Abstract
Because of their biofilm-forming capacity, invasive Staphylococcus epidermidis isolates, which cause the majority of nosocomial catheter-related bloodstream infections (BSIs), are thought to be selected at the time of catheter insertion from a population of less virulent commensal strains. This fact allows the prediction that invasive and contaminating strains can be differentiated via detection of virulence-associated genes. However, the hospital environment may pave the way for catheter-related infections by promoting a shift in the commensal bacterial population toward strains with enhanced virulence. The distribution of virulence-associated genes (icaADBC, aap, atlE, bhp, fbe, embp, mecA, IS256, and IS257), polysaccharide intercellular adhesin synthesis, and biofilm formation were investigated in S. epidermidis strains from independent episodes of catheter-related BSIs in individuals who have received bone marrow transplantation (BMT). The results were compared with those obtained for commensal S. epidermidis isolates from hospitalized patients after BMT and from healthy individuals, respectively. The clonal relationships of the strains were investigated by pulsed-field gel electrophoresis. icaADBC, mecA, and IS256 were significantly more prevalent in BSI isolates than in commensal isolates from healthy individuals. However, the prevalence of any of the genes in clonally independent, endogenous commensal strains from BMT patients did not differ from that in invasive BSI strains. icaADBC and methicillin resistance, factors important for the establishment of catheter-related infections, already ensure survival of the organisms in their physiological habitat in the hospital environment, resulting in a higher probability of contamination of indwelling medical devices with virulent S. epidermidis strains. The dynamics of S. epidermidis populations reveal that detection of icaADBC and mecA is not suitable for discriminating invasive from contaminating S. epidermidis strains.
Nosocomial bloodstream infections (BSIs) are a common and severe complication after bone marrow transplantation (BMT) (19). Coagulase-negative staphylococci, especially Staphylococcus epidermidis, which is a normal commensal organism of the skin, are encountered most frequently (19, 27). S. epidermidis BSIs typically occur in the context of implanted central venous, Hickman, or Quinton catheters. The reason for this tight pathogenic relationship is the ability of S. epidermidis to establish foreign body-adherent biofilms, which consist of multilayered bacterial communities stabilized by intercellular adhesive mechanisms (2, 8, 20). S. epidermidis biofilm formation depends on the production of the polysaccharide intercellular adhesin (PIA), synthesized by icaADBC-encoded proteins (7, 10). The important roles of icaADBC, PIA synthesis, and biofilm formation in the pathogenesis of a S. epidermidis biomaterial-related infections were documented in animal models (28, 29). icaADBC expression is controlled by the alternative sigma factor SigB (14, 16), other regulatory elements (17), and insertion sequences IS256 and IS257 (26, 35). It is assumed at present that invasive icaADBC-positive strains are selected at the time of foreign body implantation from a population of essentially icaADBC- and biofilm-negative commensal strains which are thought to be the prototype avirulent S. epidermidis subpopulation (33) due to their biofilm-forming capacity. This model is based on the observation of a higher prevalence of icaADBC in invasive strains than in commensal strains from healthy individuals (5, 6, 20). In addition, mecA, which mediates methicillin resistance, has been proposed as a marker for discriminating between invasive and contaminating strains (5). Other factors involved in foreign body colonization, like the autolysin AtlE, the fibrinogen binding protein Fbe, and the accumulation-associated protein Aap, displayed no differential distribution in invasive and commensal S. epidermidis isolates (5, 6, 31). However, in those studies data for invasive strains were compared with data for commensal strains from healthy individuals with no contact with the hospital environment. Therefore, as S. epidermidis infections are typically endogenous in character (20), the S. epidermidis populations from which invasive strains are recruited, i.e., the commensal strains of hospitalized patients, were neglected, until now. Consequently, the aim of the present study was to investigate the ecology of invasive and commensal S. epidermidis populations in patients after BMT by analyzing the distribution of icaADBC as well as those of atlE (9), fbe (25), and aap (13), all of which represent additional factors involved in the pathogenesis of S. epidermidis. Furthermore, the presence of the newly identified S. epidermidis homologue of the biofilm-associated protein Bap (called bhp) from Staphylococcus aureus (3) and the fibronectin binding protein Embp (32) and the presence of insertion sequences IS256 and IS257 were characterized. Both populations of hospital isolates were compared with commensal S. epidermidis isolates from healthy individuals with no contact with the hospital environment in order to tag the genetic virulence markers relevant in this specific clinical context and to characterize the course of selection events leading to a shift in the genetic population structure.
MATERIALS AND METHODS
Bacterial strains.
All S. epidermidis isolates were identified as S. epidermidis by negativity for the clumping factor and with the ID32Staph system (bioMerieux, Marcy l'Etoile, France). Reference strains S. epidermidis 1457 and biofilm-negative transposon mutant 1457-M10 have been described previously (21). The S. epidermidis strains were grouped according to their isolation site.
(i) Invasive strains.
Forty-one isolates were recovered from cultures of blood from 16 patients of the University Hospital Hamburg-Eppendorf after BMT (two to five isolates per patient recovered 2 to 72 days after BMT) hospitalized during 1998 and 2003. All patients (eight males; eight females; average age, 39 years; age range, 2 to 69 years) were under immune suppression and received calculated therapy (quinolones, metronidazole, vancomycin, ceftazidime, trimethoprim-sulfamethoxazole, and tobramycin; in addition, all patients received fluconazole or amphotericin B as well as acyclovir and pentamidine) when blood was drawn for culture. These S. epidermidis strains were regarded as invasive strains, as at least two clonally identical or closely related isolates (30) were obtained from cultures of blood drawn at different time points or from independent cultures of blood drawn at one time per patient. In addition, all patients exhibited systemic signs of infection (fever and elevated C-reactive protein levels), and 15 of 16 patients with a vascular access device in situ showed local inflammatory signs at the catheter insertion site.
(ii) Commensal strains from patients after BMT.
Twenty-five clonally independent S. epidermidis isolates were recovered from September 2002 to February 2003 from nose, mucous membrane, and skin swabs taken from 17 patients after BMT. All patients received prophylaxis (quinolones, fluconazole, trimethoprim-sulfamethoxazole, and acyclovir) but did not exhibit signs of infection at the time of sampling.
(iii) Commensal strains from healthy individuals.
Fifteen clonally independent S. epidermidis isolates from the noses of 15 healthy volunteers were investigated.
PFGE.
Pulsed-field gel electrophoresis (PFGE) of SmaI-cleaved DNA was carried out as described previously (22) with a CHEF-DR II system (Bio-Rad, Munich, Germany).
In vitro biofilm formation and detection of PIA synthesis.
S. epidermidis biofilm formation on polymeric surfaces was tested by the semiquantitative microtiter plate test (biofilm assay) with Trypticase soy broth (Becton Dickinson, Cockeysville, Md.) and Trypticase soy broth supplemented with 2% (wt/vol) NaCl, as described previously (15, 21). To qualitatively and semiquantitatively detect PIA synthesis, a coagglutination assay (21) using a PIA-specific antiserum and an antiserum specific for a variant PIA (PIAv) antisera was performed essentially as described previously (15, 21).
Detection of icaADBC, aap, bhp, fbe, mecA, embp, atlE, IS256, and IS257 and determination of aap repeat polymorphism.
DNA preparation and PCR were performed as described previously (26). The primers and PCR conditions are summarized in Table 1. The aap repeat region (Fig. 1) was amplified by use of the Expand Long Template PCR system (Roche, Mannheim, Germany) under the conditions recommended by the manufacturer with a forward primer (5′-GAT TTA GAT GGT GCA ACA TTG ACA T-3′) located at the 5′ end and a reverse primer (5′-TTG ACG ATT TTC ACC TGT ATC AGG T-3′) located at the 3′ end. Gel-Pro Analyzer (version 3.1) software (Media Cybernetics, Silver Spring, Md.) was used to estimate the PCR fragment sizes.
TABLE 1.
Distribution of virulence-associated genes in different S. epidermidis populations
| Genea | Prevalence (no. [%] of strains)b
|
Primers | Reference or conditions | ||
|---|---|---|---|---|---|
| Invasive strains (n = 16) | Commensal strains, BMT patients (n = 25) | Commensal strains, healthy subjects (n = 15) | |||
| icaA (U43336)c | 15 (93.8) | 20 (80) | 2 (13) | Forward, 5′-TCG ATG CGA TTT GTT CAA ACA T-3′; reverse, 5′-CTG TTT CAT GGA AAC TCC-3′ | 15 |
| aap (AJ249487)d | 15 (93.8) | 23 (92) | 13 (86.7) | Forward, 5′-AAA CGG TGG TAT CTT ACG TGA A-3′; reverse, 5′-CAA TGT TGC ACC ATC TAA ATC AGC T-3′ | 5 min, 94°C; 30 cycles of 30 s, 94°C; 30 s, 60°C; 50 s, 72°C; final extension, 4 min, 72°C |
| bhp (AY028618)e | 3 (18.8) | 4 (16) | 4 (26.7) | Forward, 5′-ATG GTA TTA GCA AGC TCT CAG CTG G-3′; reverse, 5′-AGG GTT TCC ATC TGG ATC CG-3′ | 5 min, 94°C; 30 cycles of 30 s, 94°C; 30 s, 61°C; 90 s, 72°C; final extension, 4 min, 72°C |
| fbe (Y17116)f | 16 (100) | 25 (100) | 15 (100) | Forward, 5′-CTA CAA GTT CAG GTC AAG GAC AAG G-3′; reverse, 5′-GCG TCG GCG TAT ATC CTT CAG-3′ | 5 min, 94°C; 30 cycles of 30 s, 94°C; 30 s, 60°C; 50 s, 72°C; final extension, 4 min, 72°C |
| embp (AY101364)g | 14 (87.5) | 19 (76) | 15 (100) | Forward, 5′-AGC GGT ACA AAT GTC AAT-3′; reverse, 5′-AGA AGT GCT CTAG CAT CAT CC-3′ | 32 |
| atlE (U71377)f | 16 (100) | 25 (100) | 15 (100) | Forward, 5′-CAA CTG CTC AAC CGA GAA CA-3′; reverse, 5′-TTT GTA GAT GTT GTG CCC CA-3′ | 5 min, 94°C; 30 cycles of 30 s, 94°C; 30 s, 55°C; 30 s, 72°C; final extension, 4 min, 72°C |
| mecA (X52592)h | 14 (87.5) | 24 (96) | 1 (6.7) | Forward, 5′-GAA ATG ACT GAA CGT CCG AT-3′; reverse, 5′-GCG ATC AAT GTT ACC GTA GT-3′ | 5 min, 94°C; 30 cycles of 30 s, 94°C; 30 s, 55°C; 30 s, 72°C; final extension, 4 min, 72°C |
| IS256 (AF051917)i | 15 (93.8) | 19 (76) | 0 | Forward, 5′-TGA AAA GCG AAG AGA TTC AAA GC-3′; reverse, 5′-ATG TAG GTC CAT AAG AAC GGC-3′ | 35 |
| IS257 (U40381)f | 16 (100) | 25 (100) | 15 (100) | Forward, 5′-ACG TTC ATC ATT CAA CGG TC-3′; reverse, 5′-AGT GTT CGC TTA ACT TGC TAG-3′ | 5 min, 94°C; 30 cycles of 30 s, 94°C; 30 s, 55°C; 30 s; 72°C; final extension, 4 min, 72°C |
GenBank accession numbers are given in parentheses.
icaADBC, mecA, and IS256 were significantly more prevalent in invasive strains and commensals from BMT patients than in commensals from healthy individuals (Yates' corrected chi-square test), whereas all other differences were below statistical significance.
P = 0.44 for invasive strains versus commensal strains from BMT patients; P = 0.0002 for commensal strains from BMT patients versus commensal strains from healthy subjects; P < 0.0001 for comparison of all three groups of strains.
P = 0.68 for invasive strains versus commensal strains from BMT patients; P = 1.0 for commensal strains from BMT patients versus commensal strains from healthy subjects; P = 0.95 for comparison of all three groups of strains.
P = 0.84 for invasive strains versus commensal strains from BMT patients; P = 0.68 for commensal strains from BMT patients versus commensal strains from healthy subjects; P = 0.92 for comparison of all three groups of strains.
Statistical analysis is not applicable.
P = 0.61 for invasive strains versus commensal strains from BMT patients; P = 0.1 for commensal strains from BMT patients versus commensal strains from healthy subjects; P = 0.49 for comparison of all three groups of strains.
P = 0.68 for invasive strains versus commensal strains from BMT patients; P < 0.0001 for commensal strains from BMT patients versus commensal strains from healthy subjects; P < 0.0001 for comparison of all three groups of strains.
P = 0.29 for invasive strains versus commensal strains from BMT patients; P < 0.0001 for commensal strains from BMT patients versus commensal strains from healthy subjects; P < 0.0001 for comparison of all three groups of strains.
FIG. 1.
Schematic representation of Aap from S. epidermidis 5179 as deduced from the aap nucleotide sequence (GenBank accession number AY359815). Arrows indicate the positions of the primers used for amplification of domain B (For, forward; Rev, reverse). E, export signal; A, 564-aa domain; B, domain consisting of five (B1 to B5) complete 128-aa repeat units and one (Bp) 68-aa partial repeat unit; C, collagen triple-helix motifs; L, LPXTG motif containing cell wall anchor.
RESULTS
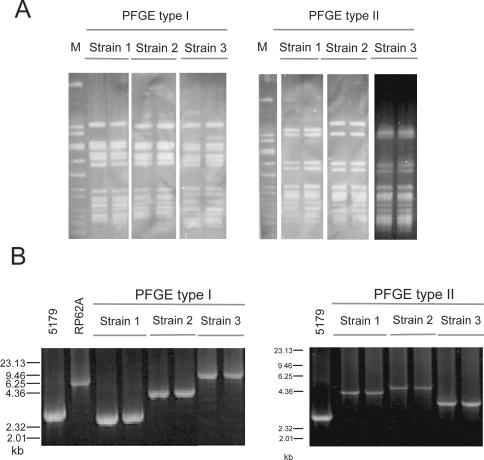
In order to rule out the possibility of infections due to one endemic S. epidermidis strain, all isolates were typed by PFGE. Comparison of the restriction patterns of strains from different BSI episodes revealed that 10 of 16 BSI episodes were due to clonally unrelated S. epidermidis strains. The remaining six BSI episodes were attributable to S. epidermidis strains belonging to two PFGE types (three strains each of PFGE types I and II; Fig. 2A). PFGE patterns that displayed two to three fragment differences were classified as different types. These clonally related strains were isolated over a period of 5 years and could be differentiated by further subtyping (Fig. 2B). Thus, all BSIs investigated were caused by distinct S. epidermidis strains not representing derivatives of an endemic strain.
FIG. 2.
(A) PFGE analysis of SmaI-digested DNA from pairs of strains isolated from three distinct BSI episodes. After separation the bands were made visible by ethidium bromide staining. Isolates of PFGE type I were obtained in 1999, 2001, and 2002; and isolates of PFGE type II were obtained in 1998, 1999, and 2001. Despite the long time interval, the strains were clonally identical according to the criteria of Tenover et al. (30) and were therefore designated PFGE type I and type II, respectively. M, marker (SmaI-digested chromosomal DNA of S. aureus NCTC 8325). (B) PCR analysis of repetitive aap domain B in clonally related PFGE type I and II S. epidermidis strains. The resulting amplicons were separated on a 0.7% agarose gel and stained with ethidium bromide. S. epidermidis 5179, which carries 5 complete repeats and 1 incomplete repeat (GenBank accession number AY359815) and RP62A (http//:www.tigr.org), which carries 12 repeats, served as controls.
The commensal S. epidermidis strains isolated from patients after BMT all displayed unique PFGE patterns and were not related to any of the invasive strains according to their PFGE types (data not shown), demonstrating that during the collection period the patients were not colonized with a single dominant S. epidermidis clone. The S. epidermidis strains isolated from healthy volunteers were all clonally independent from the other strains investigated.
icaADBC, aap, fbe, embp, atlE, mecA, IS256, and IS257 were all highly prevalent in the invasive strains, whereas bhp was present in only a minority of these isolates (Table 1). There were no differences in the distribution of genes within PFGE genotypes I and II. The gene set was identical in subsequent isolates recovered during a given BSI episode. A strain became oxacillin sensitive due to the loss of mecA only in a single episode. Twelve of 15 icaADBC-positive BSI strains expressed PIA (n = 11) or PIAv (n = 1), as detected by a specific coagglutination assay (15), and produced a biofilm in the standard biofilm assay (26) when they were grown either in TSB or in TSB supplemented with 2% NaCl. The biofilm phenotype was remarkably unstable, with subsequently cultured isolates of seven strains exhibiting differential biofilm expression. In two episodes the biofilm-negative phenotype of those phase variants was attributable to a lack of PIA synthesis. As no large structural alterations of icaADBC were found by PCR analysis (data not shown), these phase variations must rely on mechanisms other than the insertional inactivation of icaADBC by insertion elements (26, 35). PIA or PIAv synthesis was still detected in biofilm-negative phase variants of the remaining five strains, suggesting quantitative differences in PIA synthesis or alterations of the structures mediating the primary attachment. The detection of phase variants might reflect the dispersal of S. epidermidis biofilms by switching off PIA synthesis. All icaADBC-negative strains were biofilm negative.
In contrast to the invasive strains, the commensal S. epidermidis strains isolated from healthy individuals almost completely lacked icaADBC, mecA, and IS256 (Table 1). All other genes in the commensal strains displayed a distribution comparable to that in the invasive strains (Table 1). The 2 icaADBC-positive strains were biofilm positive, whereas the 13 icaADBC-negative strains did not produce a biofilm under any of the conditions tested.
Analysis of the commensal S. epidermidis strains isolated from hospitalized BMT recipients revealed that the prevalence of the genes investigated was very similar to that in the invasive BSI strains (Table 1). In particular, icaADBC, mecA, and IS256 were detected in commensal strains nearly as often as they were detected in invasive strains. Only the proportion of icaADBC-positive but biofilm-negative strains was higher in the commensal population than that in the invasive population (44 versus 20%, respectively).
The observed genotypic differences between invasive and commensal strains from healthy individuals would suggest that an inserted intravascular catheter selects for icaADBC-, mecA-, and IS256-positive strains, which are particularly virulent, making those gene loci suitable markers for invasive strains in BSIs after BMT. However, the data for the commensal strains isolated from BMT patients show that the hospital environment already exerts selective pressure on the commensal S. epidermidis population, leading to a high prevalence of icaADBC-, mecA-, and IS256-positive strains, making a differentiation between invasive and colonizing strains on the basis of the detection of these genes impossible.
A difference between invasive BSI strains and commensal S. epidermidis strains was found by detection of a length polymorphism of domain B of aap (Fig. 1; Fig. 2B), which probably resulted from variations in the number of repeats (Fig. 1). In commensal strains from healthy individuals as well as from BMT patients, an amplicon size larger than 4.2 kb, which comprised about 11 copies of the 128-amino-acid (aa) (384-bp) repeat, was found significantly more often than in the invasive strains (Fig. 3). The size of the repeat domain was stable for subsequent isolates of a strain isolated from a given BSI episode. Interestingly, the sizes of the aap repeat domains of strains of PFGE types I and II differed (Fig. 2B), allowing further subtyping.
FIG. 3.
Distribution of polymorphic aap domain B sizes in BSI strains, commensal strains from BMT patients, and commensal strains from healthy individuals. The frequency of a domain of ≤4,200 bp with 11 deduced repeats was significantly higher in BSI strains than in commensal isolates (P = 0.02; Yates' corrected chi-square test). The difference between BSI strains and commensal isolates from BMT patients was also statistically significant (P = 0.03; Yates' corrected chi-square test). The difference between BSI strains and commensal isolates from healthy individuals was near statistical significance (P = 0.06; Fisher exact test).
DISCUSSION
Biofilm formation plays the key role in S. epidermidis foreign body colonization and infection (20). PIA, synthesized by icaADBC-encoded proteins, is essential in this process. Until now, studies investigating the distribution of virulence factors in different S. epidermidis populations found a much lower prevalence of icaADBC in commensal S. epidermidis strains than in invasive strains, whereas other genes, like fbe, atlE, and aap, were all widely distributed in both populations (5, 6, 31, 34). Therefore, according to the present understanding of the pathogenesis of foreign body infections, icaADBC-positive strains, which usually represent only a small part of the mainly icaADBC- and biofilm-negative commensal S. epidermidis population, are preferentially selected at the time of foreign body insertion and colonization due to their biofilm-forming capacity. icaADBC-negative strains are therefore regarded as an avirulent S. epidermidis subpopulation (33). In conclusion, detection of icaADBC was suggested as a tool for discriminating invasive from contaminating strains in clinical specimens (5, 6). However, a general problem is the analysis of commensal strains from healthy individuals without contact with the hospital environment. As invasive S. epidermidis strains originate from the patient's own skin flora (20), which is subject to severe changes as a result of selective pressure exerted by the hospital environment (1, 15), this S. epidermidis population is unsuitable as a control group. This notion led us to compare BSI S. epidermidis isolates with skin isolates from hospitalized BMT patients. Importantly, the commensal strains from BMT recipients investigated in this study were clonally unrelated, indicating that they were not hospital-acquired, endemic S. epidermidis isolates but represented an endogenous population selected by the hospital environment. Indeed, by using this approach, no differences in the distributions of any of the genes investigated were found between invasive BSI S. epidermidis strains and commensal strains isolated from hospitalized BMT patients.
In contrast, icaADBC, mecA, and IS256 were significantly less prevalent in commensal strains from healthy individuals with no contact with the hospital environment. The significance of IS256 as a marker of S. epidermidis of hospital origin has recently been highlighted (18). This indicates that hospitalization on a BMT unit represents a selection step that preferentially allows expansion of icaADBC-, mecA-, and IS256-positive S. epidermidis strains, a hypothesis that is supported by the finding that commensal S. epidermidis strains isolated from patients hospitalized on a surgical intensive care unit displayed a similar high prevalence of icaADBC, mecA, and IS256 (H. Rohde, M. Kalitzky, and D. Mack, unpublished results). Additionally, de Silva et al. (4) also failed to find a difference in rates of icaADBC detection between S. epidermidis strains from neonates with BSIs and commensal strains from babies hospitalized in the same intensive care unit. Consequently, the detection of icaADBC and mecA appears to be by no means suitable for discriminating invasive and colonizing strains. Furthermore, identification of distinct factors promoting the high prevalence of icaADBC- and mecA-positive strains in various clinical settings is demanded in order to develop preventive strategies.
The selective pressure exerted by antibiotics reasonably explains the shift toward a mecA-positive S. epidermidis population (1, 11). The preferential detection of icaADBC- and biofilm-positive S. epidermidis strains can be explained by the linkage of oxacillin resistance expression, biofilm formation, and icaADBC transcription (23, 24) Furthermore, S. epidermidis isolates organized in a biofilm are naturally more resistant to antibiotics and alcoholic skin disinfectants (15, 23). Therefore, measures intended to prevent infections on a BMT unit would directly pave the way for hard-to-treat biofilm-related S. epidermidis foreign body infections caused by methicillin-resistant strains. With respect to catheter-related BSIs, our findings appear to be a plausible molecular rationale for Infectious Diseases Society of America guidelines recommending the restricted use of antibiotic prophylaxis in afebrile neutropenic patients (12).
It still remains an important question whether all S. epidermidis strains selected by the hospital environment finally have the ability to cause foreign body infections or if a distinct subpopulation carrying specific determinants exists. Evidence for further changes in the population profile comes from the detection of aap repeat length polymorphisms within BSI and commensal strains. In addition, the majority of invasive icaADBC-positive strains were also biofilm positive, whereas most commensal S. epidermidis isolates from BMT patients were biofilm negative, despite an icaADBC-positive genotype. de Silva et al. (4) reported similar results, interpreting this as evidence for differences in the expression control of icaADBC in those strains. Further studies by alternative genetic methods like assays with microarray technology and multilocus sequence typing are warranted in order to identify and characterize S. epidermidis subpopulations with enhanced virulence.
Acknowledgments
We thank R. Laufs for continuous support. B. Zöllner's help with statistical analysis is gratefully acknowledged.
This work is supported by grants from the Deutsche Forschungsgemeinschaft (grant SPP 1047 Ma 1522/4-3 to D.M., H.R., and J.K.-M.K. and grant SFB 470, Teilprojekt C10, to D.M.).
REFERENCES
- 1.Archer, G. L. 1991. Alteration of cutaneous staphylococcal flora as a consequence of antimicrobial prophylaxis. Rev. Infect. Dis. 13(Suppl. 10):S805-S809. [DOI] [PubMed] [Google Scholar]
- 2.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 3.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, I., and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Silva, G. D., M. Kantzanou, A. Justice, R. C. Massey, A. R. Wilkinson, N. P. Day, and S. J. Peacock. 2002. The ica operon and biofilm production in coagulase-negative staphylococci associated with carriage and disease in a neonatal intensive care unit. J. Clin. Microbiol. 40:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frebourg, N. B., S. Lefebvre, S. Baert, and J. F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galdbart, J. O., J. Allignet, H. S. Tung, C. Ryden, and N. El Solh. 2000. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182:351-355. [DOI] [PubMed] [Google Scholar]
- 7.Gerke, C., A. Kraft, R. Süssmuth, O. Schweitzer, and F. Götz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 8.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 9.Heilmann, C., M. Hussain, G. Peters, and F. Götz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 10.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 11.Hoiby, N., J. O. Jarlov, M. Kemp, M. Tvede, J. M. Bangsborg, A. Kjerulf, C. Pers, and H. Hansen. 1997. Excretion of ciprofloxacin in sweat and multiresistant Staphylococcus epidermidis. Lancet 349:167-169. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, W. T., D. Armstrong, G. P. Bodey, E. J. Bow, A. E. Brown, T. Calandra, R. Feld, P. A. Pizzo, K. V. Rolston, J. L. Shenep, and L. S. Young. 2002. 2002 guidelines for the use of antimicrobial agents in neutropenic patients with cancer. Clin. Infect. Dis. 34:730-751. [DOI] [PubMed] [Google Scholar]
- 13.Hussain, M., M. Herrmann, C. von Eiff, F. Perdreau-Remington, and G. Peters. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knobloch, J. K. M., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation of Staphylococcus epidermidis depends on RsbU, a functional activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knobloch, J. K., M. A. Horstkotte, H. Rohde, P. M. Kaulfers, and D. Mack. 2002. Alcoholic ingredients in skin disinfectants increase biofilm expression of Staphylococcus epidermidis. J. Antimicrob. Chemother. 49:683-687. [DOI] [PubMed] [Google Scholar]
- 16.Knobloch, J. K., S. Jäger, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 72:3838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knobloch, J. K., M. Nedelmann, K. Kiel, K. Bartscht, M. A. Horstkotte, S. Dobinsky, H. Rohde, and D. Mack. 2003. Establishment of an arbitrary PCR for rapid identification of Tn917 insertion sites in Staphylococcus epidermidis: characterization of biofilm-negative and nonmucoid mutants. Appl. Environ. Microbiol. 69:5812-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krüger, W., B. Rüssmann, N. Kroger, C. Salomon, N. Ekopf, H. A. Elsner, P. M. Kaulfers, D. Mack, N. Fuchs, M. Dürken, H. Kabisch, R. Erttmann, and A. R. Zander. 1999. Early infections in patients undergoing bone marrow or blood stem cell transplantation—a 7 year single centre investigation of 409 cases. Bone Marrow Transplant. 23:589-597. [DOI] [PubMed] [Google Scholar]
- 20.Mack, D. 1999. Molecular mechanisms of Staphylococcus epidermidis biofilm formation. J. Hosp. Infect. 43(Suppl.):S113-S125. [DOI] [PubMed] [Google Scholar]
- 21.Mack, D., K. Bartscht, C. Fischer, H. Rohde, C. de Grahl, S. Dobinsky, M. A. Horstkotte, K. Kiel, and J. K. M. Knobloch. 2001. Genetic and biochemical analysis of Staphylococcus epidermidis biofilm accumulation. Methods Enzymol. 336:215-239. [DOI] [PubMed] [Google Scholar]
- 22.Mack, D., H. Rohde, S. Dobinsky, J. Riedewald, M. Nedelmann, J. K. M. Knobloch, H.-A. Elsner, and H. H. Feucht. 2000. Identification of three essential regulatory gene loci governing expression of the Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 68:3799-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack, D., A. Sabottke, S. Dobinsky, H. Rohde, M. A. Horstkotte, and J. K. Knobloch. 2002. Differential expression of methicillin resistance by different biofilm-negative Staphylococcus epidermidis transposon mutant classes. Antimicrob. Agents Chemother. 46:178-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mempel, M., H. Feucht, W. Ziebuhr, M. Endres, R. Laufs, and L. Grüter. 1994. Lack of mecA transcription in slime-negative phase variants of methicillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 38:1251-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pei, L., M. Palma, M. Nilsson, B. Guss, and J. I. Flock. 1999. Functional studies of a fibrinogen binding protein from Staphylococcus epidermidis. Infect. Immun. 67:4525-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohde, H., J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2001. Correlation of biofilm expression types of Staphylococcus epidermidis with polysaccharide intercellular adhesin synthesis: evidence for involvement of icaADBC genotype-independent factors. Med. Microbiol. Immunol. (Berlin) 190:105-112. [DOI] [PubMed] [Google Scholar]
- 27.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231-243. [DOI] [PubMed] [Google Scholar]
- 28.Rupp, M. E., J. S. Ulphani, P. D. Fey, K. Bartscht, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rupp, M. E., J. S. Ulphani, P. D. Fey, and D. Mack. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 67:2656-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, B. J. Rijnders, and J. Van Eldere. 2003. Reliability of the ica, aap and atlE genes in the discrimination between invasive, colonizing and contaminant Staphylococcus epidermidis isolates in the diagnosis of catheter-related infections. Clin. Microbiol. Infect. 9:114-119. [DOI] [PubMed] [Google Scholar]
- 32.Williams, R. J., B. Henderson, L. J. Sharp, and S. P. Nair. 2002. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 70:6805-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]
- 34.Ziebuhr, W., C. Heilmann, F. Götz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziebuhr, W., V. Krimmer, S. Rachid, I. Löβner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]