnosis of N. gonorrhoeae infection and the detection of ciprofloxacin susceptibility were performed directly with untreated clinical samples. Furthermore, a survey of fluoroquinolone-resistant N. gonorrhoeae strains isolated in 2002 and 2003 in Anhui Province, People's Republic of China, and the frequency and patterns of mutations involving the gyrA and parC genes in 78 ciprofloxacin-resistant strains were further analyzed by use of the biochip assay.
MATERIALS AND METHODS
Bacterial strains.
Reference (wild-type) strain N. gonorrhoeae ATCC 49226 was a kind gift from the Institute of Dermatology, Chinese Academy of Medical Sciences. Eighty-seven urethral and cervical swab samples were collected from patients with suspected N. gonorrhoeae infection receiving treatment at the First Affiliated Hospital of Anhui Medical University (Hefei, People's Republic of China) during October 2002 and May 2003. The age range of the patients was from 20 to 63 years (mean, 31.3 years), and the ratio of male to female patients was 1.3:1. Samples from each patient were divided into two parts; one was used for bacterial culture and antibiotic susceptibility testing, and the other was stored at −70°C for use in the biochip assay.
Determination of MICs.
Antibiotic susceptibilities were determined by the agar plate dilution method recommended by the National Committee for Clinical Laboratory Standards (19). Ciprofloxacin was purchased from Bayer, Inc. (Leverkusen, Germany). We defined the ciprofloxacin susceptibilities of the N. gonorrhoeae isolates as fully susceptible (Cips; MICs, less than or equal to 0.03 μg/ml), less (intermediately) susceptible (Cipi; MICs, 0.06 to 0.5 μg/ml), resistant (Cipr; MICs, 1.0 to 2.0 μg/ml), or highly resistant (Ciph; MICs, equal to or greater than 4.0 μg/ml).
Sample DNA preparation.
A total of 100 μl of lysis solution (10 mM NaOH, 0.5 mM EDTA [pH 8.0], 0.1 mM Tris-HCl [pH 8.0], 20 mM proteinase K, 1% Nonidet P-40, 1% Triton X-100) was added to each tube containing a swab sample, and the DNA in the solution was incubated at 55°C for 1 h and then at 95°C for 5 min. The tubes were then centrifuged at 14,000 rpm with a Peltier Thermal Cycle-225 instrument for 5 min. The DNA supernatant was carefully collected and directly used as the template DNA for target PCR amplification.
Duplicate PCR amplification of gyrA and parC genes.
Primers were discriminated and synthesized at TaKaRa Biotechnology, Inc. (Dalian, People's Republic of China), according to the sequences of the N. gonorrhoeae gyrA and gyrC genes from the GenBank database. The following primers were used to specifically generate gyrA and parC amplicons from N. gonorrhoeae for DNA sequencing: primers NG-gyrA-A (5′-ATCCGCCACGACCACAAAT-3′; forward) and NG-gyrA-B (5′-CGTCCACCGATCCGAAGTT-3′; reverse) were used to amplify the gyrA gene in N. gonorrhoeae, covered nucleotides 16 through 364, and produced a fragment of 349 bp. Primers NG-parC-A (5′-GCACGCTTCCCATACCGA-3′; forward) and NG-parC-B (5′-TCCACCGTCCCCTGATTG-3′; reverse) targeted the region between nucleotides 15 and 443 of the parC gene in N. gonorrhoeae and produced a fragment of 429 bp.
Amplicons containing the gyrA and parC mutations were prepared by the PCR method after extraction of the chromosomal DNA of each N. gonorrhoeae strain. Each PCR mixture (20 μl) contained 50 to 100 ng of unpurified bacterial DNA, 1× PCR buffer (TaKaRa Biotechnology, Inc.), 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate (Boehringer Mannheim, Mannheim, Germany), 200 nM each primer, and 1 U of Taq DNA polymerase (AmpliTaq; Perkin-Elmer Co.). PCR amplification was performed with a thermal cycler (PCR Express; Thermohybaid), and PCR was initiated at 95°C for 3 min, followed by 35 amplification cycles. The cycling protocol included denaturation at 94°C for 60 s, followed by annealing at 52°C for 50 s and extension at 72°C for 55 s. A final extension was done at 72°C for 5 min. The PCR products were checked by electrophoresis on a 1.8% agarose gel and were stained with ethidium bromide.
Sequencing analysis.
Both directions of the PCR amplicons from the 87 clinical isolates were sequenced by Bo-Ya Biotechnology, Inc. (Shanghai, People's Republic of China) with an ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin-Elmer, Applied Biosystems, Foster City, Calif.) and an ABI 377A automated DNA sequencer; and the sequences were analyzed with Sequence Navigator software (Perkin-Elmer, Applied Biosystems).
Probe design and tiling strategy for oligonucleotide biochip.
All the mutation sites selected for detection in the gyrA and parC genes in this study have been reported previously (1, 2, 7, 8, 16, 17, 23). The sequences of the gyrA and parC genes from different N. gonorrhoeae isolates were aligned by use of Lasergene software (DNAStar, Madison, Wis.). The oligonucleotide probes immobilized on the biochips and used for the genotyping of the bacterial gyrA and parC genes were designed and discriminated by use of Premier software (version 5.0; Primer Biosoft International, Palo Alto, Calif.) according to the sequences available in the GenBank database (accession numbers U08817 and U08907, respectively). Probes were synthesized by TaKaRa Biotechnology, Inc. Probe lengths ranged from 13 to 21 nucleotides. The probes specific for a given mutation site had identical sequences except for a base substitution in the middle of the probes that discriminated the wild-type and mutant genotypes. The predicted melting temperatures of the probes were limited to between 48 and 52°C. The sequences of the oligonucleotide probes are shown in Table 1. All of these probes contained amino linkers at their 5′ ends so that they could covalently attach to aldehyde-coated slides. A spacer arm with a poly(T) 16-mer was inserted between the probe sequence and the 5′ end of the amino linker molecule to decrease steric interference on the biochips.
TABLE 1.
Probe sequences used in the oligonucleotide biochip assaya
| Gene and probe | Oligonucleotide sequence | Tm (°C) |
|---|---|---|
| gyrA | ||
| Probe1 (Ser91; wild type) | 5′-NH2-(T)16-CGATTCCGCAGTTTA-3′ | 49.5 |
| Probe2 (Phe91; mutation) | 5′-NH2-(T)16-CGATTTCGCAGTTTA-3′ | 49.1 |
| Probe3 (Asp95; wild type) | 5′-NH2-(T)16-AGTTTACGACACCATC-3′ | 46.8 |
| Probe4 (Asn95; mutation) | 5′-NH2-(T)16-AGTTTACAACACCATCGT-3′ | 50.2 |
| Probe5 (Ala95; mutation) | 5′-NH2-(T)16-TTACGCCACCATCG-3′ | 49.3 |
| Probe6 (Gly95; mutation) | 5′-NH2-(T)16-TTACGGCACCATCG-3′ | 49.3 |
| parC | ||
| Probe7 (Asp86/Ser87; wild type) | 5′-NH2-(T)16-CACGGCGACAGTTCC-3′ | 49.2 |
| Probe8 (Asn86; mutation) | 5′-NH2-(T)16-GCACGGCAACAGTTCC-3′ | 51.3 |
| Probe9 (Arg87; mutation) | 5′-NH2-(T)16-CGACCGTTCCGCCT-3′ | 50.7 |
| Probe10 (Asn87; mutation) | 5′-NH2-(T)16-CGACAATTCCGCCTAT-3′ | 48.9 |
| Probe 11 (Ile 87; mutation) | 5′-NH2-(T)16-CGACATTTCCGCCTAT-3′ | 47.9 |
| Probe 12 (Arg 87; mutation) | 5′-NH2-(T)16-CGACAGGTCCGCCTA-3′ | 49.2 |
| Probe 13 (Glu91; wild type) | 5′-NH2-(T)16-CTATGAGGCGATGGTG-3′ | 52.4 |
| Probe 14 (Lys91; mutation) | 5′-NH2-(T)16-CCTATAAGGCGATGGTG-3′ | 54.6 |
| Probe 15 (Gly91; mutation) | 5′-NH2-(T)16-CCTATGGGGCGATG-3′ | 52.8 |
| Probe 16 (Ala 91; mutation) | 5′-NH2-(T)16-CCTATGCGGCGATG-3′ | 52.8 |
| Probe 17 (Gln91; mutation) | 5′-NH2-(T)16-CTATCAGGCGATGGTG-3′ | 50.6 |
Probe lengths were limited to between 13 and 21 nucleotides. The probes specific for a given mutation were designed so that the base substitution was in the middle of the probe sequences. The predicted melting temperatures (Tm) of the probes were limited to between 48 and 52°C. All probes contained amino linkers at their 5′ ends for covalent attachment to aldehyde-coated slides. A spacer arm with a poly(T) 16-mer was inserted between the specific probe sequence and amino linker molecule at the 5′ end to decrease steric interference on the biochips.
Biochip preparation.
The probes were diluted to a final concentration of 10 μM with spotting solution (TeleChem) and spotted onto aldehyde-coated glass slides (Sigma, St. Louis, Mo.) with a robot (Cartesian Pixsys 7500). Four replicate spots of each probe were aligned in the same rows of the biochips. A universal probe (5′-TAGGCGGTGCTGGTGTTTA-3′) was used as an internal positive control and was spotted on the first left column on the biochips. After the spotting process, the biochips were fixed at 48°C for 72 h and were dried for further use.
Hybridization and analysis.
The primers used for preparation of the targets from the gyrA and parC genes of N. gonorrhoeae for the biochip assay were the same as those used for DNA sequencing, except that the forward primers were modified by attachment of a fluorescent dye, Cy5, to their 5′ ends. Hybridization of the Cy5-labeled DNA targets with the oligonucleotide probes immobilized on the biochip was carried out as follows: 2 μl of Cy5-labeled unpurified PCR product was mixed with 10 μl of DIG Easy Hyb (Roche Molecular Biochemicals, Laval, Quebec, Canada). The hybridization mixture was preheated at 95°C for 30 s to denature the double-stranded DNA and was then quickly quenched on ice. The hybridization mixture was applied to the array and covered with a glass coverslip (18 by 18 mm2) to prevent evaporation of the mixture during hybridization. The biochips were incubated at 45°C for 30 min in a moist incubator. After hybridization, the biochips were washed once for 10 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.2% sodium dodecyl sulfate solution at 25°C, followed by another wash in 2× SSC solution for 1 min, and were then dried with a nitrogen stream to remove any remaining solution. After the slides were washed, the fluorescence signal intensities from all spots on the biochips were captured and calculated by use of a fluorescence signal reader (Scanarray 3000; Axon Instruments, Foster City, Calif.). Positive fluorescence signals were defined as those for which there was a factor of 3 difference between the target and the reference signals. All hybridization experiments were repeated two times.
Sensitivity of oligonucleotide biochip.
The sensitivity of the oligonucleotide biochip was determined as follows: the extracted DNA was quantified with a spectrophotometer. A serial dilution of one of the N. gonorrhoeae nucleic acid targets (10 to 10−5 μg/ml) was prepared and assayed on the biochip simultaneously. The average densities of the fluorescence signals of four replicates from each dilution were calculated. If it is assumed that an average N. gonorrhoeae cell yields 4.5 × 10−15 g of genomic DNA, 10 to 10−5 μg/ml is equivalent to 5 × 10−6 N. gonorrhoeae cells (5 genome copies) per PCR mixture. This indicates that the biochips were sufficiently sensitive to detect genes from untreated specimens containing only 5 N. gonorrhoeae genome copies.
RESULTS
Susceptibility to ciprofloxacin.
Seventy-eight of the 87 N. gonorrhoeae isolates detected in this study were divided into three groups according to their ciprofloxacin resistance: Cipi, Cipr, and Ciph. Nine of the 78 strains (11.54%) were Cips (MICs, 0.002 to 0.03 μg/ml). The numbers of isolates in the other groups were as follows: Cipi, 20 isolates (25.64%; MICs, 0.06 to 0.5 μg/ml); Cipr, 12 isolates (15.38%; MICs, 1.0 to 2.0 μg/ml), and Ciph, 46 isolates (58.97%; MICs, 4.0 to 16.0 μg/ml).
Sensitivity of oligonucleotide biochip assay.
The oligonucleotide biochip assay was significantly more sensitive than electrophoresis. The template DNA concentrations detectable by the biochip assay ranged from 10 to 10−5 μg/ml; and the biochip assay had a 1,000-fold higher sensitivity than agarose gel electrophoresis, by which the template DNA concentrations that were detectable ranged from 10 to 10−2 μg/ml.
Detection of mutations in N. gonorrhoeae gyrA and parC genes by oligonucleotide biochip assay.
Two products of 349 and 429 bp from the gyrA and parC genes, respectively, obtained by duplicate PCRs with bacterial DNA samples were amplified and were further checked by electrophoresis on 1.8% agarose gels and stained with ethidium bromide. The oligonucleotide biochip assays were successfully performed with the 87 clinical samples. Nine isolates were wild type; 14 isolates had double mutations, mainly in the Ser91 and Asp95 codons of the GyrA protein; and 64 isolates were identified as having multiple mutations because they simultaneously contained two or more mutations in both the GyrA and ParC proteins. All ciprofloxacin-resistant strains had double mutations at codons 91 and 95 in the GyrA protein and single or double mutations in the ParC protein. The biochip assay results are shown in Fig. 1 and were completely in agreement with the results of DNA sequencing analysis.
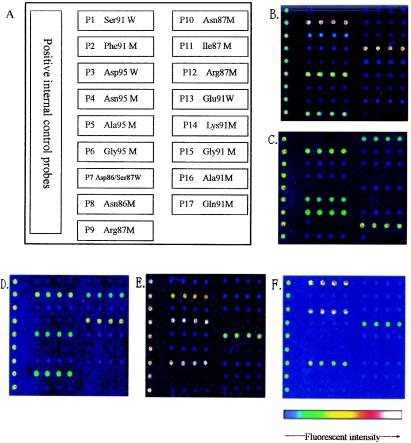
FIG. 1.
Layout of probes on biochips and fluorescence images of clinical isolates. (A) Probe layout on the oligonucleotide biochip. Following hybridization of Cy5-labeled fragmented DNA, as described in Materials and Methods, the fluorescence signals of mutations in the gyrA and parC genes from four clinical isolates were detected, as follows: Ser91→Phe/Asp95→Gly/Ser87→Arg (B), Ser-91→Phe/Asp95→Gly/Ser87→Asn/Glu91→Gln (C), Ser91→Phe/Asp95→Ala/Asp86→Asn/Ser87→Ile (D), and Ser91→Phe/Asp95→Asn/Glu91→Lys (E). (F) Wild-type sequence in gyrA and parC (Ser91/Asp95/Asp86/Ser87/Glu91).
The mutations within the QRDRs of GyrA and ParC proteins from 87 isolates of N. gonorrhoeae are shown in Table 2. Cipi isolates exhibited four patterns of alterations: two patterns with alterations only in gyrA and two patterns with alterations in both gyrA and parC. Most Cipi isolates (12 of 20; 60%) exhibited alterations at amino acids 91 and 95 in GyrA. Cipr isolates exhibited six patterns of alterations that included one pattern with an alteration in gyrA alone and five patterns involving alterations in both gyrA and parC in combination. Additionally, five patterns of alterations in gyrA and parC were observed in the Ciph group.
TABLE 2.
Alterations in gyrA and parC genes of 87 N. gonorrhoeae strains
| Patterna | Resistance phenotype | Amino acid substitution in:
|
Ciprofloxacin MIC (mg/liter) | No. (%) of strains | ||||
|---|---|---|---|---|---|---|---|---|
| GyrA
|
ParC
|
|||||||
| Ser-91 (TCC) | Asp-95 (GAC) | Asp-86 (GAC) | Ser-87 (AGT) | Glu-91 (GAG) | ||||
| M1 | Cips | 0.0075 | 9 | |||||
| M2 | Cipi | Phe (TTC) | Gly (GGC) | 0.125 | 2 (2.56) | |||
| M3 | Cipi | Phe (TTC) | Ala (GCC) | Asn (AAC) | 0.125-0.25 | 6 (7.69) | ||
| M4 | Cipi | Phe (TTC) | Ala (GCC) | 0.125-0.25 | 10 (12.82) | |||
| M5 | Cipi | Phe (TTC) | Gly (GGC) | Gln (CAG) | 0.5 | 2 (2.56) | ||
| M6 | Cipi | Phe (TTC) | Asn (AAC) | Gly (GGG) | 1 | 1 (1.28) | ||
| M7 | Cipr | Phe (TTC) | Asn (AAC) | Lys (AAG) | 1 | 1 (1.28) | ||
| M8 | Cipr | Phe (TTC) | Asn (AAC) | 1 | 2 (2.56) | |||
| M9 | Cipr | Phe (TTC) | Ala (GCC) | Asn (AAT) | 1-2 | 4 (5.12) | ||
| M10 | Cipr | Phe (TTC) | Ala (GCC) | Ala (GCG) | 2 | 3 (3.84) | ||
| M11 | Cipr | Phe (TTC) | Ala (GCC) | Gly (GGG) | 2 | 1 (1.28) | ||
| M12 | Ciph | Phe (TTC) | Asn (AAC) | Arg (AGG) | 4 | 1 (1.28) | ||
| M13 | Ciph | Phe (TTC) | Gly (GGC) | Arg (CGT) | 4-8 | 16 (20.51) | ||
| M14 | Ciph | Phe (TTC) | Asn (AAC) | Arg (CGT) | 4-8 | 9 (11.54) | ||
| M15 | Ciph | Phe (TTC) | Ala (GCC) | Asn (AAC) | Ilc (ATT) | 16 | 2 (2.56) | |
| M16 | Ciph | Phe (TTC) | Ala (GCC) | Arg (CGT) | Ala (GCG) | 4-16 | 18 (23.07) | |
The biochip assay identified 15 different alteration patterns involved in the formation of ciprofloxacin resistance. M1, wild type; M2 to M16, mutation types.
Correlation of ciprofloxacin MICs with QRDR sequence changes.
The associations between the results of mutation analysis by the biochip assay and those of MIC susceptibility testing are shown in Table 2. Among the 87 clinical strains tested by the biochip technique, 9 wild-type strains were determined to be susceptible to ciprofloxacin. The others (78 strains) contained more or fewer mutations. Fifteen different patterns of alterations involved in the formation of ciprofloxacin resistance were identified. The most prevalent were patterns M16 (23.07%), M13 (20.51%), M4 (12.82%), and M14 (11.54%). Twelve strains with double mutations in the gyrA gene were defined to be less sensitive. Fifty-six strains with multiple mutations in both the gyrA and parC genes were defined as resistant and/or high-level-resistant strains. In comparison, mutations of Ser91 and Asp95 in the GyrA protein were predominant. The mutations of Glu91 and Ser87 in the ParC protein were consistent with resistance and high-level resistance, respectively.
DISCUSSION
The occurrence of ciprofloxacin resistance affects the treatment of N. gonorrhoeae infection and is closely associated with point mutations in the gyrA and parC genes. The mechanisms involved in antimicrobial resistance could be related to either reduced access of an antibiotic to the target sites or alteration of the antibiotic target. Multiple determinants of resistance may occur simultaneously in a single isolate. A comprehensive understanding of the genetic events that lead to drug resistance by isolates in clinical specimens is of importance to elucidate the effects of antimicrobial agents (2, 16, 18).
Traditionally, culture was a useful method for the clinical identification of N. gonorrhoeae infection, but it is quite time-consuming. Several PCR-based methods for genotyping have been reported (2, 7, 8, 16); however, the main disadvantage of these methods is that the sizes and the restriction endonuclease patterns of the DNA amplified from gyrA and parC are potentially variable and limited. Thus, a better sequence-specific approach is needed. The development of a rapid, sensitive, and accurate method for the identification of point mutations in the gyrA and parC genes is therefore valuable and reasonable. Oligonucleotide biochips offer a very useful tool that can be used to quickly inspect clinical specimens for the presence of biological agents and efficiently analyze the nucleotide sequences of those agents for point mutations (3, 4, 10, 12, 26, 28). Knowledge of the sequence changes that result in a resistant phenotype allows the development of probe-based assays for the detection of antibiotic resistance. In the present study we developed a combined PCR-biochip method that substantially improved the specificity of the biochip array.
In this study, immobilization of oligonucleotide probes onto an aldehyde-coated glass surface produced a high-density monolayer that was easily accessible for hybridization with DNA samples, which made the reaction between the fluorescence-labeled DNA targets and oligonucleotide probes more efficient and quick and which reduced the overall assay time to less than 4 h. Due to the high degree of allele specificity of the oligonucleotide probes for the bacterial target genes (gyrA and parC) and the highly stringent conditions that were used, only the target specifically complementary to the probe could be detected. To increase the hybridization efficiency between the probes and the targets and the sensitivity of the assay and to obtain higher signal-to-noise ratios, the lengths of the probes and the spacer arms and the hybridization conditions were optimized (data not shown). The results for the detection of single base substitutions in the gyrA and parC genes obtained by the biochip assay were in good accordance with those obtained by DNA sequencing. Therefore, the validity of the assay for the detection of point mutations in the gyrA and parC genes in association with decreased susceptibilities to ciprofloxacin in N. gonorrhoeae was confirmed.
In this study, we demonstrated the frequency of N. gonorrhoeae isolates with intermediate or full resistance to ciprofloxacin, which has become of significant concern in Anhui Province of the People's Republic of China. Of the 87 clinical isolates tested, only 9 (10.34%) isolates were found to be susceptible to ciprofloxacin (MICs, ≤0.03 μg/ml). Among the ciprofloxacin-resistant isolates (MICs, ≥1 μg/ml), 46 were highly resistant (MICs, ≥4 μg/ml). Thus, it should be kept in mind that the resistance of N. gonorrhoeae to ciprofloxacin is a common clinical event in Anhui. The development of personalized therapeutic strategies would be more efficient in patients with N. gonorrhoeae infection.
Our research also revealed the characteristics of the patterns of alterations in the GyrA and ParC QRDRs of N. gonorrhoeae strains in Anhui Province: (i) N. gonorrhoeae isolates without alterations in the gyrA and parC genes were quite susceptible to ciprofloxacin, (ii) all isolates that were less susceptible to ciprofloxacin had mutations in GyrA (alterations of Ser91 and Asp95), and (iii) all of the strains with ciprofloxacin resistance had a combination of alterations at positions 91 and 95 of the GyrA protein and/or a single or a double ParC alteration, indicating that GyrA mutations are responsible for the development of ciprofloxacin resistance.
It has been reported that the level of ciprofloxacin resistance appears to correlate with the location and the number of mutations in the gyrA and parC genes (7, 8). However, in most published studies, those alteration patterns do not always correlate to MICs because of the few isolates tested. In this study, associations between the results of mutation analysis by the biochip assay and those of MIC susceptibility testing were shown, and 15 different alteration patterns involved in the formation of ciprofloxacin resistance were identified by biochip assay. Of these, the double alteration at both positions 91 and 95 in the GyrA protein accompanied by a single and/or double ParC alterations in combination were more predominant, indicating that the increasing level of resistance to ciprofloxacin was associated with the number of alterations in the GyrA and ParC QRDRs. Simultaneous mutations in both the parC and the gyrA genes might exert a combined effect in increasing the level of resistance. Additionally, some novel alteration patterns were found among the 87 isolates, including M4, M10, M12, M13, and M16 (Table 2). These differences probably reflect a geographical variation among N. gonorrhoeae isolates and/or a temporal evolution of the mutations in the QRDRs of the GyrA and ParC proteins, since the strains examined in this study were isolated 10 years later (2002 and 2003) than those detected in Japan (1991 to 1993) (24).
In conclusion, the highly sensitive and synchronous oligonucleotide biochip assay is a promising tool for the analysis of point mutations in N. gonorrhoeae strains causing infections. In this study, only the known mutations in N. gonorrhoeae were detected. A main advantage of the oligonucleotide biochip method is that it can be used with clinical samples without purification and culture, and reliable results can be obtained much more efficiently and conveniently in a shorter period. The oligonucleotide biochip technology could be applied not only to the rapid and reliable identification of point mutations in resistance genes but also to the development of personalized therapeutic strategies. A target for the detection of any new mutations in N. gonorrhoeae with the biochip technology remains to be determined.
Acknowledgments
This work was supported by grants from the Chinese Academy of Sciences Orientation Research Project (STZ-00-03) and the Chinese Academy of Sciences Intellective Innovation Great Project (KSCX-06).
We thank Scott Venners for helpful comments on the manuscript.
REFERENCES
- 1.Alcala, B., L. Arreaza, C. Salcedo, I. Antolin, N. Borrell, J. Cacho, C. De Las Cuevas, L. Otero, G. Sauca, F. Vazquez, H. Villar, and J. A. Vazquez. 2003. Molecular characterization of ciprofloxacin resistance of gonococcal strains in Spain. Sex. Transm. Dis. 30:395-398. [DOI] [PubMed] [Google Scholar]
- 2.Belland, R. J., S. G. Morrison, C. Ison, and W. M. Huang. 1994. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol. Microbiol. 14:371-380. [DOI] [PubMed] [Google Scholar]
- 3.Booth, S. A., M. A. Drebot, I. E. Martin, and L. K. Ng. 2003. Design of oligonucleotide arrays to detect point mutations: molecular typing of antibiotic resistant strains of Neisseria gonorrhoeae and hantavirus infected deer mice. Mol. Cell. Probes 17:77-84. [DOI] [PubMed] [Google Scholar]
- 4.Call, D. R., M. K. Borucki, and F. J. Loge. 2003. Detection of bacterial pathogens in environmental samples using DNA microarrays. J. Microbiol. Methods 53:235-243. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, V. G., M. Morley, F. Aguilar, A. Massimi, R. Kucherlapati, and G. Childs. 1999. Making and reading microarrays. Nat. Genet. 21:15-19. [DOI] [PubMed] [Google Scholar]
- 6.Corkill, J. E., A. Percival, and M. Lind. 1991. Reduced uptake of ciprofloxacin in a resistant strain of Neisseria gonorrhoeae and transformation of resistance to other strains. J. Antimicrob. Chemother. 28:601-604. [DOI] [PubMed] [Google Scholar]
- 7.Deguchi, T., M. Yasuda, M. Asano, K. Tada, H. Iwata, H. Komeda, T. Ezaki, I. Saito, and Y. Kawada. 1995. DNA gyrase mutations in quinolone-resistant clinical isolates of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 39:561-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deguchi, T., M. Yasuda, M. Nakano, S. Ozeki, T. Ezaki, I. Saito, and Y. Kawada. 1996. Quinolone-resistant Neisseria gonorrhoeae: correlation of alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV with antimicrobial susceptibility profiles. Antimicrob. Agents Chemother. 40:1020-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du, W., C. Marsac, M. Kruschina, F. Ortigao, and C. Florentz. 2003. Functionalized self-assembled monolayer on gold for detection of human mitochondrial tRNA gene mutations. Anal. Biochem. 322:14-25. [DOI] [PubMed] [Google Scholar]
- 10.Du, W., Z. Xu, X. Ma, L. Song, and E. M. Schneider. 2003. Biochip as a potential platform of serological interferon alpha2b antibody assay. J. Biotechnol. 106:87-100. [DOI] [PubMed] [Google Scholar]
- 11.Fodde, R., and M. Losekoot. 1994. Mutation detection by denaturing gradient gel electrophoresis (DGGE). Hum. Mutat. 3:83-94. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima, M., K. Kakinuma, H. Hayashi, H. Nagai, K. Ito, and R. Kawaguchi. 2003. Detection and identification of Mycobacterium species isolates by DNA microarray. J. Clin. Microbiol. 41:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, Z., R. A. Guilfoyle, A. J. Thiel, R. Wang, and L. M. Smith. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 22:5456-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacia, J. G., L. C. Brody, M. S. Chee, S. P. A. Fodor, and F. S. Collins. 1996. Detection of heterozygous mutations in BRCA1 using high density oligonucleotide array and two-colour fluorescence analysis. Nat. Genet. 14:441-447. [DOI] [PubMed] [Google Scholar]
- 15.Hacia, J. G., and F. S. Collins. 1999. Mutational analysis using oligonucleotide microarrays. J. Med. Genet. 36:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, D. C. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kam, K. M., S. S. Kam, D. T. Cheung, V. W. Tung, W. F. Au, and M. M. Cheung. 2003. Molecular characterization of quinolone-resistant Neisseria gonorrhoeae in Hong Kong. Antimicrob. Agents Chemother. 47:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knapp, J. S., K. K. Fox, D. L. Trees, and W. L. Whittington. 1997. Fluoroquinolone resistance in Neisseria gonorrhoeae. Emerg, Infect. Dis. 3:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 1993. Method for dilution antimicrobial susceptibility test for bacteria that grow aerobically, 3rd ed. Approved standard M7-A3. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 20.Schena, M., D. Shalon, R. Heller, et al. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93:457-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shultz, T. R., J. W. Tapsall, and P. A. White. 2001. Correlation of in vitro susceptibilities to newer quinolones of naturally occurring quinolone-resistant Neisseria gonorrhoeae strains with changes in GyrA and ParC. Antimicrob. Agents Chemother. 45:734-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su, X., and I. Lind. 2001. Molecular basis of high-level ciprofloxacin resistance in Neisseria gonorrhoeae strains isolated in Denmark from 1995 to 1998. Antimicrob. Agents Chemother. 45:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka, M., H. Fukuda, K. Hirai, M. Hosaka, T. Matsumoto, and J. Kumazawa. 1994. Reduced uptake and accumulation of norfloxacin in resistant strains of Neisseria gonorrhoeae isolated in Japan. Genitourin. Med. 70:253-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trees, D. L., A. L. Sandul, V. Peto-Mesola, M. R. Aplasca, H. B. Leng, W. L. Whittington, and J. S. Knapp. 1999. Alterations within the quinolone resistance-determining regions of GyrA and ParC of Neisseria gonorrhoeae isolated in the Far East and the United States. Int. J. Antimicrob. Agents 12:325-332. [DOI] [PubMed] [Google Scholar]
- 26.Troesch, A., H. Nguyen, C. G. Miyada, S. Desvarenne, T. R. Gingeras, P. M. Kaplan, P. Cros, and C. Mabilat. 1999. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J. Clin. Microbiol. 37:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. 1998. Western Pacific Gonococcal Antimicrobial Surveillance Programme. Resistance in gonococci isolated in the Western Pacific Region to various antimicrobials used in the treatment of gonorrhoea, 1997. Commun. Dis. Intell. 22:288-291. [DOI] [PubMed] [Google Scholar]
- 28.Ye, R. W., T. Wang, L. Bedzyk, and K. M. Croker. 2001. Applications of DNA microarrays in microbial systems. J. Microbiol. Methods 47:257-272. [DOI] [PubMed] [Google Scholar]