Abstract
A multiplex PCR toxigenic culture approach was designed for simultaneous identification and toxigenic type characterization of Clostridium difficile isolates. Three pairs of primers were designed for the amplification of (i) a species-specific internal fragment of the tpi (triose phosphate isomerase) gene, (ii) an internal fragment of the tcdB (toxin B) gene, and (iii) an internal fragment of the tcdA (toxin A) gene allowing distinction between toxin A-positive, toxin B-positive (A+B+) strains and toxin A-negative, toxin B-positive (A−B+) variant strains. The reliability of the multiplex PCR was established by using a panel of 72 C. difficile strains including A+B+, A−B−, and A−B+ toxigenic types and 11 other Clostridium species type strains. The multiplex PCR assay was then included in a toxigenic culture approach for the detection, identification, and toxigenic type characterization of C. difficile in 1,343 consecutive human and animal stool samples. Overall, 111 (15.4%) of 721 human samples were positive for C. difficile; 67 (60.4%) of these samples contained A+B+ toxigenic isolates, and none of them contained A−B+ variant strains. Fifty (8%) of 622 animal samples contained C. difficile strains, which were toxigenic in 27 (54%) cases, including 1 A−B+ variant isolate. Eighty of the 721 human stool samples (37 positive and 43 negative for C. difficile culture) were comparatively tested by Premier Toxins A&B (Meridian Bioscience) and Triage C. difficile Panel (Biosite) immunoassays, the results of which were found concordant with toxigenic culture for 82.5 and 92.5% of the samples, respectively. The multiplex PCR toxigenic culture scheme described here allows combined diagnosis and toxigenic type characterization for human and animal C. difficile intestinal infections.
Clostridium difficile was recognized as a potential enteric pathogen in the late 1970s (5, 6). Toxigenic strains of C. difficile are responsible for 10 to 25% of antibiotic-associated diarrhea and for virtually all cases of pseudomembranous colitis (8, 23). Most pathogenic strains are toxin A-positive, toxin B-positive (A+B+) strains, although toxin A-negative, toxin B-positive (A−B+) variant isolates have been recognized as pathogenic (2, 21, 27).
Results of several studies support the opinion that both direct detection of toxins in stool samples and isolation of toxigenic strains (“toxigenic culture”) contribute to the optimal diagnosis of C. difficile-associated disease (12, 22). Although detection of toxin B by cell culture cytotoxicity assay (CA) is considered the “gold standard,” it is not routinely performed by clinical microbiology laboratories because it requires the use of cell culture and a reliable antitoxin for neutralization. Toxin enzyme immunoassays (EIAs) or toxigenic culture are thus used as alternative methods. Toxigenic culture has been shown to be as sensitive as CA and sometimes allows the detection of toxigenic isolates in CA-negative stool specimens (3, 12), even for patients with clinical evidence of C. difficile-associated disease (16). In addition, stool culture provides isolates suitable for toxigenic typing and for molecular epidemiological analysis of nosocomial outbreaks. Toxigenic typing allows the differentiation of A−B+ variants (in which the 3′ end of the toxin A gene is deleted) from A+B+ strains. Of note, these A−B+ variant strains are not detected by toxin A EIAs largely used by microbiology laboratories (4, 15), and their retrospectively estimated prevalence varies widely depending upon the study (4, 9, 18, 24, 28).
The purpose of this study was to design a toxigenic culture approach using multiplex PCR for simultaneous identification and toxigenic type characterization of C. difficile isolates. This approach is based on specific culture of spore-forming bacteria followed by multiplex PCR targeting a species-specific internal fragment of the triose phosphate isomerase (tpi) housekeeping gene (13), an internal fragment of the toxin B (tcdB) gene, and the 3′ region, deleted or not, of the toxin A (tcdA) gene.
MATERIALS AND METHODS
Multiplex PCR for C. difficile identification and toxigenic type characterization.
A collection of 72 C. difficile isolates from a previous study (20) and the type strains of 12 different species (Clostridium perfringens ATCC 13124T, C. ramosum ATCC 25582T, C. innocuum ATCC 14501T, C. difficile ATCC 9689T, C. butyricum ATCC 19398T, C. sporogenes ATCC 3584T, C. clostridioforme ATCC 25537T, C. bifermentans ATCC 638T, C. sordellii ATCC 9714T, C. septicum ATCC 12464T, C. cadaveris ATCC 25783T, and C. tertium ATCC 14573T) were first used to test the ability of the multiplex PCR (i) to differentiate C. difficile from other Clostridium species and (ii) to distinguish the three main toxigenic types (A+B+, A−B+, and A−B−) of C. difficile. To prepare a DNA sample for PCR amplification, a bacterial colony was taken from blood agar culture and resuspended in 1 ml of distilled water in a microcentrifuge tube. The sample was then boiled for 20 min prior to being centrifuged for 2 min to settle bacterial debris, and 10 μl of supernatant, containing the genomic DNA, was used for subsequent PCR amplification.
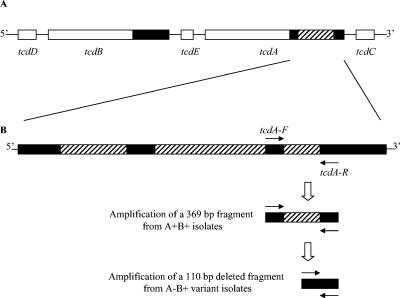
Three pairs of primers were designed. The tpi-specific primers (tpi-F [5′-AAAGAAGCTACTAAGGGTACAAA-3′] and tpi-R [5′-CATAATATTGGGTCTATTCCTAC-3′]) were deduced from alignments of internal fragments of the tpi gene. This housekeeping gene was used in a previous work to differentiate 12 Clostridium species including C. difficile (13); here it generated a 230-bp amplified fragment specific for C. difficile. The tcdB-specific primers (tcdB-F [5′-GGAAAAGAGAATGGTTTTATTAA-3′] and tcdB-R [5′-ATCTTTAGTTATAACTTTGACATCTTT-3′]) were designed from the conserved 5′ region of tcdB and generated a 160-bp fragment. The tcdA-specific primers (tcdA-F [5′-AGATTCCTATATTTACATGACAATAT-3′] and tcdA-R [5′-GTATCAGGCATAAAGTAATATACTTT-3′]) were designed to flank the smallest of the three deletions in the 3′ region of tcdA characterized in A−B+ variant strains (27) (Fig. 1) and generated a 369-bp fragment for A+B+ strains and a 110-bp fragment for A−B+ strains. The tcdA primers target regions which are not identically repeated elsewhere along the tcdA gene.
FIG. 1.
Positions of tcdA primers allowing differentiation between A+B+ and A−B+ isolates. (A) Partial map of the pathogenicity locus with tcdA and tcdB genes and their adjacent accessory genes tcdD, tcdE, and tcdC. Black regions represent the 3′ repetitive sequences characteristic of C. difficile tcdA and tcdB genes. Hatched region represents the 1.8-kb deletion characteristic of most A−B+ variant strains. (B) Details of the 1.8-kb deletion, resulting in combination of three small deletions (hatched regions). The tcdA primers flank the smallest deletion (in the 3′ end of the gene) and generate a 369-bp amplified fragment from A+B+ strains but a 110-bp amplified fragment from A−B+ variant strains.
PCRs were performed on a GeneAmp System 2400 thermal cycler (Applied Biosystems) in a final volume of 25 μl containing 10% (vol/vol) glycerol, 1 μM each primer (except for tpi-F and tpi-R [0.5 μM]), 200 μM each deoxynucleoside triphosphate, and 0.5 U of Taq DNA polymerase in a 1× amplification buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2). The PCR mixtures were denatured (3 min at 95°C), and then a touchdown procedure was implemented, consisting of 30 s at 95°C, annealing for 30 s at temperatures decreasing from 65 to 55°C during the first 11 cycles (with 1°C decremental steps in cycles 1 to 11), and a final extension step at 72°C for 30 s. A total of 40 cycles were performed. PCR products were resolved by electrophoresis on a 2% agarose gel stained with ethidium bromide.
Development of multiplex PCR toxigenic culture for human and animal clinical samples.
A total of 721 human stool samples (from 502 patients) and 622 animal stool samples (from 559 animals) sent to our laboratory between July 2002 and June 2003 for investigation of C. difficile infection were submitted to multiplex PCR toxigenic culture. A portion of each stool sample was mixed with an equal volume of ethanol (ethanol shock) for 1 h at room temperature. One hundred microliters of stool suspension was then inoculated onto Trypticase soy agar supplemented with 5% sheep blood, lysozyme (10 mg/liter), and cefoxitin (10 mg/liter). Plates were incubated in an anaerobic chamber for 48 h at 37°C. Bacterial DNA was extracted from colonies suspected of being C. difficile (on the basis of morphology, odor, and fluorescence under 365-nm UV illumination), and PCRs were performed as described above.
Eighty of the 721 human stool samples (corresponding to the first 37 patients positive for C. difficile culture and the first 43 patients negative for C. difficile culture) were also tested by two immunoassays: the Premier Toxins A&B test (Meridian Bioscience), which detects both toxin A and toxin B, and the Triage C. difficile Panel (Biosite), which detects both C. difficile toxin A and the C. difficile Gdh (glutamate dehydrogenase) antigen. These assays were performed and interpreted according to the manufacturer's recommendations.
RESULTS
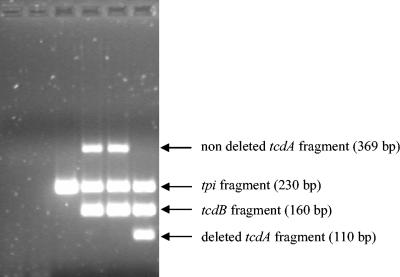
The first step of this work was to test the ability of the multiplex PCR (i) to distinguish C. difficile from other Clostridium species and (ii) to distinguish between the three main toxigenic types (A+B+, A−B+, and A−B−) of C. difficile. The species-specific internal fragment of the tpi gene was detected from all of the 72 C. difficile isolates used in this work, despite their various host and geographic origins. On the other hand, no amplification signal was detected from the type strains of 11 other Clostridium species, thus confirming the specificity of this PCR assay (data not shown). The toxigenic types of all of the 72 C. difficile isolates determined by the multiplex PCR were concordant with the results of previous PCRs targeting each of the two toxin genes separately (20): the 52 A+B+ isolates showed tpi, 379-bp tcdA, and tcdB amplification signals; the 12 nontoxigenic (A−B−) isolates showed only the tpi signal; and all of the 8 A−B+ variants showed tpi, deleted tcdA (110 bp), and tcdB amplicons. An example of the three different toxigenic types revealed by this PCR assay is given in Fig. 2. None of the 11 other Clostridium species tested generated an amplification signal for tcdA or tcdB, including C. sordellii, which carries a closely related lethal toxin gene (10) (data not shown).
FIG. 2.
Characterization of the three main C. difficile toxigenic types by multiplex PCR. First lane, C. bifermentans ATCC 638T; second lane, C. sordellii ATCC 9714T; third lane, nontoxigenic (A−B−) C. difficile strain; fourth and fifth lanes, toxigenic (A+B+) C. difficile strains; sixth lane, A−B+ C. difficile variant strain.
The second step of this work was to include the multiplex PCR in a toxigenic culture approach for detection, identification, and toxigenic type characterization of C. difficile in human and animal stool samples. The results for the 721 human feces samples and the 622 animal samples analyzed are summarized in Table 1. Of the 721 human feces samples tested, 111 (15.4%) were positive for the presence of C. difficile (amplification of tpi), 67 (60.4%) contained A+B+ toxigenic isolates, and none corresponded to A−B+ variant isolates. Of the 622 animal samples tested, 50 (8%) contained C. difficile strains; 27 (54%) of these were toxigenic isolates, among which 1 A−B+ variant isolate was detected (from a horse).
TABLE 1.
Multiplex PCR toxigenic culture results for 721 human and 622 animal samples
| Sample origin | No. (%) of positive cultures with:
|
No. (%) of negative cultures | Total | ||
|---|---|---|---|---|---|
| A+B + toxigenic strains | Nontoxigenic strains | A−B+ variant strains | |||
| Human | 67 (9.3) | 44 (6.1) | 0 (0) | 610 (84.6) | 721 |
| Animal | 26 (4.2) | 23 (3.7) | 1 (0.2) | 572 (91.9) | 622 |
Eighty of the 721 human stool samples (37 positive and 43 negative for C. difficile culture) were also tested by two immunoassays: the Premier Toxins A&B test (Meridian Bioscience), which detects both toxin A and toxin B, and the Triage C. difficile Panel (Biosite), which detects both C. difficile toxin A and C. difficile Gdh antigen. The results are summarized in Table 2. Good agreement between multiplex PCR toxigenic culture and the Premier Toxins A&B test was obtained for 66 of the 80 samples (82.5%). Of note, among the 14 discordant cases, 10 samples were positive by the Premier Toxins A&B test and negative by toxigenic culture and the Triage test and were finally considered negative for C. difficile. On the other hand, good agreement between the Triage C. difficile Panel and toxigenic culture was obtained for 74 (92.5%) of the 80 samples. Of note, 6 samples containing strains that were toxigenic according to multiplex PCR toxigenic culture were detected only by the common antigen (Gdh) on the Triage C. difficile Panel (false toxin A-negative results).
TABLE 2.
Comparison of multiplex PCR toxigenic culture, the Premier Toxins A&B test, and the Triage C. difficile Panel on the basis of analysis of 80 human samples for C. difficile diagnosis
| No. (%) of samples | Result by:
|
|||||
|---|---|---|---|---|---|---|
| Multiplex PCR toxigenic culture
|
Premier Toxins A&B test | Triage C. difficile Panel
|
||||
| tpi | tcdA | tcdB | Gdh | ToxA | ||
| 33 (41.2) | − | − | − | − | − | − |
| 14 (17.5) | + | + | + | + | + | + |
| 7 (21.3) | + | − | − | − | + | − |
| 2 (2.5) | + | + | + | + | + | − |
| 4 (5.0) | + | + | + | − | + | − |
| 10 (12.5) | − | − | − | + | − | − |
DISCUSSION
The aim of this work was to design an optimized toxigenic culture scheme for detection and combined identification and toxigenic type characterization of C. difficile from stool samples. Indeed, as long as toxinogenic C. difficile strains are recognized as the primary cause of nosocomial gastrointestinal disease, a good management approach includes optimal diagnostic methods, which so far include EIAs, CA, or toxigenic culture.
The presence of C. difficile in stool cultures is easily suspected on the basis of phenotypic characteristics (such as colony morphology, odor, and fluorescence under UV light). However, for accurate identification, additional tests, such as biochemical test panels or even gas-liquid chromatographic analysis (17), are needed. In the present work, we propose to identify C. difficile by PCR amplification of an internal fragment of the tpi housekeeping gene, which was found more discriminatory than 16S ribosomal DNA for the identification of 12 species within the genus Clostridium (13). This tpi fragment was successfully amplified from all the 72 C. difficile strains previously selected from various hosts and geographic sources and was not amplified from the other 11 species tested, including C. sordellii and C. bifermentans, which are phylogenetically closely related to C. difficile (11). Furthermore, the combined detection of tpi, tcdA, and tcdB fragment genes by multiplex PCR provides a one-step method for identification and toxigenic type characterization of C. difficile. Of note, A−B+ variant strains are sometimes reported only on the basis of lack of tcdA amplification, or even on the basis of lack of toxin A detection by EIA (1, 28). This approach may be unreliable, because PCR inhibitors or any variations in the tcdA sequence (other than those characterizing these A−B+ variants) can result in lack of tcdA fragment amplification or lack of toxin A detection by EIA. The primers designed in this work allow the detection of a partially deleted tcdA fragment in A−B+ variant strains and thus avoid false toxigenic type characterization.
At present, many C. difficile variant strains are characterized on the basis of their tcdA and tcdB toxin genes in comparison to the reference strain VPI 10463 (26), but A−B+ variant strains (especially those belonging to toxinotype VIII and serogroup F) are of particular clinical significance (2, 21, 27). Epidemiological studies have reported various prevalence rates: 0.2% in the United States (24), 2.5 to 3% in European countries (4, 9), and 6.7 to 39% in Japan (18, 19). Higher rates, such as 56.5% reported in an Israeli hospital (28), suggest local epidemic events. However, almost all these results were obtained from retrospective studies, while at the time of diagnosis, A−B+ variants were only suspected or even not detected. The present multiplex PCR allows the characterization of toxigenic types at the time of diagnosis and is thus more efficient for diagnosis of intestinal infections involving A−B+ variants. The detection of nontoxigenic isolates by tpi amplification could also contribute to a better knowledge of the global epidemiology of this species.
The C. difficile detection rate for human samples in the present work was comparable to that of previous studies (22, 29). However, few data are available about C. difficile detection rates for animal samples. In the present study, 8% of animal samples (corresponding to 23 of 245 horses, 20 of 272 cattle, 5 of 22 dogs, and 2 of 10 cats) were positive for C. difficile culture, but only 4.3% were positive for toxinogenic strains (13 horses, 11 cattle, 2 dogs, and 1 cat). It was suggested that 21 and 30% of domestic pets (dogs and cats, respectively) could be infected or colonized by C. difficile (25). Other authors reported that 6 to 42% of horses that developed acute colitis or diarrhea were positive for C. difficile culture and that horses that developed acute colitis during or immediately after antimicrobial treatment had the highest recovery rates (7, 14). In fact, our results agree with the data mentioned above, since only a few horses had a history of antibiotic treatment associated with diarrhea or colitis.
Although CA and many EIAs are available for the diagnosis of C. difficile-associated disease, culture of C. difficile remains essential (12). The epidemiology of an outbreak can be monitored only by bacterial culture, and macroepidemiology or antimicrobial susceptibility studies also require strain isolation. The multiplex PCR-toxigenic culture scheme described here provides both strain isolation and toxigenic type characterization. It may also be compared to the Triage C. difficile Panel, since both methods target a species-specific marker (the tpi gene in multiplex PCR, the Gdh antigen in Triage) and toxins of C. difficile (tcdA and tcdB genes in multiplex PCR, toxin A in Triage). Indeed, since Triage detects only toxin A, it cannot characterize A−B+ variants, which appear as nontoxigenic strains (requiring complementary tests in the case of Gdh-positive and toxin A-negative results to avoid a mistaken diagnosis). Rapid diagnosis of C. difficile-associated disease is needed in order to initiate specific treatment and to take adequate measures to control nosocomial spread, but monitoring pathogenic variant strains is also essential in order to better evaluate the relevance of the diagnostic tests in each hospital laboratory. The present multiplex PCR-toxigenic culture scheme may be proposed as a reliable diagnostic method, since it provides detection and toxigenic type results within 36 to 48 h. An EIA such as the Triage C. difficile Panel provides results within 1 h and is thus well suited for urgent situations, as when there is a suspicion of pseudomembranous colitis. Finally, since CA is not routinely performed in most clinical microbiology laboratories, EIAs are largely used for the diagnosis of C. difficile-associated disease. Since toxigenic culture is also an alternative diagnostic method (which also provides isolates suitable for epidemiological analysis) (12), the multiplex PCR-toxigenic culture scheme may be proposed as an improved diagnostic approach for human and animal C. difficile intestinal infections, offering combined species identification and toxigenic type characterization.
Acknowledgments
We thank Anne Collignon (Chatenay-Malabry, France), Frederic Barbut (Paris, France), Haru Kato (Tokyo, Japan), and Francine Mory (Nancy, France) for supplying strains and Martine Pestel-Caron for helpful reading of the manuscript.
REFERENCES
- 1.al-Barrak, A., J. Embil, B. Dyck, K. Olekson, D. Nicoll, M. Alfa, and A. Kabani. 1999. An outbreak of toxin A negative, toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can. Commun. Dis. Rep. 25:65-69. [PubMed] [Google Scholar]
- 2.Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barrak, G. K. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbut, F., C. Kajzer, N. Planas, and J. C. Petit. 1993. Comparison of three enzyme immunoassays, a cytotoxicity assay, and toxigenic culture for diagnosis of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 31:963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbut, F., V. Lalande, B. Burghoffer, H. V. Thien, E. Grimprel, and J. C. Petit. 2002. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J. Clin. Microbiol. 40:2079-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett, J. G., T. W. Chang, M. Gurwith, S. L. Gorbach, and A. B. Onderdonk. 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N. Engl. J. Med. 298:531-534. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett, J. G., N. Moon, T. W. Chang, N. Taylor, and A. B. Onderdonk. 1978. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75:778-782. [PubMed] [Google Scholar]
- 7.Baverud, V., A. Gustafsson, A. Franklin, A. Aspan, and A. Gunnarsson. 2003. Clostridium difficile: prevalence in horses and environment, and antimicrobial susceptibility. Equine Vet. J. 35:465-471. [DOI] [PubMed] [Google Scholar]
- 8.Brazier, J. S. 1998. The diagnosis of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl. C):29-40. [DOI] [PubMed] [Google Scholar]
- 9.Brazier, J. S., S. L. Stubbs, and B. I. Duerden. 1999. Prevalence of toxin A negative/B positive Clostridium difficile strains. J. Hosp. Infect. 42:248-249. [PubMed] [Google Scholar]
- 10.Chaves-Olarte, E., P. Low, E. Freer, T. Norlin, M. Weidmann, C. von Eichel-Streiber, and M. Thelestam. 1999. A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large clostridial cytotoxins. J. Biol. Chem. 274:11046-11052. [DOI] [PubMed] [Google Scholar]
- 11.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 12.Delmee, M. 2001. Laboratory diagnosis of Clostridium difficile disease. Clin. Microbiol. Infect. 7:411-416. [DOI] [PubMed] [Google Scholar]
- 13.Dhalluin, A., L. Lemee, M. Pestel-Caron, F. Mory, G. Leluan, J. F. Lemeland, and J. L. Pons. 2003. Genotypic differentiation of twelve Clostridium species by polymorphism analysis of the triosephosphate isomerase (tpi) gene. Syst. Appl. Microbiol. 26:90-96. [DOI] [PubMed] [Google Scholar]
- 14.Donaldson, M. T., and J. E. Palmer. 1999. Prevalence of Clostridium perfringens enterotoxin and Clostridium difficile toxin A in feces of horses with diarrhea and colic. J. Am. Vet. Med. Assoc. 215:358-361. [PubMed] [Google Scholar]
- 15.Frey, S. M., and T. D. Wilkins. 1992. Localization of two epitopes recognized by monoclonal antibody PCG-4 on Clostridium difficile toxin A. Infect. Immun. 60:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerding, D. N., M. M. Olson, L. R. Peterson, D. G. Teasley, R. L. Gebhard, M. L. Schwartz, and J. T. Lee, Jr. 1986. Clostridium difficile-associated diarrhea and colitis in adults. A prospective case-controlled epidemiologic study. Arch. Intern. Med. 146:95-100. [PubMed] [Google Scholar]
- 17.Holdeman, L. V., E. P. Cato, and W. E. C. Moore (ed.). 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 18.Kato, H., N. Kato, K. Watanabe, N. Iwai, H. Nakamura, T. Yamamoto, K. Suzuki, S. M. Kim, Y. Chong, and E. B. Wasito. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu, M., H. Kato, M. Aihara, K. Shimakawa, M. Iwasaki, Y. Nagasaka, S. Fukuda, S. Matsuo, Y. Arakawa, M. Watanabe, and Y. Iwatani. 2003. High frequency of antibiotic-associated diarrhea due to toxin A-negative, toxin B-positive Clostridium difficile in a hospital in Japan and risk factors for infection. Eur. J. Clin. Microbiol. Infect. Dis. 22:525-529. [DOI] [PubMed] [Google Scholar]
- 20.Lemee, L., A. Dhalluin, M. Pestel-Caron, J. F. Lemeland, and J. L. Pons. 2004. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 42:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limaye, A. P., D. K. Turgeon, B. T. Cookson, and T. R. Fritsche. 2000. Pseudomembranous colitis caused by a toxin A− B+ strain of Clostridium difficile. J. Clin. Microbiol. 38:1696-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozniewski, A., C. Rabaud, E. Dotto, M. Weber, and F. Mory. 2001. Laboratory diagnosis of Clostridium difficile-associated diarrhea and colitis: usefulness of Premier Cytoclone A+B enzyme immunoassay for combined detection of stool toxins and toxigenic C. difficile strains. J. Clin. Microbiol. 39:1996-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyerly, D. M., H. C. Krivan, and T. D. Wilkins. 1988. Clostridium difficile: its disease and toxins. Clin. Microbiol. Rev. 1:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyerly, D. M., L. M. Neville, D. T. Evans, J. Fill, S. Allen, W. Greene, R. Sautter, P. Hnatuck, D. J. Torpey, and R. Schwalbe. 1998. Multicenter evaluation of the Clostridium difficile TOX A/B TEST. J. Clin. Microbiol. 36:184-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley, T. V., J. E. Adams, G. L. O'Neill, and R. A. Bowman. 1991. Gastrointestinal carriage of Clostridium difficile in cats and dogs attending veterinary clinics. Epidemiol. Infect. 107:659-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmee. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambol, S. P., M. M. Merrigan, D. Lyerly, D. N. Gerding, and S. Johnson. 2000. Toxin gene analysis of a variant strain of Clostridium difficile that causes human clinical disease. Infect. Immun. 68:5480-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samra, Z., S. Talmor, and J. Bahar. 2002. High prevalence of toxin A-negative toxin B-positive Clostridium difficile in hospitalized patients with gastrointestinal disease. Diagn. Microbiol. Infect. Dis. 43:189-192. [DOI] [PubMed] [Google Scholar]
- 29.Turgeon, D. K., T. J. Novicki, J. Quick, L. Carlson, P. Miller, B. Ulness, A. Cent, R. Ashley, A. Larson, M. Coyle, A. P. Limaye, B. T. Cookson, and T. R. Fritsche. 2003. Six rapid tests for direct detection of Clostridium difficile and its toxins in fecal samples compared with the fibroblast cytotoxicity assay. J. Clin. Microbiol. 41:667-670. [DOI] [PMC free article] [PubMed] [Google Scholar]