Abstract
Staphylococcus intermedius is a zoonotic organism that can be associated with human disease. We report two separate cases of S. intermedius infection in which a false-positive rapid penicillin binding protein 2a latex test in conjunction with the phenotypic properties of β-hemolysis and coagulase positivity allowed the clinical isolates to masquerade as methicillin-resistant Staphylococcus aureus. 16S rRNA gene sequencing and the absence of mecA revealed the strains to be methicillin-susceptible S. intermedius.
Staphylococcus intermedius is a coagulase-positive zoonotic organism found in pigeons, dogs, foxes, mink, and horses (6). Initially, all coagulase-positive staphylococci were identified as Staphylococcus aureus, until Hájek in 1976 established the unique identity of a group of organisms, originally identified as S. aureus biotypes E and F, as S. intermedius (6). The name of this species reflects the fact that while the organisms possess some phenotypic properties of S. aureus, they also exhibit some properties of Staphylococcus epidermidis.
S. intermedius is a common commensal of oral, nasal, and skin flora in healthy dogs, where it can also cause invasive disease (6, 15, 16). In humans, it is recognized as an invasive zoonotic pathogen and has been isolated from 18% of canine-inflicted wounds (9). In three cases it was also isolated in pure growth from non-canine-inflicted wounds: two elderly patients with infected varicose ulcers and a 13 year old with an infected suture line, each with a history of exposure to dogs (9). Other reports of invasive human disease include pneumonia following coronary artery bypass grafting and catheter-related bacteremia in a patient with lung cancer (3, 17). We report two cases of non-canine-inflicted S. intermedius wound infection from two separate hospitals which occurred over an 8-month period. History regarding the association with animals was not available in either case. Using protocols for identification and detection of methicillin resistance routinely used in the two laboratories, both isolates were initially misidentified and reported as methicillin-resistant Staphylococcus aureus (MRSA).
In the first case, a 60-year-old female undergoing chemotherapy for breast cancer was noted to have onycholysis involving the distal half of her fingernails. One of her nail beds was noted to be inflamed with greenish discoloration and was cultured. Gram's stain revealed 3+ gram-positive cocci, 3+ gram-positive rods, and 1+ gram-negative rods amidst a few inflammatory cells (3+ indicates 6 to 30 organisms, 2+ indicates 1 to 5 organisms, and 1+ indicates <1 organism per high-power field using an oil immersion lens at 100×). The culture grew a coagulase-positive Staphylococcus isolate (isolate 1) that was identified as S. aureus. Pseudomonas aeruginosa, diphtheroids, and coagulase-negative staphylococcus were also present in the initial culture. Penicillin binding protein 2a (PBP2′, PBP2a) latex agglutination testing of isolate 1, performed and interpreted in accordance with the manufacturer's instructions (Oxoid Limited, Denka Seiken, Ltd., Basingstoke, England), gave a strong positive result, and the isolate was reported as methicillin-resistant S. aureus. Susceptibility testing with a Vitek I GPS-107 card (bioMérieux, Durham, N.C.) showed that isolate 1 was susceptible to oxacillin, ampicillin/sulbactam, ampicillin/clavulanic acid, cefazolin, ciprofloxacin, levofloxacin, erythromycin, clindamycin, gentamicin, nitrofurantoin, tetracycline, trimethoprim-sulfamethoxazole, and vancomycin. The strain was resistant to penicillin and was β-lactamase positive.
In the second case, a 37-year-old male sustained a laceration on his right leg 2 days prior to his initial hospital outpatient visit. He had developed localized cellulitis of the lower leg without evidence of systemic symptoms. He was treated with a single intravenous dose of cefazolin and was discharged on oral cephalexin. He returned to the outpatient clinic 3 weeks later and was noted to have a nonhealing leg wound with a foul-smelling discharge, which was cultured. Gram's stain revealed 3+ gram-positive cocci and 2+ gram-negative rods amid small numbers of inflammatory cells. The culture grew a coagulase-positive staphylococcus, isolate 2, identified as S. aureus. Group B streptococci and Escherichia coli were also present in the initial wound culture. PBP2a latex agglutination testing gave a weak positive result for this isolate, which was reported as methicillin-resistant S. aureus. Susceptibility testing with a Vitek I GPS-107 card showed that isolate 2 was susceptible to oxacillin, ampicillin/sulbactam, ampicillin/clavulanic acid, cefazolin, ciprofloxacin, levofloxacin, erythromycin, clindamycin, gentamicin, nitrofurantoin, trimethoprim-sulfamethoxazole, and vancomycin. The strain was resistant to penicillin and tetracycline and was β-lactamase positive.
Both isolates, 1 and 2, failed to grow on oxacillin-salt agar performed in accordance with NCCLS guidelines (12). Due to the discrepant oxacillin susceptibility results between the Vitek 1, oxacillin-salt agar screening test, and PBP2a latex agglutination test for the two isolates, further testing to confirm methicillin susceptibility and identification of the isolates was performed.
Microbiological identification
Both isolates grew equally well on blood and MacConkey agar incubated aerobically at 35°C. After 24 h of incubation, the isolates formed white, entire, convex, glistening colonies, 5 to 6 mm in diameter and surrounded by a zone of β-hemolysis on blood agar. Both isolates were tube coagulase, pyrrolidonyl arylamidase, and o-nitrophenyl-β-d-galactopyranoside positive, and both hydrolyzed urea. The Voges-Proskauer reaction was negative, and the isolates failed to ferment mannitol on mannitol salt agar. Isolate 1 was also slide coagulase positive and gave a weak positive result with the BACTi Staph latex agglutination kit (REMEL, Lenexa, Kans.). These additional phenotypic characteristics suggested the isolates were most likely S. intermedius. Molecular identification of isolates 1 and 2 was performed by PCR and sequencing of the first 500 bp of the 16S rRNA gene by using a previously described method (14). Both isolates had 100% sequence identity to the sequence of S. intermedius strain MAFF 911388 (GenBank accession no. D83369; ATCC 29663T) (487 of 487 bp sequenced for isolate 1 and 515 of 515 bp sequenced for isolate 2).
Methicillin susceptibility testing
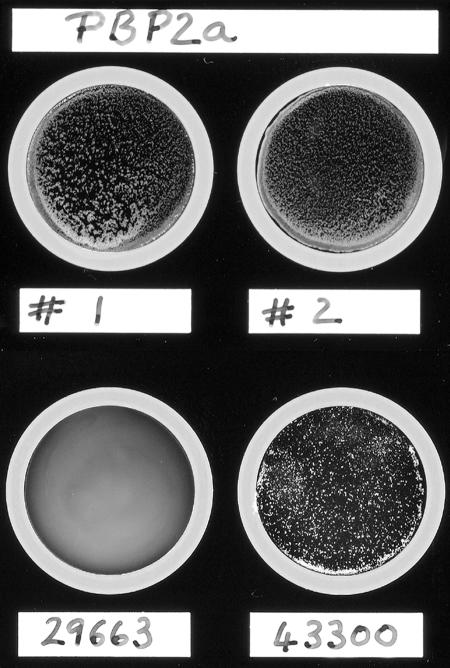
Oxacillin (methicillin) susceptibility testing of isolates 1 and 2 was performed by the broth macrodilution method, oxacillin disk diffusion method, and oxacillin-salt screening agar in accordance with NCCLS standards as well as by the Vitek method (12, 13). The S. intermedius ATCC 29663 strain was also tested for comparison. All three isolates, the two clinical strains and the ATCC strain, were oxacillin susceptible by both MIC and disk diffusion methods using NCCLS criteria. There was no interpretative difference noted when either the S. aureus or the coagulase-negative staphylococcus criteria were used for either the MIC or disk diffusion methods (12, 13). All three isolates failed to grow on the oxacillin-salt screening agar. Repeat PBP2a testing of the two clinical isolates duplicated the initial results, with a strong positive agglutination for isolate 1 and a weak positive agglutination for isolate 2 within the recommended 3-min interval. No difference in the PBP2a result was noted after induction of isolates 1 and 2 with oxacillin. S. intermedius ATCC 29663 was PBP2a negative, and S. aureus ATCC 43300 (oxacillin resistant) was PBP2a positive (Fig. 1; Table 1). For all four strains, no agglutination was seen with the control latex.
FIG. 1.
Results of a PBP2a latex agglutination test. A PBP2a latex agglutination test was performed and interpreted according to the manufacturer's guidelines. Shown are clinical isolates 1 (#1) and 2 (#2), where isolate 1 had a strong positive reaction and isolate 2 had a weak positive reaction. The lower disks are of oxacillin-resistant S. aureus ATCC 43300 (43300) showing a strong positive reaction and S. intermedius ATCC 29663 (29663) showing a negative reaction.
TABLE 1.
Summary of methods used to detect methicillin susceptibility in two clinical isolates of S. intermedius
| Isolate tested | Detection of methicillin resistance
|
|||||
|---|---|---|---|---|---|---|
| Oxacillin broth macro- dilution MIC (μg/ml) | Oxacillin disk diffusion (mm) | Oxacillin screen agar | Vitek GPS 107 card MIC (μg/ml) | PBP2a latex agglutination testa | mecA PCRb | |
| Isolate 1 | 0.125 | 24 | Negative | <0.25 | Strong positive | Negative |
| Isolate 2 | 0.125 | 27 | Negative | <0.25 | Weak positive | Negative |
| S. intermedius ATCC 29663 | 0.125 | 27 | Negative | <0.25 | Negative | Negative |
| S. aureus ATCC 43300 | —c | —c | Positive | —c | Strong positive | Positive |
PBP2a latex agglutination test results were interpreted in accordance with the manufacturers instructions (Denka Seiken Ltd.).
mecA PCR performed using two primer sets amplifying 188-bp internal and 533-bp flanking fragments of the mecA gene (4, 5).
—, quality control organisms as recommended were used as controls. For oxacillin broth macrodilution MIC and Vitek GPS 107 card, S. aureus ATCC 29213 was used; for oxacillin disk diffusion, S. aureus ATCC 25923 was used.
mecA gene PCR
A 188-bp fragment within the mecA gene and a 178-bp fragment within the S. aureus-specific Sa442 gene were amplified for isolates 1 and 2 in separate reactions on a LightCycler (Roche Diagnostics GmbH, Roche Molecular Biochemicals, Mannheim, Germany) using SYBR green with primers Mec-S and Mec-A or Sa442-F and Sa442-RS, respectively, as previously described (5). mecA-positive isolates generate SYBR green melting temperatures of 79°C. Sa442-positive isolates also generate a SYBR green melting temperature of 79°C. S. aureus ATCC 29213 (oxacillin susceptible) and S. aureus ATCC 43300 (oxacillin resistant) were used as controls for the Sa442 gene and as negative and positive controls, respectively, for the mecA gene. In contrast to the controls, no mecA or Sa442 product was generated for isolates 1 and 2. An additional PCR was performed to detect a larger 533-bp fragment of the mecA gene (4). No product was generated in isolates 1 and 2 or S. aureus ATCC 29213, but a product of the expected size was generated from S. aureus ATCC 43300. The 16S rRNA gene, used as a positive control, was amplified from all strains (data not shown).
Accurate and timely detection of methicillin resistance in S. aureus is an important function of the clinical microbiology laboratory (20). While detection of the mecA gene by PCR is the gold standard, the PBP2a latex agglutination test has been shown to be a simple and rapid test, with good performance characteristics to detect methicillin resistance in both S. aureus and coagulase-negative staphylococcal species, notwithstanding the need for prior induction in some cases (1, 21). The PBP2a assay uses latex particles sensitized with monoclonal antibodies raised against PBP2a (11). The specificity of this test reportedly approaches 100% (18, 19). Although there are no reports regarding the performance of PBP2a latex agglutination tests for S. intermedius isolates, false-positive PBP2a results have been noted with two S. warneri isolates, one at 1 min and the other at 6 min, which were both mecA PCR negative with an oxacillin MIC of ≤0.5 μg/ml (21). According to the manufacturer's package insert, false-positive reactions are generally weak agglutinations that are often negative on repeat testing with a fresh culture. However, we found that our two isolates were repeatedly positive after multiple subcultures, with one being strongly positive. The S. intermedius ATCC 29663 strain was, in fact, negative by PBP2a test but was also phenotypically different from the two clinical strains with regard to positive mannitol fermentation, β-lactamase negativity, and penicillin sensitivity (results not shown).
Coagulase positivity is commonly used to attribute clinical significance and pathogenicity to isolates of Staphylococcus spp. While S. aureus is the most common coagulase-positive staphylococcus isolated in the clinical laboratory, S. intermedius, S. delphini, S. schleiferi subsp. coagulans, S. lutrae, and some strains of S. hyicus are also coagulase positive (1). Laboratories often use a combination of tests to detect free coagulase or clumping factor with and without protein A to identify coagulase-positive staphylococci. S. intermedius isolates are positive for free coagulase but are negative for protein A, and 14% of the isolates have a clumping factor, which would account for the positive slide coagulase and weak positive BACTi Staph latex for isolate 1 (6). As described previously, we found pyrrolidonyl arylamidase positivity to be a rapid way to determine that a coagulase-positive staphylococcus is not S. aureus (10). We believe that our cases suggest that the true incidence of S. intermedius in human wound infections is probably underestimated, because all coagulase-positive staphylococci are often lumped together as S. aureus (17). In the absence of definitive identification of S. intermedius by phenotypic or biochemical methods, sequencing of the 16S rRNA gene has been found to be useful for taxonomic classification of Staphylococcus and Macrococcus species (8).
An initial study in 1989 showed 72% of S. intermedius isolates from canine gingiva and canine-inflicted wound infections were susceptible to penicillin, and none were resistant to oxacillin (16). A more recent study revealed that oxacillin resistance is an increasing problem in S. intermedius isolates, with 60 to 85% greater oxacillin resistance rates noted for isolates from nose, eyes, and abscesses of dogs compared to those from the other sites (7). In the first human infection due to methicillin-resistant S. intermedius, where it was the causative agent of pneumonia, the oxacillin MIC was 32 μg/ml (3). Both of our isolates were resistant to penicillin and were β-lactamase positive; the oxacillin MIC for these isolates was 0.125 μg/ml. The penicillin resistance in these isolates, in the absence of oxacillin resistance with a negative mecA PCR and susceptibility to β-lactam/β-lactamase inhibitor combinations, can be explained by the positive finding of the β-lactamase assay. Using the primer set targeting a 533-bp fragment of the mecA gene, Gortel et al. confirmed that the mecA gene confers methicillin resistance in staphylococci isolated from dogs (4). These investigators found that among 10 coagulase-positive staphylococcal strains carrying mecA, 9 were S. aureus and 1 was S. intermedius. They hypothesized that the difference in prevalence of methicillin resistance in S. intermedius compared to that of S. aureus was due to a difference in mecA regulation between the species or the lack of intense antibiotic selection pressure on S. intermedius (4). While no specific NCCLS interpretative criteria exist for coagulase-positive staphylococci other than S. aureus, the results of oxacillin susceptibility testing by phenotypic and genotypic methods (mecA gene PCR using two primer sets targeting 188- and 533-bp fragments), combined with low oxacillin MICs compared to those previously reported for oxacillin-resistant S. intermedius isolates (MIC > 4 μg/ml), establishes isolates 1 and 2 in this study to be oxacillin sensitive (3, 4).
It is well accepted that methicillin resistance in S. aureus is mainly due to the acquisition of an additional penicillin-binding protein, PBP2a, encoded by the mecA gene. A search for the origin of the mecA gene led to the identification of a possible evolutionary precursor in S. sciuri, a commensal of animals. The mecA homologue in S. sciuri showed 79.5% DNA sequence similarity to the mecA gene of S. aureus and 88% amino acid identity with PBP2a. The S. sciuri mecA homologue does not confer resistance to methicillin in nature. However, Couto et. al identified methicillin-resistant S. sciuri isolates from humans, where overexpression of the mecA homologue resulting from insertion of an IS256 element upstream of the structural gene or single-nucleotide alterations in the promoter region resulted in the production of a protein functionally resembling PBP2a (2). Similar to the S. sciuri isolates, the S. intermedius isolates described here may contain a mecA homologue encoding a PBP2a-like protein, which cross-reacts with the PBP2a latex agglutination test but does not confer methicillin resistance. Antigenic mimicry with a completely unrelated protein is also possible. In either case, we hope to raise awareness in the microbiological community that clinical isolates exist that may challenge the specificity of the PBP2a latex agglutination test.
In conclusion, we provide the first report of false-positive PBP2a results for two strains of S. intermedius which, combined with an initial error in the interpretation of phenotypic tests, led to misidentification as MRSA. False-positive PBP2a results could lead to misdirected infection control attempts, unnecessary use of antibiotics like vancomycin, and an overall increase in hospital expenses (20). These cases underscore the importance of actively pursuing discrepant laboratory test results to avoid reporting errors. Accurate species identification of the coagulase-positive staphylococci is essential to determine the pathogenicity, clinical significance, susceptibility patterns, and epidemiology of the clinical isolates.
ADDENDUM
Since submission of this manuscript, we have recovered two additional staphylococcal isolates from unrelated patients that were also initially mistaken for S. aureus due to their phenotypic characteristics and weak positive PBP2a latex agglutination tests. The oxacillin MIC for these isolates was 0.25 μg/ml, with a 25-mm-diameter zone of inhibition for the oxacillin disk, and their mecA PCR was negative. 16S rRNA gene sequences definitively identified the isolates as S. intermedius.
Acknowledgments
We thank Rajyashree Gupta and Monica M. Fujii for their excellent technical assistance.
REFERENCES
- 1.Bannerman, T. L. 2003. Staphylococcus, Micrococcus, and other catalase-positive cocci that grow aerobically, p. 384-404. In P. R. Murray, E. J. Baron, M. A. Pfaller, J. H. Jorgensen, and R. H. Yolken (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, D.C.
- 2.Couto, I., S. W. Wu, A. Tomasz, and H. de Lencastre. 2003. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. J. Bacteriol. 185:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerstadt, K., J. S. Daly, M. Mitchell, M. Wessolossky, and S. H. Cheeseman. 1999. Methicillin-resistant Staphylococcus intermedius pneumonia following coronary artery bypass grafting. Clin. Infect. Dis. 29:218-219. [DOI] [PubMed] [Google Scholar]
- 4.Gortel, K., K. L. Campbell, I. Kakoma, T. Whittem, D. J. Schaeffer, and R. M. Weisiger. 1999. Methicillin resistance among staphylococci isolated from dogs. Am. J. Vet. Res. 60:1526-1530. [PubMed] [Google Scholar]
- 5.Grisold, A. J., E. Leitner, G. Mühlbauer, E. Marth, and H. H. Kessler. 2002. Detection of methicillin-resistant Staphylococcus aureus and simultaneous confirmation by automated nucleic acid extraction and real-time PCR. J. Clin. Microbiol. 40:2392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hájek, V. 1976. Staphylococcus intermedius, a new species isolated from animals. Int. J. Syst. Bacteriol. 26:401-408. [Google Scholar]
- 7.Hoekstra, K. A., and R. J. L. Paulton. 2002. Clinical prevalence and antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus intermedius in dogs. J. Appl. Microbiol. 93:406-413. [DOI] [PubMed] [Google Scholar]
- 8.Kwok, A. Y. C., and A. W. Chow. 2003. Phylogenetic study of Staphylococcus and Macrococcus species based on partial hsp60 gene sequences. Int. J. Syst. Bacteriol. 53:87-92. [DOI] [PubMed] [Google Scholar]
- 9.Lee, J. 1994. Staphylococcus intermedius isolated from dog-bite wounds. J. Infect. 29:105. [DOI] [PubMed] [Google Scholar]
- 10.Mahoudeau, I., X. Delabranche, G. Prevost, H. Monteil, and Y. Piemont. 1997. Frequency of isolation of Staphylococcus intermedius from humans. J. Clin. Microbiol. 35:2153-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatomi, Y., and J. Sugiyama. 1998. A rapid latex agglutination assay for the detection of penicillin-binding protein 2′. Microbiol. Immunol. 42:739-743. [DOI] [PubMed] [Google Scholar]
- 12.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 6th ed. NCCLS document M7-A6. NCCLS, Wayne, Pa.
- 13.NCCLS. 2003. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 8th ed. NCCLS document M2-A8. NCCLS, Wayne, Pa.
- 14.Pottumarthy, S., A. P. Limaye, J. L. Prentice, Y. B. Houze, S. R. Swanzy, and B. T. Cookson. 2003. Nocardia veterana, a new emerging pathogen. J. Clin. Microbiol. 41:1705-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raus, J., and D. N. Love. 1983. Characterization of coagulase-positive Staphylococcus intermedius and Staphylococcus aureus isolated from veterinary clinical specimens. J. Clin. Microbiol. 18:789-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talan, D. A., D. Staatz, A. Staatz, E. J. C. Goldstein, K. Singer, and G. D. Overturf. 1989. Staphylococcus intermedius in canine gingival and canine-inflicted human wound infections: laboratory characterization of a newly recognized zoonotic pathogen. J. Clin. Microbiol. 27:78-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandenesch, F., M. Célard, D. Arpin, M. Bes, T. Greenland, and J. Etienne. 1995. Catheter-related bacteremia associated with coagulase-positive Staphylococcus intermedius. J. Clin. Microbiol. 33:2508-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Griethuysen, A., M. Pouw, N, van Leeuwen, M. Heck, P. Willemse, A. Buiting, and J. Kluytmans. 1999. Rapid slide latex agglutination test for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 37:2789-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Leeuwen, W. B., C. van Pelt, A. Luijendijk, H. A. Verbrugh, and W. H. Goessens. 1999. Rapid detection of methicillin resistance in Staphylococcus aureus isolates by the MRSA-screen latex agglutination test. J. Clin. Microbiol. 37:3029-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Versalovic, J. 2003. Is real time detection of drug-resistant Staphylococcus aureus worth considering? Arch. Pathol. Lab. Med. 127:784-785. [DOI] [PubMed] [Google Scholar]
- 21.Yamazumi, T., I. Furuta, D. J. Diekema, M. A. Pfaller, and R. N. Jones. 2001. Comparison of the Vitek gram-positive susceptibility 106 card, the MRSA-screen latex agglutination test, and mecA analysis for detecting oxacillin resistance in geographically diverse collection of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 39:3633-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazumi, T., S. A. Marshall, W. W. Wilke, D. J. Diekema, M. A. Pfaller, and R. N. Jones. 2001. Comparison of the Vitek gram-positive susceptibility 106 card and the MRSA-screen latex agglutination test for determining oxacillin resistance in clinical bloodstream isolates of Staphylococcus aureus. J. Clin. Microbiol. 39:53-56. [DOI] [PMC free article] [PubMed] [Google Scholar]