Abstract
Similar to other segmented RNA viruses, influenza viruses can exchange genome segments and form a wide variety of reassortant strains upon coreplication within a host cell. Therefore, the mapping of genome segments of influenza viruses is essential for understanding their phenotypes. In this work, we have developed an oligonucleotide microarray hybridization method for simultaneous genotyping of all genomic segments of two highly homologous strains of influenza B virus. A few strain-specific oligonucleotide probes matching each of the eight segments of the viral genomes of the B/Beijing/184/93 and B/Shangdong/7/97 strains were hybridized with PCR-amplified fluorescently labeled single-stranded DNA. Even though there were a few mismatches among the genomes of the studied virus strains, microarray hybridization showed highly significant and reproducible discrimination ability and allowed us to determine the origins of individual genomic segments in a series of reassortant strains prepared as vaccine candidates. Additionally, we were able to detect the presence of at least 5% of mixed genotypes in virus stocks even when conventional sequencing methods failed, for example, for the NS segment. Thus, the proposed microarray method can be used for (i) rapid and reliable genome mapping of highly homologous influenza B viruses and (ii) extensive monitoring of influenza B virus reassortants and the mixed genotypes. The array can be expanded by adding new oligoprobes and using more quantitative assays to determine the origin of individual genomic segments in series of reassortant strains prepared as vaccine candidates or in mixed virus populations.
Influenza viruses A and B possess segmented genomes consisting of eight separate RNA molecules, each coding for one or more viral proteins (15, 19). Upon coreplication in a cell, the viruses can exchange segments, leading to diversity of reassortant strains. Together with accumulation of point mutations, segment reassortment is the basis for evolution and maintenance of diversity for many viral pathogens. It provides them with the ability to rapidly adapt to the pressure of the host immune system and leads to the continuous emergence of new virus variants that cause seasonal outbreaks of influenza (17). Because of this ability, segmented viruses can exist in numerous genotypes and serotypes, presenting a challenge to the creation of protective vaccines. On the other hand, it also provides the basis for a rational approach to the development of influenza vaccines (2, 4, 21, 23).
Gradual antigenic drift and intermittent occurrences of more substantial antigenic shifts make the creation of new prospective seed virus strains for both inactivated and live attenuated influenza vaccines a continuous process that must be completed within a short period before each epidemic season. To be effective, prospective vaccine strains must not only possess the antigenic properties of the currently circulating strains but also replicate well in the substrate used for their manufacture. Some naturally occurring influenza virus isolates do not grow well in substrates used for vaccine production (e.g., embryonated chicken eggs). Adaptation of influenza viruses for optimal growth by passaging them in a target substrate is a time-consuming process that may also allow the accumulation of random mutations that can alter antigenic specificity. An alternative strategy is to create reassortants between the currently circulating strains and another strain previously adapted for high growth in vitro (2, 21). The reassortants should contain genes coding for the protective antigens (hemagglutinin [HA] and neuraminidase [NA]), as well as genes determining the optimal replicative properties of the virus. Selection of appropriate reassortants is aided by reverse transcription (RT)-PCR and restriction fragment length polymorphism (RFLP) tests of multiple candidate clones (13, 18) and multiplex RT-PCR with single-strand conformation polymorphism analysis (1). However, none of these methods is fully adequate, and the creation of new rapid, highly specific, sensitive, and reproducible segment-mapping techniques remains highly desirable.
Hybridization with microarrays of immobilized oligonucleotides is a rapid alternative to DNA sequence analysis (6, 12) and has been used for the genotyping of viruses, bacteria, and genes of higher organisms (7, 9, 10, 20). Recently, a microarray method has been used for the successful genotyping of the VP7 gene of rotaviruses (8) and for analysis of intermolecular recombinants of poliovirus (5), as well as for detection and discrimination of orthopoxviruses (14). Recently, the potential of microarray hybridization for detection and discrimination between types and subtypes of influenza A viruses has been shown by using 24 long oligoprobes (average, 500 nucleotides [nt] [16]). Shorter oligoprobe microarrays (between 4 and 128 nt) have been developed for the detection of influenza viruses (22). However, in the latter study, the intensities of probe spots completely complementary to target sequences lacked definitive boundaries between significant hybridization and nonspecific background binding, and most of the monospecific NA and HA subtype probes have not been discriminated by the array.
For better discrimination of closely related segmented RNA viruses, we report a new high-throughput oligonucleotide microarray-based method to map the origins of all segments of two closely related influenza B viruses, B/Beijing/184/93 and B/Shangdong/7/97, which were used as the models for development of the method. The two influenza B viruses were chosen for the study because B/Shangdong/7/97 is a naturally occurring reassortant with three segments (PA, NP, and M) derived from a precursor virus phylogenetically related to B/Beijing/184/93 and therefore presented a challenge by requiring the ability to distinguish between single-base substitutions. In addition, the strains possess different growth characteristics in embryonated chicken eggs (23) and may serve as a model for the reassortment approach to the creation of influenza B vaccines. The proposed microarray approach enabled us to successfully discriminate all eight segments of the Beijing and Shangdong strains despite minimal differences between them.
MATERIALS AND METHODS
Experimental methods. (i) Viruses and reassortants.
The strains used to generate virus reassortants were previously described (23). The potential vaccine candidate influenza viruses Beijing/184/93 and Shangdong/7/97 passaged only in embryonated chicken eggs were received from the Centers for Disease Control and Prevention, Atlanta, Ga. The strains used to generate virus reassortants were previously described (23). The viruses were grown in 11-day-old embryonated chicken eggs. The allantoic cavities of the embryonated eggs were inoculated with the original allantoic fluids diluted 1:1,000 in phosphate-buffered saline containing 10 μg of gentamicin/ml. Each egg was inoculated with 0.2 ml of diluted allantoic fluid and incubated at 33°C for 72 h. To make reassortants, eggs were coinoculated with 1,000 50% egg infective doses (each) of the B/Beijing/184/93 and B/Shangdong/7/97 virus strains/0.2 ml. The mixed progeny from reassortment passage were harvested and neutralized with postinfection ferret anti-B/Shangdong/7/97 serum (1 volume of a 1:10 dilution of virus added to 1 volume of a 1:10 dilution of serum) for 1 h at room temperature. Subsequently, the embryonated eggs were inoculated with 10-fold serial dilutions of the virus-serum mixture. Viruses recovered from this passage that had HA titers of 1:128 to 1:256 were selected for cloning by the terminal-dilution method in two further passages in eggs.
(ii) RNA preparation and reverse transcription.
Viral RNA was extracted from allantoic fluid using the Viral RNA Mini kit (QIAGEN, Valencia, Calif.), and cDNA was synthesized using Thermoscript RT polymerase (ThermoScript RT-PCR System; Invitrogen, Carlsbad, Calif.) with an AGCAGAAGC primer complementary to the conserved 3′ ends of all influenza B virus genome segments, either alone or in a mixture with a random hexanucleotide (dN6). The resulting cDNA was used for multiplex PCR amplification.
(iii) Nucleic acid sequencing and analysis.
The nucleotide sequences of all eight segments of B/Beijing/184/93 were obtained from GenBank. The sequences of genome segments of B/Shangdong/7/97 were determined in our laboratory (23).
cDNA samples were prepared by 30 cycles of PCR amplification (HotStarTaq Master Mix; QIAGEN) using primers specific for partial sequences of eight genome segments. Amplification products were separated by electrophoresis in 1% agarose gel, extracted with the QIAquick Gel Extraction kit (QIAGEN), and sequenced using a dRhodamine Terminator Cycle Sequencing kit (PE Biosystems, Warrington, United Kingdom) and an ABI Prizm model 377 (Applied Biosystems, Foster City, Calif.).
(iv) Multiplex PCR and synthesis of Cy5-labeled hybridization samples.
Amplicons of all eight influenza B virus genome segments were prepared in two multiplex PCRs; the primer pair for the PB2, PB1, PA, and HA segments in one multiplex PCR was B1-S1, and the primer pair for the NP, NA, M, and NS segments the other multiplex PCR was B2-S2 (B and S refer to Beijing and Shangdong, respectively). The sequences of the PCR primers are listed in Table 1. One primer in each pair was 5′ biotinylated for strand separation and preparation of Cy5 fluorescently labeled single-stranded DNA samples as previously described (8). Briefly, 30 μl of a reaction mixture containing 1× AmpliTaq PCR buffer with 1.5 mM MgCl2; a 200 nM concentration of each primer; 20 μM Cy5-dCTP; 20 μM dCTP; 200 μM (each) dATP, dGTP, and dTTP; and 0.8 U of HotStartTaq DNA Polymerase (QIAGEN) was used. Amplification was performed by 35 PCR cycles consisting of 30 s at 94°C, 30 s at 45°C, and 60 s at 72°C. Single-stranded DNA was prepared by the separation of DNA strands using streptavidin-coated magnetic beads (Dynal ASA, Oslo, Norway) according to the manufacturer's protocol. Fluorescently labeled single-stranded DNA was eluted from the magnetic beads by washing them with 50 μl of 0.1 M NaOH purified through CentriSep spin columns (Princeton Separation, Adelphia, N.J.), concentrated by vacuum drying, and finally resuspended in water.
TABLE 1.
PCR primers used for amplification of genomic segments of influenza B virus
| Primer name | Sequence | Tm (°C) (basic)a |
|---|---|---|
| PB2-541POS-BIOT | CCAAGAGAATCTACTTGGATACATA | 53 |
| PB2-844NEG | AGTCTTGTTTGCAATTTCTACAGC | 52 |
| PB-1-790POS-BIOT | GAAAATCTAGAACAAAGTGGTTTGC | 53 |
| PB-1-990NEG | CAAAAATCCCGGAACCAAATTGG | 53 |
| PA-1845POS-BIOT | CAGTATTGGGACTCAAGAAGG | 52 |
| PA-2061NEG | CAAGGGTTGTTACTAATACATTCT | 51 |
| HA-327NEG | TGTTCTGTCGTGCATTATAGGAAAG | 54 |
| HA-160-POS-BIOT | TCTCATTTTGCAAATCTCAAAGGAACA | 54 |
| NP-4POS | TCCAACATGGATATTGACGGT | 50 |
| NP-273NEG-BIOT | GTTATAGAATTCACCCAGTT | 46 |
| NA-1006POS | CAGATGATGGAAGCATAACAG | 50 |
| NA-1156NEG-BIOT | CTCCATCATACTTGACATACAGTT | 52 |
| M-894-POS-BIOT | AACAGAGAGGTATCAATTTTGAG | 50 |
| M-1053-NEG | TTAATGCAATTCTTCTACCTCCAA | 51 |
| NS-264POS | CCTATTTATGAACCCATCTGCT | 51 |
| NS-485NEG-BIOT | CCTTCTTTCTGAGTGTTTACTTCC | 54 |
Tm, melting temperature.
(v) Preparation of oligoprobes and microchips and microarray hybridization.
Oligoprobes (17 to 22 nt) were designed to match the regions of each segment that were previously found to contain one or more nucleotide differences between the RNA sequences of the B/Beijing/184/93 and B/Shangdong/7/97 strains. The selected oligoprobe pairs differed by 1 to 7 nt (Table 2). The 5′ end of each oligoprobe contained an aminolink group (TFA Aminolink CE reagent; PE Applied Biosystems) added during the synthesis. The probes were covalently immobilized on the surfaces of aldehyde-coated glass slides. Additional spots of oligonucleotides complementary to one of the primers for PCR amplification were used as qualitative positive controls for sample preparation and hybridization (Table 2). These controls were not used for the normalization of array signals and therefore were not reflected in the results (i.e., they are not shown in Fig. 1).
TABLE 2.
Oligoprobes used for discrimination between Beijing and Shangdong strains of influenza B virusa
| Oligoprobe | Sequence | Tm (°C) (basic)b |
|---|---|---|
| PB2 | ||
| PB2-B-625 | CTTGAGAGGGAATTGGTTGC | 52 |
| PB2-S-625 | CTTGAAAGAGAACTGGTTGC | 50 |
| PB2-B-643 | CCAGAAGAAGGTTCCTGCC | 53 |
| PB2-S-643 | CCCGAAGAAGATTCTTGCC | 51 |
| PB2-B-591 | GAACGATGATAACTCCCATT | 50 |
| PB2-S-590 | AACAATGATAACTCCAATT | 49 |
| PB2-665-F-cont | TGGCAGGAGCAACATCAGC | 53 |
| PB1 | ||
| PB1-B-929 | AATGAATGCTTAAATCCAAGAA | 46 |
| PB1-S-929 | AATGAATGTTTAAACCCAAGAA | 46 |
| PB1-B-823 | GGAAATGAAAAGAAGGCCAA | 48 |
| PB1-S-823 | GGAAACGAAAAGAAAGCCAA | 48 |
| PB1-990-F-cont | CCAATTTGGTTCCGGGATTTTTG | 53 |
| PA | ||
| PA-B-1954 | AGGGGTTTAGTGCCGAGT | 50 |
| PA-S-1954 | AGGGATTTAGTGCAGAGT | 46 |
| PA-B-1979 | GGAGACTTCTACTGTTG | 45 |
| PA-S-1979 | GGAGACTTTTACTGTTG | 42 |
| PA-B-2034 | GACTTAGAGGGAATGTA | 42 |
| PA-S-2034 | GACTTAGAAGGAATGTA | 40 |
| PA-1107-F-cont | GTTCAAACAGAGATGAATCTATTG | 51 |
| HA | ||
| HA-B-253 | CCAATGTGTGTGGGGACCAC | 56 |
| HA-S-253 | CCAAAATGCACGGGGAACAT | 52 |
| HA-B-207 | CCCAAACTGTCTCAACTG | 48 |
| HA-S-207 | CCCAAAATGCCTCAACTG | 48 |
| HA-327-F-cont | CTTTCCTATAATGCACGACAGAA | 52 |
| NP | ||
| NP-B-30 | TTGTCAGTTGTCCCAGTG | 48 |
| NP-S-30 | TTGTCAATTGTTCCAGTG | 43 |
| NP-B-37 | CGGTGTTTTGTCAGTTGT | 46 |
| NP-S-37 | CGGTGCTTTGTCAATTGT | 46 |
| NP-B-188 | CTTTTGGGCTTTCCTTC | 45 |
| NP-S-188 | CTTTTGGGTTTTCCTTC | 42 |
| NP-4-R-cont | ACCGTCAATATCCATGTTGGA | 50 |
| NA | ||
| NA-B-1051 | TGATGCCACCACGCCCTT | 53 |
| NA-S-1051 | CTTGATACCTCCATTCCCTT | 50 |
| NA-B-1108 | GTTCGAGAATACCACCTTC | 49 |
| NA-S-1108 | GTTCGAAAGTACCATCTTC | 47 |
| NA-1006-R-cont | CTGTTATGCTTCCATCATCTG | 50 |
| M | ||
| M-B-970 | ACAACATGGAGGTATTGA | 43 |
| M-S-970 | ACAACATAGAAGTATTGA | 39 |
| M-B-1004 | GAGACACAGTTACCAAAA | 43 |
| M-S-1004 | GAGACACAATTACCAAAA | 41 |
| M-1053-F-cont | TTGGAGGTAGAAGAATTGCATTAA | 51 |
| NS | ||
| NS-B-304 | GCTATTGGAGGGATTTTTCA | 48 |
| NS-S-304 | GCTATTTGAGGAACTTTTCA | 46 |
| NS-B-337 | CCAATTGCAGTCTGGACA | 48 |
| NS-S-337 | CCAATTGTATTTCGGACAG | 47 |
| NS-B-403 | GTCACCAACATTCTCCGGTTC | 54 |
| NS-S-403 | GTCATCAACATCCTCTGTTTC | 50 |
| NS-264-R-cont | AGCAGATGGGTTCATAAATAGG | 51 |
PB2, PB1, PA, HA, NP, NA, M, and NS, segments of influenza B virus. F-cont and R-cont indicate the oligoprobes complementary to forward primers and to reverse primers, respectively, which were used for PCR amplification. The homotypic hybridization signals of these oligoprobes were used as the qualitative indicators (positive controls) of sample preparation and hybridization.
Tm, melting temperature.
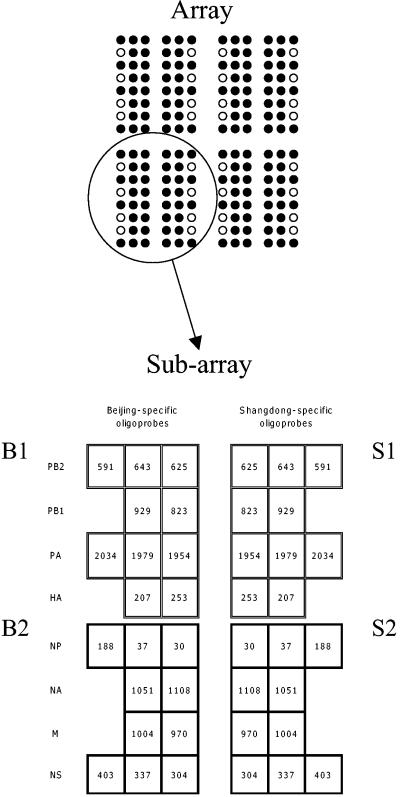
FIG. 1.
Influenza B virus array design. The array consists of four identical subarrays. A solid circle indicates a spot containing printed oligoprobe; an open circle indicates a control spot containing no printed oligoprobe to assess background sample binding. Each subarray contained 40 oligoprobes (Table 1). The left part of each subarray contained 20 oligoprobes complementary to the Beijing strain; the right part contained 20 oligoprobes complementary to the Shangdong strain. The pairs of homologous oligoprobes of the Beijing and Shangdong strains were printed symmetrically across the vertical axis of a subarray. Each subarray consisted of four areas. Each area contained oligoprobes complementary to one of the four fluorescently labeled cDNA samples: B1, B2, S1, or S2 (see “Multiplex PCR and synthesis of Cy-5-labeled hybridization samples”). To assist in the visual presentation, the areas of oligoprobes complementary to the B1 and S1 samples are separated from the B2 and S2 samples.
Microarray fabrication and hybridization were described in detail earlier (8). The concentration of oligonucleotides in 0.25 M acetic acid was adjusted to 75 mM. After the oligonucleotides were spotted using a robotic arrayer (Cartesian Technologies Inc., Ann Arbor, Mich.), the slides were dried for 20 min at 84°C and treated with a 0.25% aqueous solution of NaBH4 for 5 min. Single-stranded Cy5-labeled influenza B virus DNA samples were mixed with equal volumes of 2× hybridization buffer, denatured for 1 min at 95°C, and chilled on ice for 1 min. The final concentration of each fluorescent sample in hybridization solution was typically >0.1 μM; 3-μl aliquots from each sample were applied to the array area. Each array was covered with a 4- by 7-mm plastic coverslip, and hybridization was allowed to proceed for 30 min at 42°C. After hybridization, the slides were washed sequentially with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2× SSC, and 1× SSC and dried in an air stream.
(vi) Array design.
Figure 1 shows the layout of oligoprobes on the microarray. Each array consisted of four identical subarrays, and each subarray contained 20 oligoprobes redundantly representing eight segments of the B/Beijing/184/93 strain and 20 oligoprobes representing eight segments of the B/Shangdong/7/97 strain. Each subarray consisted of four areas containing oligoprobes complementary to one of four fluorescently labeled cDNA samples designated B1, B2, S1, and S2 (see “Multiplex PCR and synthesis of Cy-5-labeled hybridization samples” above).
Eight identical arrays were printed on each slide. Therefore, each of the four fluorescently labeled cDNA samples could be hybridized twice on each slide. This design enabled us to produce eight replicates of the homotypic hybridization signal and eight replicates of the heterotypic hybridization signal (used as the negative control) for each of the 20 unique oligoprobes spotted on a slide. For example, the Beijing sample B1 could hybridize to eight homotypic Beijing-specific oligoprobes (PB2-B-591) and to eight heterotypic Shangdong-specific oligoprobes (PB2-S-591).
(vii) Microchip scanning and data analysis.
Microchip images were captured using a ScanArray 5000 confocal fluorescent scanner (Perkin-Elmer, Boston, Mass.) with two HeNe lasers (632 nm for excitation of Cy5 and 543 nm for Cy3). The fluorescent images were analyzed using QuantArray software (Perkin-Elmer).
(viii) Restriction enzyme digestion.
Three micrograms of each NS segment amplicon was digested at 30°C for 65 min in 20 μl of reaction mixture containing 2 U of the HgaI restriction enzyme (New England BioLabs Inc., Beverly Mass.). The products of digestion were separated by electrophoresis in 9.5% polyacrylamide gels.
Statistical methods: background subtraction and signal normalization.
Let A represent the value of the intensity of the hybridization signal of a given fluorescently labeled cDNA sample hybridized with the homotypic oligoprobe on the array; let B represent the value of the intensity of the hybridization signal of the sample hybridized with the heterotypic oligoprobe on the same array. Let K be the value of the background intensity of the signal measured in mock spots on this array (printed without oligoprobe) (Fig. 1). We measured K values in 64 mock spots on each array.
To reduce the effect of background (nonspecific binding of the fluorescently labeled cDNA sample to the array) and to compare the homotypic hybridization signals across arrays and different slides, we estimated the relative level of homotypic hybridization signal for a given oligoprobe by the following formula:
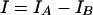 |
(1) |
where IA = (A − K̄)/(Ā − K̄) and IB = (B − K̄)/(Ā − K̄). K̄ is the mean value of background intensities averaged over the 64 signals measured in mock spots on the array (printed without oligoprobe [Fig. 1]). Ā is the mean value of the homotypic signal averaged over four subarrays on the array. Because the oligoprobes of the array were hybridized to only one of the four labeled cDNA samples, the local (on the array) normalization of the hybridization signal on factor Ā − K̄ allowed us to compare the interarray and interslide variabilities of the specific signals for a given oligoprobe.
The four pairs of IA and IB values on each array were compared by Wilcoxon's signed-rank pair test. We also used the Kruskal-Wallis test for multiple comparisons of the arrays in the replicated experiments and a paired t test and a one-way analysis of variance test to compare the IA and IB values in the replicated experiments. Estimation of the number of replicates of arrays and the confidence probabilities of the false-negative rate for hybridizations is described in the appendix.
The MLAB (Civilized Software, Inc., Silver Spring, Md.), Statistica-5 (StatSoft, Inc., Tulsa, Okla.), and JMP (SAS Institute Inc.) programs were used for statistical analysis.
Nucleotide sequence accession numbers.
The sequences of genome segments of B/Shangdong/7/97 determined in our laboratory (23) were deposited in GenBank under accession numbers AF 484967, AY044173, AF486835, AF486836, AY044169, AY044172, AY044171, AY044170, AF101974, AF101992, AF102009, AF050061, AF100367, AF100381, and AF100399.
RESULTS
Analysis of hybridization patterns of Beijing and Shangdong strains.
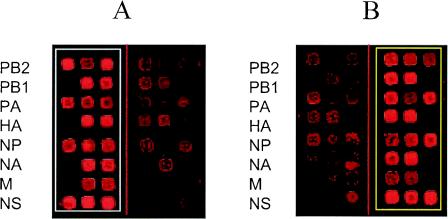
Figure 2 shows typical hybridization patterns for the B/Beijing/184/93 and B/Shangdong/7/97 strains (only one of four identical subarrays is shown). All segments were unambiguously identified as either Beijing or Shangdong, even though for some oligoprobe pairs there was some cross-hybridization.
FIG. 2.
Typical hybridization for reference virus strains B/Beijing/184/93 and B/Shangdong/7/97 on a subarray. For purposes of visual presentation, hybridization patterns obtained for multiplex PCR products containing PB2, PB1, PA, and HA segments and NP, NA, M, and NS segments were combined in one image. (A) Hybridization spots for labeled single-stranded DNA samples of Beijing virus homotypically hybridized with 20 Beijing-complementary oligoprobes and heterotypically hybridized with 20 Shangdong-complementary oligoprobes (control). (B) Hybridization spots for labeled single-stranded DNA samples of Shangdong virus heterotypically hybridized with 20 Beijing-complementary oligoprobes (control) and homotypically hybridized with 20 Shangdong-complementary oligoprobes. The hybridization spots in the rows of a subarray represent hybridizations of a Cy-5 fluorescently labeled single-stranded DNA sample with different oligoprobes printed in the row. For purposes of visual presentation and inspection, the vertical line going through the subarray separates three columns containing the Beijing-complementary oligoprobes (on the left side of the subarray) and the three columns containing the Shangdong-complementary oligoprobes (on the right side of the subarray). Each row on a subarray represents oligoprobes of one genome segment of influenza virus. The oligoprobes were printed in a mirror symmetry manner relative to this line (see details in Fig. 1). The identifiers of genome segments are showed on the left in panels A and B. The PB2, PA, NP, and NS genome segments of the Beijing and Shangdong strains are represented by three different oligoprobes and by three observed hybridization spots. The PB1, NA, HA, and M genome segments of these strains are represented by two different oligoprobes and by the two hybridization spots. In the last case, two “empty” spots on the rows of the subarray are observed.
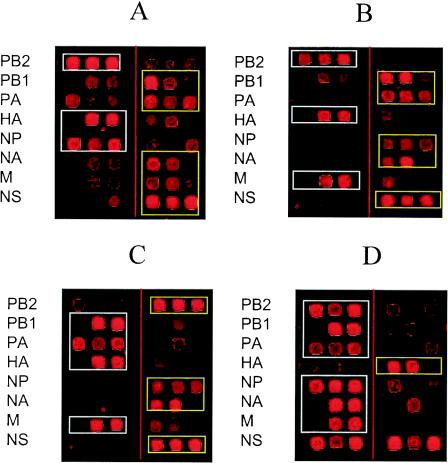
We also obtained several hybridization patterns for reassortant samples. Representative examples are shown in Fig. 3. In reassortant A, the segments PB2, HA, and NP were derived from the Beijing parent while the other five segments were derived from the Shangdong parent. In reassortant B, the segments PB2, HA, and M were derived from the Beijing parent while the other five segments were derived from the Shangdong parent. In reassortant C, the segments PB1, PA, HA, and M were derived from the Beijing parent while the other four segments were derived from the Shangdong parent. These results are in complete agreement with the sequence analysis data (not shown) and thus demonstrate that microarray hybridization of amplicons of reassortants between the Beijing and Shangdong strains can reveal the origins of the segments. However, in some instances, identification of the segment origin was not as clear. One such example is shown in Fig. 3: reassortant D consisted of segments PB2, PB1, PA, NP, NA, and M derived from the Beijing parent, while segment HA was derived from the Shangdong parent. However, the hybridization intensities of all Beijing- and Shangdong-specific oligoprobes binding the NS segment were comparable. This pattern could be a result of the mixture of two NS segment genotypes in the sample. To test this possibility and to determine the limits of the ability of the microarray to detect genetic heterogeneity, we performed a detailed quantitative analysis of microarray hybridization results (see “Identification of mixed genotype samples: microarray hybridization is superior to conventional nucleotide sequencing analysis” below). To do this, we had to determine the statistical significance of the microarray hybridization data.
FIG. 3.
Discrimination of hybridization patterns of four different reassortants on a subarray. See details in the text. Microarray elements specific to the prevailing RNA segment are boxed.
Statistical analysis of microarray hybridization data.
Seven independent hybridization experiments were performed and analyzed for evaluation of the discriminating power of individual oligoprobes. Each experiment included eight replicates of homotypic and heterotypic hybridization signals (fluorescence intensity) obtained from 20 pairs of oligoprobes (spotted on the slide) specific to B/Beijing/184/93 and B/Shangdong/7/97. Relative hybridization intensities were calculated by background subtraction and normalization as described in Materials and Methods. For each of the 40 different oligoprobes, the normalized signal intensities did not differ significantly for the matching spot pairs located on the same slide (P > 0.1 by the Wilcoxon and paired t tests) and for all seven slides (P > 0.1 by the Wilcoxon and Kruscal-Wallis tests). Hence, the background subtraction and normalization procedures allowed us to treat specific hybridization signals for the same oligoprobes on a slide and across different slides as samples chosen from the same population. The statistical comparisons of the 56 combined hybridization experiments (7 hybridization experiments, each including eight replicates) implies that the microarray method is highly sensitive, specific, and reproducible (Table 3). Comparison of homotypic and heterotypic signals by the t and F tests showed that the P values for homotypic signal intensities were statistically significant (0.000001 < P < 0.01) for 40 oligoprobes.
TABLE 3.
Significance of homotypic hybridizations for oligonucleotides of influenza B virus chipa
| Oligonucleotide | E(I) | SD | t | F |
|---|---|---|---|---|
| PB2-B-643 | 0.935 | 0.188 | 37.3 | 1,284 |
| PB2-B-625 | 0.886 | 0.169 | 39.2 | 1,342 |
| PB2-B-591 | 0.895 | 0.177 | 37.9 | 1,225 |
| PB1-B-929 | 0.811 | 0.235 | 25.8 | 576.7 |
| PB1-B-823 | 0.803 | 0.249 | 24.1 | 487.9 |
| PA-B-1979 | 0.864 | 0.244 | 26.5 | 660.9 |
| PA-B-1954 | 0.708 | 0.388 | 13.6 | 150.8 |
| PA-B-2034 | 0.886 | 0.206 | 32.1 | 948.8 |
| HA-B-207 | 0.602 | 0.173 | 26 | 575.4 |
| HA-B-253 | 0.864 | 0.148 | 48.8 | 1,647 |
| NP-B-30 | 0.866 | 0.268 | 24.1 | 484.1 |
| NP-B-37 | 0.992 | 0.409 | 18.2 | 326.2 |
| NP-B-1108 | 0.779 | 0.292 | 20 | 250.7 |
| NA-B-1051 | 0.923 | 0.193 | 35.7 | 1,141 |
| NA-B-1108 | 0.995 | 0.186 | 39.9 | 1,566 |
| M-B-970 | 0.979 | 0.229 | 32 | 986.1 |
| M-B-1004 | 0.943 | 0.371 | 19 | 337.3 |
| NS-B-304 | 0.98 | 0.135 | 54.2 | 2,809 |
| NS-B-337 | 0.993 | 0.16 | 46.4 | 2,139 |
| NS-B-403 | 0.978 | 0.16 | 45.8 | 2,001 |
| PB2-S-643 | 0.947 | 0.174 | 40.7 | 1,531 |
| PB2-S-625 | 0.801 | 0.171 | 35.1 | 936.2 |
| PB2-S-591 | 0.349 | 0.23 | 11.4 | 73.29 |
| PB1-S-929 | 0.575 | 0.277 | 15.5 | 187.7 |
| PB1-S-823 | 0.864 | 0.181 | 35.7 | 1,052 |
| PA-S-1979 | 0.657 | 0.303 | 16.2 | 206.4 |
| PA-S-1954 | 0.889 | 0.254 | 26.2 | 601.7 |
| PA-S-2034 | 0.654 | 0.277 | 17.6 | 261.3 |
| HA-S-207 | 0.808 | 0.16 | 37.7 | 1,124 |
| HA-S-253 | 0.996 | 0.12 | 62 | 3,823 |
| NP-S-30 | 0.45 | 0.29 | 13.4 | 88.11 |
| NP-S-37 | 0.68 | 0.533 | 9.54 | 85.12 |
| NP-S-1108 | 0.657 | 0.329 | 14.4 | 178 |
| NA-S-1051 | 0.986 | 0.141 | 52.4 | 2,674 |
| NA-S-1108 | 0.906 | 0.24 | 28.3 | 714.3 |
| M-S-970 | 0.937 | 0.241 | 29.1 | 782.5 |
| M-S-1004 | 0.848 | 0.375 | 16.9 | 279.5 |
| NS-S-304 | 0.913 | 0.116 | 59.1 | 3,308 |
| NS-S-337 | 0.994 | 0.257 | 29 | 801.2 |
| NS-S-403 | 0.992 | 0.14 | 53.2 | 2,708 |
E(I), mean value of the difference between normalized homotypic and heterotypic signals; SD, standard deviation of the difference; t, paired-sample t test value; F, F statistic value (analysis of variance test). The 56 observations were from seven hybridization experiments in which eight replicates were produced. All t and F values are statistically significant at P values ranging between 0.01 and 0.000001.
Sample replication within each slide and redundancy of oligoprobes for each segment of the influenza B virus genome increase the reliability of the method. In particular, performance of the seven hybridization experiments reduces the likelihood of making false-negative determinations of the presence of the segment from a given strain (Table 4). The maximum probabilities of false-negative determinations for each segment were calculated as described in the appendix. For all eight influenza B virus segments, the probabilities ranged from 0.027 to 0.00014, indicating the high power of the influenza B virus microarray to discriminate between the Beijing and Shangdong viruses.
TABLE 4.
Characteristics of reliability of influenza B virus chipa
| Oligonucleotide | No. of unlikely events | pB | PS |
|---|---|---|---|
| PB2-B-643 | 0 | 0.052 | 0.00014 |
| PB2-B-625 | 0 | 0.052 | |
| PB2-B-591 | 0 | 0.052 | |
| PB1-B-929 | 0 | 0.052 | 0.027 |
| PB1-B-823 | 0 | 0.052 | |
| PA-B-2034 | 0 | 0.052 | 0.00014 |
| PA-B-1979 | 0 | 0.052 | |
| PA-B-1954 | 0 | 0.052 | |
| HA-B-207 | 0 | 0.052 | 0.027 |
| HA-B-253 | 0 | 0.052 | |
| NP-B-30 | 0 | 0.052 | 0.00014 |
| NP-B-37 | 0 | 0.052 | |
| NP-B-188 | 0 | 0.052 | |
| NA-B-1051 | 0 | 0.052 | 0.027 |
| NA-B-1108 | 0 | 0.052 | |
| M-B-970 | 0 | 0.052 | 0.027 |
| M-B-1004 | 0 | 0.052 | |
| NS-B-304 | 0 | 0.052 | 0.00014 |
| NS-B-337 | 0 | 0.052 | |
| NS-B-403 | 0 | 0.052 | |
| PB2-S-643 | 0 | 0.052 | 0.00033 |
| PB2-S-625 | 0 | 0.052 | |
| PB2-S-591 | 3 | 0.123 | |
| PB1-S-929 | 0 | 0.052 | 0.027 |
| PB1-S-823 | 0 | 0.052 | |
| PA-S-2034 | 1 | 0.078 | 0.00032 |
| PA-S-1979 | 1 | 0.078 | |
| PA-S-1954 | 0 | 0.052 | |
| HA-S-207 | 0 | 0.052 | 0.027 |
| HA-S-253 | 0 | 0.052 | |
| NP-S-30 | 0 | 0.052 | |
| NP-S-37 | 4 | 0.143 | 0.00038 |
| NP-S-1108 | 0 | 0.052 | |
| NA-S-1051 | 0 | 0.052 | 0.027 |
| NA-S-1108 | 0 | 0.052 | |
| M-S-970 | 0 | 0.052 | 0.027 |
| M-S-1004 | 1 | 0.052 | |
| NS-S-304 | 0 | 0.052 | 0.00014 |
| NS-S-337 | 0 | 0.052 | |
| NS-S-403 | 0 | 0.052 |
Estimations of the probability of unlikely events, e.g., of the probability of rare false-negative hybridizations for each oligonucleotide probe and for each segment. pB is the estimate of a maximum probability of unlikely events for a given oligonucleotide based on all 56 replicated hybridizations. PS is the estimate of a maximum probability of unlikely events for a given genomic segment (see the appendix). PS is the multiplication of pB for a given segment.
Identification of mixed genotype samples: microarray hybridization is superior to conventional nucleotide sequencing analysis.
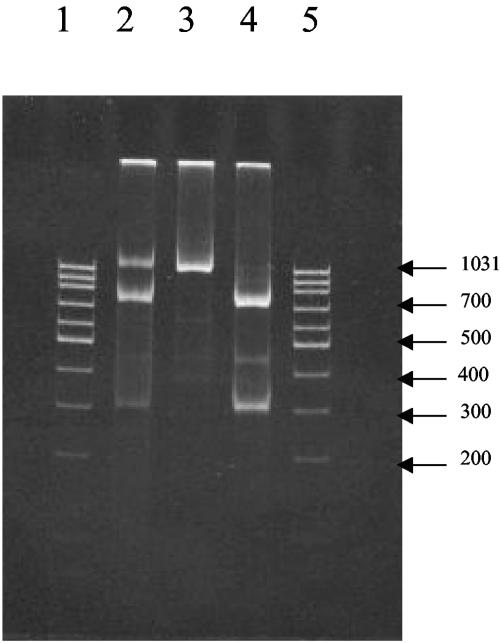
We compared the discriminating abilities of microarray hybridization and conventional nucleotide sequencing analysis. We tested the origins of all eight segments of six reassortant strains. Table 5 shows that for 35 of the 44 comparisons both methods provided consistent results. In the remaining nine instances, the results of hybridization revealed the presence of two genotypes similar to pattern D in Fig. 3 for the NS segment. To obtain an independent confirmation that more than one genotype was present in these samples and to rule out possible cross-hybridization, we used the RFLP method. The HgaI restriction enzyme is expected to cut the NS segment of the Shangdong amplicon, producing two fragments of 730 and 300 bp, but not to cut the NS segment of the Beijing amplicon. Figure 4 shows the restriction patterns for the reassortant strain analyzed by microarray hybridization. The pattern of DNA bands in Fig. 4 indicates that both Beijing and Shangdong sequences of the NS gene segment were present. The RFLP analysis suggests that the microarray hybridization multiple pattern was not the result of excessive cross-hybridization but was due to the presence of a mixture of NS gene segments from the Beijing and Shangdong strains. Therefore, microarray hybridization could be more sensitive than conventional nucleotide sequencing for detecting mixed influenza B virus genotypes.
TABLE 5.
Comparison of sequencing and microarray methods by mapping of influenza B virus reassortants on all influenza B virus genomic segmentsa
| Reassortant | Method | PB2 | PB1 | PA | HA | NP | NA | M | NS |
|---|---|---|---|---|---|---|---|---|---|
| 18 | Sequencing | B | S | S | B | M | M | M | S |
| Microarray | M | S | S | B | M | M | M | M | |
| 19 | Sequencing | B | S | S | B | S | M | M | S |
| Microarray | M | S | S | B | M | M | M | M | |
| 21 | Sequencing | B | S | NA | B | B | NA | S | S |
| Microarray | B | S | S | B | B | M | S | S | |
| 22 | Sequencing | B | S | S | B | B | S | S | S |
| Microarray | B | M | M | B | B | M | S | S | |
| 23 | Sequencing | NA | S | NA | B | B | S | S | S |
| Microarray | B | S | S | B | B | M | S | S | |
| 24 | Sequencing | B | S | S | B | B | S | S | S |
| Microarray | B | S | S | B | B | S | S | S |
B, Beijing strain; S, Shangdong strain; M, mixed B-S genotype; NA, not applicable. For reassortant 24, the microarray hybridization pattern is presented in Fig. 3A. The boldface underlined letters indicate the inconsistent observations derived by sequencing and microarray methods.
FIG. 4.
RFLP of NS segments digested by HgaI. Lane 2, reassortant (the size of the band undigested by HgaI is 1,030 bp, and the sizes of the digested bands are 730 and 300 bp); lane 3, Beijing (the size of the band undigested by HgaI is 1,030 bp); lane 4, Shangdong (the sizes of the bands digested by HgaI are 730 and 300 bp); lanes 1 and 5, DNA ladders.
Quantitative analysis of segment mixtures.
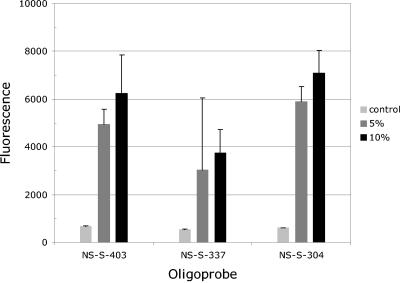
The ability of microarray hybridization to detect the presence of mixed genotypes suggests that it can be used for quantitative assessment of the relative abundances of segment variants in strain mixtures. To determine the sensitivity of the microarray for detection of mixed genotypes, we spiked an NS segment sample prepared from the Beijing strain (175 μM) with 5 and 10 or 50% of the NS segment of the Shangdong strain. Figure 5 shows that for all three NS oligoprobe pairs, the signals produced by a Beijing sample spiked with 5% Shangdong DNA significantly exceeded that with the unspiked sample. This result suggests that microarray hybridization is sensitive to the presence of at least a 5% alternative genotype. However, despite the trend seen in Fig. 5, the difference between spikes with 5 and 10% and with 5 and 50% (not shown) was not statistically significant, meaning that quantification of genotype mixtures was unreliable. This may be due either to saturation of the probe or to a nonlinear dose-response of hybridization intensities.
FIG. 5.
Hybridization of mixed samples with NS segment oligoprobes. The initial Shangdong sample (175 mM) was diluted to final concentrations of 10 and 5% in a Beijing sample (175 mM) and then hybridized with Shangdong-specific oligonucleotides. The heterotypic hybridization signal of the Beijing sample to Shangdong-specific oligoprobes was set as the control. The hybridization was repeated five times. The error bars represent standard deviations.
DISCUSSION
Segment reassortment is an essential feature of many viral pathogens, including influenza viruses. More than one type of reassortant virus can be present simultaneously in natural populations, further complicating their analysis. Detailed genome segment mapping of virus isolates is needed to elucidate the mechanisms leading to the emergence and evolution of viral pathogens and to facilitate the creation of new protective vaccines.
The rational design of vaccines against influenza includes the generation of reassortants between a recent field isolate and a reference strain adapted for efficient growth in the substrate used for vaccine production. After a variety of reassortant strains is generated in vitro by growing two parental strains together, a prospective vaccine strain is selected by genotyping all relevant genomic segments to identify the right combination of segments coding for immunogenic proteins and those determining high-growth characteristics. Traditionally, this time-critical task of selection of prospective vaccine strains has been accomplished by RFLP, sequencing of cDNA, or other approaches (1, 13, 18). The recently developed high-throughput microarray analysis method appears to be ideally suited for one-step mapping of segmented viruses.
Microarray hybridization was previously used to genotype individual segments of influenza A virus (16, 22). In our work, we have developed and used an improved oligonucleotide microarray method to discriminate viruses with high levels of genomic homology through identification of minor genetic differences, including single-nucleotide variations, simultaneously in all segments of influenza B viruses. We observed unambiguous discrimination between the B/Beijing/184/93 and B/Shangdong/7/97 influenza strains. These strains were discriminated with statistical certainty by using different oligoprobes, including PA-1979, PA-2034, NP-188, and M-1004, differing at only 1 nucleotide and representing the highly homologous segments PA, NP, and M of both viruses. The high level of nucleotide homology between the PA, NP, and M segments of B/Shangdong/7/97 and B/Beijing/184/93 has been an obstacle to the genotyping of reassortants derived from the two strains by RT-PCR and RFLP because of the lack of suitable restriction sites (23).
Microarray hybridization, coupled with statistical interpretation of the results, allowed us to estimate the effects of cross-hybridization and to account for nonspecific sample binding (background). Our statistical analysis demonstrated high levels of sensitivity and reproducibility for all individual oligoprobes to all influenza B virus segments; P values for specific hybridization signals for all of the 40 selected oligoprobes were highly significant (P < 0.01).
The statistical planning of the number of hybridization experiments developed in this work can be used in the future in the design of similar oligonucleotide microchip experiments to assess the discriminating power of individual oligoprobes and to select only those that produce the clearest results. Identification of the origin of each segment based on hybridization with two or more specific oligoprobes significantly increased the reliability of identification. To this end, we estimated the maximum probability of false-negative identification of each segment based on multiple results for individual oligoprobes in 56 hybridization experiments. The P values for segments of different origin were calculated by multiplication of P values for homotypic hybridization with oligoprobes specific to each segment. They were statistically significant and demonstrated high overall discriminating power of the microarray for all eight segments of influenza B virus.
Note that our approaches for estimating the minimum number of replicate experiments and for estimating the false-negative error rate for a given gene segment seem novel for microarray genotyping. However, depending on the specific designs of microarray experiments, other statistical approached can also be useful (22a).
Interestingly, the influenza B virus microarray can detect the presence in mixed viral stocks of as little as 5% of a second closely related influenza B virus strain (Fig. 4). The greater ability to detect mixed viral populations is an important advantage of the method over conventional methods based on nucleotide sequencing, since natural isolates often contain more than one genotype. Although the restriction pattern of DNA shown in Fig. 4 indicates that the NS segments of both the Beijing and Shangdong strains were present, similar analyses of other segments and other reassortant strains prepared as vaccine candidates need to be studied in the future. Our preliminary experiments presented here showed that the method could not be used to quantify the components of a mixture without further refinement. The ability to detect as little as 5% of a second strain in a mixture on one hand and the lack of statistically significant hybridization between the values obtained for 5% and increasing spike concentrations on the other suggest that the dose-response curve may be nonlinear. There may a number of reasons for this, including saturation of immobilized probes during hybridization and nonlinear response of the fluorescence detector. Further experiments are needed to make this method more quantitative and to accurately assess the detection limit for mixtures.
The data presented in this paper suggest that the microarray method can be used for high-throughput screening of influenza B virus reassortants, substantially reducing the time needed for vaccine development. The influenza B virus array described in this paper contains only a few oligoprobes specific to each segment of two influenza B virus strains, Beijing and Shangdong. The microarray method can be expanded by adding new oligoprobes, including those specific to other strains, possibly covering the entire repertoire of currently circulating influenza B virus strains. Such extension of the array might allow us to use a more quantitative assay to determine the origins of individual genomic segments in a series of reassortant strains prepared as vaccine candidates or mixed virus populations.
Our preliminary analysis of the repertoire of vaccine reassortants suggests a highly biased (nonrandom) generation of certain segment combinations. Extensive studies of a large number of reassortant strains may help to reveal the preferred combination of segments and to examine the reason for this nonrandomness of the reassortment process.
It is known that rates of homologous recombination in negative-sense RNA viruses, including influenza virus, are substantially lower than their point mutation rates (24). However, patterns of sequence variations compatible with the action of recombination were recently observed in several RNA viruses, including influenza A virus and influenza B virus (3). A search for the evidence of within-segment recombination in influenza viruses could be done by a microarray method similar to the one developed to analyze recombinants in poliovirus (5).
The ability to perform rapid and comprehensive genomic analysis of multiple field isolates can help in efforts to recognize and respond to the emergence of new pandemic strains of influenza virus by providing a better understanding of the processes of viral evolution in populations of susceptible hosts. Similar approaches can also be developed for analysis of influenza A viruses and for other viruses with segmented genomes and can substantially advance our understanding of their evolution and help to create new effective vaccines.
Acknowledgments
We thank David Asher for critical reading of the manuscript, Gennady Rezapkin for his helpful suggestions in planning the experiments, and Valery Pickalov for creation of a database of microarray experiments.
The work was supported by grants from the Defense Advanced Research Projects Agency (DARPA) and the National Vaccine Program Office (NVPO).
APPENDIX
Estimating the number of replicate experiments.
To design an optimal microarray experiment, we need to know how large a sample size n (i.e., the number of replicate hybridizations [or experiments] for a homotypic oligoprobe) should be to give 95% confidence that <5% of specific hybridizations will produce false-negative results. In other words, we consider the question of how large the sample size n must be so that we can be at least 95% sure that the further use of the microarray gives false-negative values of index I (index IA or IB in equation 11) in ≤5% of homotypic hybridizations.
Let us plan to draw random samples X1, X2,…, Xn of the n hybridizations for a given homotypic oligoprobe. Let A represent the homotypic hybridizations showing a positive value of I (true event). Let B represent the homotypic hybridizations showing a negative or zero value of I (false-negative homotypic hybridization). We assume that the events B are rare or unlikely events. Let p represent the probabilities of event B, and let (1 − p) represent the probability of event A. These A and B events can be considered mutually independent and identically distributed events. Let α represent the level of significance for maximum probability of rejection of a null hypothesis, which is ascribed to a confidence probability of the events B after n true binomial experiments having probability (1 − p) in each of the n experiments. Let pB represent a maximum value of a probability of the event B for a given oligoprobe. Then, using a binomial probability distribution function BIN(0;n,pB) (11), we have P (events B in n true homotypic hybridizations) = (1 − pB)n.
Then, assuming that P = α = 1 − β, we obtain:
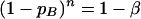 |
(2) |
and
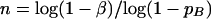 |
(3) |
Let α = 1 − β = 1 − 0.95 = 0.05; then, using equation 3 we obtain n = log0.05/log(0.95) ≈ 58. Thus, we expect that ∼58 replicated hybridization experiments for each homotypic oligoprobe should be sufficient to get an expected reliability for the purpose of planning experiments. In actuality, we analyzed seven slides containing 56 replicates of each of the 40 distinct homotypic oligonucleotides.
Estimating the reliability of homotypic hybridizations for genomic segments.
When no error is observed in all 56 of the replicated experiments, we can estimate the maximum probability of false-negative events (homotypic hybridizations), called B, by equation 2: pB = 1 − (1 − β)1/n. Then, pB = 1 − (1 − 0.95)1/56 = 0.05209.
When I, 2, …, k,… errors are found, the maximum probability of false-negative events, pB, can be estimated by the binomial probability distribution BIN(k;n,pB) (11):
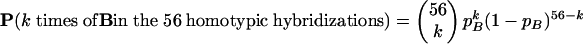 |
(4) |
We can estimate pB (k = 0) = 0.05209; pB (k = 1) = 0.0781; pB (k = 2) = 0.104; pB (k = 3) = 0.1432; pB (k = 4) = 0.1532; pB (k = 5) = 0.1829 for k = 0, 1, 2, 3, 4, 5, which have been observed in all slides.
We selected from two to four homotypic oligoprobes for each of the eight genomic segments to use in hybridization experiments. Let pB1, pB2,… represent the observed frequencies of false-negative events B1, B2,…associated with homotypic hybridization on the first, second, etc. distinct oligoprobes representing a given genome segment. Analysis of hybridization with two or more distinct oligoprobes covering a given segment in the same microarray experiment allowed us to increase the reliability of genomic-segment identification by estimating the joint probability function of false-negative events for a given genomic segment by the formula Ps = pB1pB2. … For example, when two oligoprobes used for the identification of a given segment had pB1 = 0.0781 and pB2 = 0.0108, respectively, the maximum probability of false-negative identification of the given segment was estimated to be Ps = pB1pB2 = 0.0079.
REFERENCES
- 1.Cha, T. A., J. Zhao, E. Lane, M. A. Murray, and D. S. Stec. 1997. Determination of the genome composition of influenza virus reassortants using multiplex reverse transcription-polymerase chain reaction followed by fluorescent single-strand conformation polymorphism analysis. Anal. Biochem. 252:24-32. [DOI] [PubMed] [Google Scholar]
- 2.Chanock, R. M., and B. R. Murphy. 1980. Use of temperature-sensitive and cold-adapted mutant viruses in immunoprophylaxis of acute respiratory tract disease. Rev. Infect. Dis. 2:421-432. [DOI] [PubMed] [Google Scholar]
- 3.Chare, E. R., E. A. Gould, and E. C. Holmes. 2003. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J. Gen. Virol. 84:2691-2703. [DOI] [PubMed] [Google Scholar]
- 4.Chen, H., Y. Matsuoka, D. Swayne, Q. Chen, N. J. Cox, B. R. Murphy, and K. Subbarao. 2003. Generation and characterization of a cold-adapted influenza A H9N2 reassortant as a live pandemic influenza virus vaccine candidate. Vaccine 21:4430-4436. [DOI] [PubMed] [Google Scholar]
- 5.Cherkasova, E., M. Laassri, V. Chizhikov, E. Korotkova, E. Dragunsky, V. I. Agol, and K. Chumakov. 2003. Microarray analysis of evolution of RNA viruses: new evidence of circulation of virulent highly divergent vaccine-derived polioviruses. Proc. Natl. Acad. Sci. USA 100:9398-9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chetverin, A. B., and F. R. Kramer. 1993. Sequencing of pools of nucleic acids on oligonucleotide arrays. Biosystems 30:215-231. [DOI] [PubMed] [Google Scholar]
- 7.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group a rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hacia, J. G. 1999. Resequencing and mutational analysis using oligonucleotide microarrays. Nat. Genet. 21:42-47. [DOI] [PubMed] [Google Scholar]
- 10.Hacia, J. G., J. B. Fan, O. Ryder, L. Jin, K. Edgemon, G. Ghandour, R. A. Mayer, B. Sun, L. Hsie, C. M. Robbins, L. C. Brody, D. Wang, E. S. Lander, R. Lipshutz, S. P. Fodor, and F. S. Collins. 1999. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 22:164-167. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, N. L., S. Kotz, and A. W. Kemp. 1993. Univariative discrete distributions, 2nd ed. John Wiley and Sons, Inc., New York, N.Y.
- 12.Khrapko, K. R., P. Y. Lysov, A. A. Khorlyn, V. V. Shick, V. L. Florentiev, and A. D. Mirzabekov. 1989. An oligonucleotide hybridization approach to DNA sequencing. FEBS Lett. 256:118-122. [DOI] [PubMed] [Google Scholar]
- 13.Klimov, A. I., and N. J. Cox. 1995. PCR restriction analysis of genome composition and stability of cold-adapted reassortant live influenza vaccines. J. Virol. Methods 52:41-49. [DOI] [PubMed] [Google Scholar]
- 14.Laassri, M., V. Chizhikov, M. Mikheev, S. Shchelkunov, and K. Chumakov. 2003. Detection and discrimination of orthopoxviruses using microarrays of immobilized oligonucleotides. J. Virol. Methods 112:67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb, R. A., and R. M. Krug. 2001. Orthomixoviridae: the viruses and their replication, p. 1488-1524. In D. M. Knipe and P. M. Howley (ed.), Field's virology, 4th ed. Lippincott, Williams, and Wilkins, Philadelphia, Pa.
- 16.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCullers, J. A., G. C. Wang, S. He, and R. G. Webster. 1999. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J. Virol. 73:7343-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Offringa, D. P., V. Tyson-Medlock, Z. Ye, and R. A. Levandowski. 2000. A comprehensive systematic approach to identification of influenza A virus genotype using RT-PCR and RFLP. J. Virol. Methods 88:15-24. [DOI] [PubMed] [Google Scholar]
- 19.Palese, P. 1977. The genes of influenza virus. Cell 10:1-10. [DOI] [PubMed] [Google Scholar]
- 20.Proudnikov, D., E. Kirillov, K. Chumakov, J. Donlon, G. Rezapkin, and A. Mirzabekov. 2000. Analysis of mutations in oral poliovirus vaccine by hybridization with generic oligonucleotide microchips. Biologicals 28:57-66. [DOI] [PubMed] [Google Scholar]
- 21.Richman, D. D., B. R. Murphy, R. B. Belshe, H. M. Rusten, R. M. Chanock, N. R. Blacklow, T. A. Parrino, F. B. Rose, M. M. Levine, and E. Caplan. 1977. Temperature-sensitive mutants of influenza A virus. XIV. Production and evaluation of influenza A/Georgia/74-ts-1[E] recombinant viruses in human adults. J. Infect. Dis. 136:256-262. [DOI] [PubMed] [Google Scholar]
- 22.Sengupta, S., K. Onodera, A. Lai, and U. Melcher. 2003. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 41:4542-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Simon, R. 2003. Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data. Br. J. Cancer 89:1599-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vodeiko, G. M., J. McInnis, V. Chizhikov, and R. A. Levandowski. 2003. Genetic and phenotypic analysis of reassortants of high growth and low growth strains of influenza B virus. Vaccine 21:3867-3874. [DOI] [PubMed] [Google Scholar]
- 24.Worobey, M., A. Rambaut, O. G. Pybus, and D. L. Robertson. 2002. Questioning the evidence for genetic recombination in the 1918 “Spanish flu” virus. Science 296:211. [DOI] [PubMed] [Google Scholar]