Abstract
Pseudomonas aeruginosa is a gram-negative rod that is ubiquitous in nature. P. aeruginosa is also the quintessential opportunistic pathogen, causing a wide variety of infections in compromised hosts. In cystic fibrosis patients, P. aeruginosa is the leading cause of death. In this study, the evolutionary genetic relationships among 17 P. aeruginosa isolates were examined by comparative sequence analysis of the housekeeping gene encoding malate dehydrogenase and the chaperone groEL. The P. aeruginosa isolates examined included the sequenced strain PAO1, 11 strains recovered from cystic fibrosis patients in Ireland, 4 environmental isolates recovered from a hospital environment, and 1 isolate recovered from a plant rhizosphere. Phylogenetically, clinical and environmental isolates clustered together with one another on the mdh gene tree. At the groEL locus, among the 17 isolates examined, only two polymorphic sites were observed, highlighting the close genetic relationship between isolates from these different environments. Phenotypic analysis of 12 traits among our isolates, however, found that only clinical isolates produced phenazines and elastase. Furthermore, molecular analysis of the distribution of 15 regions associated with virulence showed that two of the environmental isolates examined lacked the majority of regions. Among the clinical isolates examined, the 15 virulence regions were variably present. The distribution of two prophages (Bacto1, Pf1) was also determined, with most isolates encoding both these regions. Of the four genomic islands (the flagellum island and PAGI-1, -2, and -3) examined, only two isolates contained the flagellum island, and PAGI-1, -2, and -3 were absent from all isolates tested. Our data demonstrate the significant role horizontal gene transfer and recombination, together with gene loss, play in the evolution of this important human pathogen.
The remarkable ability of Pseudomonas aeruginosa to adapt and thrive in a wide variety of environments is due in part to its extensive genetic versatility, which contributes significantly to its potential as a pathogen. P. aeruginosa is an opportunistic pathogen that is a common cause of hospital-acquired infections, particularly infecting patients with predisposing factors, such as burn victims, immunocompromised hosts, or those with metabolic disorders. In cystic fibrosis (CF) patients, P. aeruginosa is believed to be a major contributory factor to chronic lung infections and is thought to form biofilms and to adhere to human mucin in the lower respiratory tract (46, 53). Once P. aeruginosa colonizes the CF patient's lung, it cannot be eradicated even by the most aggressive antibiotic therapy. It is likely that as part of colonization, invasion, and survival in the human host, P. aeruginosa utilizes a unique range of genes; however, the precise repertoire of virulence genes required is unclear.
Several studies have examined the genetic diversity of P. aeruginosa isolates from CF patients (9, 13, 30, 40, 44, 49). In these studies, it was determined that individual patients were colonized by unique clonal lineages. Indeed, it has been proposed that P. aeruginosa possesses an epidemic population structure (1). With the availability of the whole-genome sequence of P. aeruginosa PAO1, a number of studies have examined genetic differences between P. aeruginosa clinical and environmental isolates and have determined that variable regions, some of which encode virulence genes, are not conserved among clinical isolates (13, 28, 29, 49, 50, 54).
The pathogenesis of P. aeruginosa is due to the production of both extracellular and cell-associated virulence factors. One of these extracellular compounds, hydrogen cyanide (HCN), has been found at relatively high concentrations in patients with freshly infected burns (36). Cyanide is a potent inhibitor of cytochrome c oxidase, the terminal component of the aerobic respiratory chain in many organisms. Proteases are also thought to play a role in the pathogenesis of some P. aeruginosa infections (48). Strains of P. aeruginosa produce three proteases (33). Elastase is a metalloprotease that degrades elastin and collagen and inactivates human immunoglobulin G, serum alpha-1, proteinase inhibitor, and several complement components (19). The production of elastase protein in P. aeruginosa is regulated by several factors. One of these factors is the growth rate of P. aeruginosa. Higher levels of elastase are produced when cells are at the late-logarithmic phase of growth or when the cell density is high (32).
Cell-associated virulence factors include pili, flagella, lipopolysaccharide, type III system effector proteins, a type III secretion system, and alginate. To date, four type III effector proteins have been identified in P. aeruginosa: ExoS, ExoT, ExoU, and ExoY. ExoS and ExoT possess N-terminal domains that encode a GTPase-activating protein activity (16). ExoY is an adenylate cyclase secreted by the type III secretion system (55). Intoxication with ExoS, ExoT, and ExoY causes cell rounding and detachment and may contribute to infection by inhibiting or preventing bacterial uptake and phagocytosis. ExoU has a unique cytotoxic effect which is rapid and potent, is associated with accelerated lung injury, and plays a role in the development of septic shock (2, 27). Secreted products include phenazines. Ninety to 95% of P. aeruginosa isolates produce pyocyanin (47, 48), and the presence of high concentrations of pyocyanin in the sputa of CF patients has suggested that this compound plays a role in pulmonary tissue damage observed with chronic lung infection (31).
Bacteriophages play an important role in the acquisition of new genetic information encoding novel functions including virulence in a wide range of bacterial species (4, 51). In the P. aeruginosa PAO1 genome, two prophages, Bacto1 and Pf1, have been identified (50). In addition, φCTX, which encodes a cytotoxin, has been identified in a strain of P. aeruginosa (35). Many bacterial genomes are arranged in blocks of core chromosomal regions, which are present in all isolates of a species, and variable chromosomal regions termed genomic islands (GEIs), which are present only in some isolates (18). GEIs have many features in common with pathogenicity islands; their G+C content differs from that of the host genome, and they encode a phage-like integrase and genes required for nitrogen fixation, iron uptake, or xenobiotic or sugar metabolism (17, 18). Four GEIs have been identified in P. aeruginosa: P. aeruginosa genomic island 1 (PAGI-1), PAGI-2, PAGI-3, and the flagellum island; their role in pathogenesis, if any, is unknown (3).
In this study, we examined the evolutionary genetic relationships among 17 P. aeruginosa clinical and environmental isolates by comparative sequence analysis at the housekeeping gene mdh (encoding malate dehydrogenase) and groEL loci. Phylogenetically, both clinical and environmental isolates clustered together on the mdh gene tree. The finding of only two polymorphic sites among the 17 P. aeruginosa groEL sequences examined also indicates a close relationship between these isolates and the conserved nature of the core genome. To elucidate whether there were phenotypic differences between isolates, the production of 12 factors associated with virulence was assayed. We found little variation between clinical and environmental strains in the phenotypes assayed, with the exception of phenazine and elastase production, which was confined to clinical isolates. Molecular analysis of the distribution of 15 regions associated with virulence indicated that some of the environmental isolates lacked the majority of regions examined whereas others had a pattern similar to that of clinical isolates. We found that none of the P. aeruginosa isolates examined contained PAGI-1, PAGI-2, or PAGI-3. However, two P. aeruginosa strains, CF93 and BR642, did contain the flagellum island. These data demonstrate the significant role that horizontal gene transfer and recombination, together with gene loss, play in the evolution of this important human pathogen.
MATERIALS AND METHODS
Bacterial isolates.
A total of 17 P. aeruginosa isolates were examined in this study (Table 1). They were recovered from Ireland and Brussels over a 6-year period (1993 to 1998). Of the 17 strains examined, 11 were clinical isolates, 4 were hospital isolates, 1 was a plant rhizosphere isolate, and the sequenced strain PAO1 was used as a positive control (Table 1). The 11 clinical isolates came from seven CF patients treated at a CF center in southern Ireland and are a subset from an original collection of 250 isolates (1). The 11 strains were chosen to represent the strain diversity in the original collection based on the arbitrarily primed PCR profile and patient disease status. P. aeruginosa clinical isolates were a kind gift from Claire Adams (1). P. aeruginosa environmental isolates were a kind gift from Jean-Paul Pirnay and Pierre Cornelis (37, 38). P. aeruginosa strain X24509 was a kind gift from Stephen Lory, P. aeruginosa strain C and strain SG17 M were kind gifts from Burkhard Tümmler, and P. aeruginosa strain PAK was a kind gift from Shiwani K. Arora. All P. aeruginosa strains were grown in Luria-Bertani (LB) broth and stored at −70°C in LB broth with 20% (vol/vol) glycerol.
TABLE 1.
P. aeruginosa strains used in this study
| Strain (patient no. or environmental origin) | Country or state of isolation | Yr of isolation | Reference |
|---|---|---|---|
| Clinical strains | |||
| CF93 (CFP17) | Ireland | 1993 | 1 |
| CF95 (CFP35) | Ireland | 1993 | 1 |
| CF139 (CFP9) | Ireland | 1994 | 1 |
| CF144 (CFP16) | Ireland | 1994 | 1 |
| CF175 (CFP50) | Ireland | 1994 | 1 |
| CF177 (CFP26) | Ireland | 1994 | 1 |
| CF194 (CFP39) | Ireland | 1994 | 1 |
| CF195 (CFP26) | Ireland | 1994 | 1 |
| CF198 (CFP50) | Ireland | 1994 | 1 |
| CF208 (CFP35) | Ireland | 1994 | 1 |
| CF242 (CFP39) | Ireland | 1994 | 1 |
| Environmental strains | |||
| BR220 (hospital sink) | Brussels | 1997 | 37 |
| BR227 (hospital sink) | Brussels | 1997 | 37 |
| BR257 (plant rhizosphere) | Brussels | 1997 | 37 |
| BR642 (bedpan) | Brussels | 1997 | 37 |
| BR764 (hospital water) | Brussels | 1997 | 37 |
| Positive controls | |||
| PAO1 (wound isolate) | Australia | 51 | |
| Strain C (clinical) | Germany | 28 | |
| SG17M (river water) | Germany | 28 | |
| X24509 (clinical) | Washington | 29 | |
| PAK (laboratory strain) | Florida | 3 |
Phenotypic characteristics.
The motility assay protocol was adapted from that of Deziel et al. (12). Motility was assessed qualitatively by examining the circular turbid zone formed by the bacterial cells migrating away from the point of inoculation. Swarm medium was composed of 0.5% Bacto Agar, 0.5% tryptone, 0.5% beef extract, and 0.3% glucose. Plates were dried overnight at room temperature. Cells were point inoculated, and the plates were incubated at 37°C overnight. Twitching medium was composed of 0.5% Bacto Agar, 0.8% Caso Bouillon nutrient broth, and 0.5% glucose. Cultures were stab inoculated through a thin layer (approximately 3 mm thick) of medium to the bottom of the petri dish. After incubation at 37°C overnight under humid conditions, a hazy zone of growth at the interface between the agar and the polystyrene surface was observed.
Production of exoproducts.
The phenazine assay protocol was adapted from that of Frank and DeMoss (15). The pyoverdine assay protocol was adapted from that of Hofte et al. (23). Protease production was determined by using a variation on the method of Brown and Foster (8). Protease medium contained 1% agar, 2% skim milk dissolved in 3 mM CaCl2, and 50 mM Tris-HCl at pH 7.5. The test medium was inoculated with 100 μl of an overnight culture in holes made on the plates. Clear zones around the holes are indicative of protease production following overnight incubation at 37°C. DNase test agar was used to identify bacteria capable of producing the exoenzyme DNase. The test agar contained an emulsion of DNA and peptides as a nutrient source. Bacterial colonies that secrete DNase will hydrolyze the DNA in the medium into smaller fragments, which results in clearing around the bacterial growth observed by flooding the plates with HCl. The rhamnolipid assay was carried out according to the protocol of Siegmund and Wagner (45). Elastolytic activity of bacterial supernatants was determined by the elastin Congo red assay adapted from that of Hamood et al. (19). Following overnight growth of cultures in liquid LB medium, centrifugation at 13,000 rpm was carried out for 15 min in Eppendorf centrifuge 5417C. A 100-μl volume of filter-sterilized supernatant was added to 900 μl of 100 mM Tris-1 mM CaCl2 (pH 7.5) containing 1 mg of elastin Congo red, and the mixture was incubated with shaking at 37°C for 24 h. Centrifugation was repeated as before. The absorption of the supernatants was measured as optical density at 495 nm. Lipase medium contained 40 g of Caso Bouillon agar and 10 ml of glycerol-tributyrate per liter. Zones of hydrolysis around the point of inoculation were indicative of lipase production. The HCN test was carried out in accordance with the protocol of Castric (11).
DNA isolation.
Chromosomal DNA was extracted from each P. aeruginosa isolate by using the G-nome DNA isolation kit from Bio 101 (Vista, Calif.). Briefly, a single colony of each isolate was inoculated into 3 ml of LB broth and incubated overnight at 37°C with shaking at 150 rpm. Bacterial cells were pelleted at 3,000 rpm for 5 min, the supernatant was discarded, and the pellet was brought to a final volume of 1.85 ml in a cell suspension solution. The cells were lysed and treated with RNase and protease. DNA was ethanol extracted and resuspended in Tris-EDTA buffer.
PCR amplification and nucleotide sequencing.
PCR primers to amplify the chromosomal housekeeping genes mdh (encoding malate dehydrogenase) and groEL (encoding a chaperone) were designed from the genome sequence of P. aerguinosa strain PAO1 (50). The following PCR cycle was used to amplify the mdh gene for each isolate: an initial denaturation step at 96°C for 5 min followed by 30 cycles of denaturation at 94°C for 30 s, 30 s of primer annealing at 62.4°C, and 1.5 min of primer extension at 72°C. The primer pair mdh1F/mdh2R amplified an 887-bp fragment, representing 87% of the mdh gene. The following PCR cycle was used to amplify the groEL gene for each isolate: an initial denaturation step at 96°C for 5 min followed by 30 cycles of denaturation at 94°C for 30 s, 30 s of primer annealing at 62°C, and 1.5 min of primer extension at 72°C. The primer pair groEL1F/groEL2R amplified a 1,434-bp fragment, representing 94% of the groEL gene. PCR products were purified by using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. After purification, a 10-μl aliquot was used as a sequencing template. The mdh and groEL gene sequences were determined by MWG-Biotech based on the dye deoxy terminator method.
Phylogenetic analyses.
The mdh and groEL gene sequences were aligned by using the CLUSTALW multiple sequence alignment program (22). From the mdh sequence alignments, a 714-bp region was further analyzed by using the Molecular Evolutionary Genetics Analysis (MEGA) suite of programs, version 2.1 (26). Phylogenetic gene trees were constructed by the neighbor-joining method by use of the Jukes-Cantor distance method (24, 42). Bootstrap values were calculated for 1,000 trees. The proportions of synonymous (silent) substitutions per synonymous site (kS) and nonsynonymous (replacement) substitutions per nonsynonymous site (kN) were calculated.
PCR analysis.
PCR was used to assay 17 P. aeruginosa isolates for the presence of 15 regions associated with P. aeruginosa virulence, as well as 4 GEIs and 2 prophages. Of the 15 virulence regions examined, 2 regions comprised 3 or more genes: the phzI (ORF PA4210 to PA4216) and phzII (ORF PA1899 to PA1905) gene clusters. A total of 15 primer pairs were used to determine the distribution of these regions among the 17 P. aeruginosa isolates (Table 2). To determine the presence of the flagellum island, two primer pairs were used (FlagF-FlagR and ORF FF-ORF FR). Three primer pairs were used to assay for PAGI-1 (ORF 3F-ORF 3R, ORF 18F-ORF 18R, and ORF 42F-ORF 42R), and two primer pairs each were used to assay for PAGI-2 (C22F-C22R and C105F-C105R) and PAGI-3 (SG8F-SG8R and SG100F-SG100R) (Table 2). Gene fragments were amplified from chromosomal DNA isolated from the P. aeruginosa strains. PCR was performed in a 20-μl reaction mixture by using the following cycles: an initial denaturation step at 96°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, 30 s of primer annealing at 47 to 64°C, and 1 to 4 min of primer extension at 72°C (Table 2). Low-stringency PCR experiments were also carried out in an attempt to amplify regions that gave negative PCR results. The optimum annealing temperatures for each primer pair were reduced by 1 to 5°C. No additional regions were amplified by this method.
TABLE 2.
PCR primers used in this studya
| Primer | Sequence (5′-3′) | Predicted PCR product size (bp) | Annealing temp (°C) |
|---|---|---|---|
| Housekeeping gene primers | |||
| mdh1 | AATGACGCTGGACGAGGTCC | 887 | 62.4 |
| mdh2 | CCTGTTCGACGATGCCTTC | ||
| groEL1 | ATGGCTGCCAAAGAAGTTAAG | 1,434 | 62 |
| groEL2 | GTAGTCTGGCGGCTACCTC | ||
| Virulence gene primers | |||
| exoTF | CAATCATCTCAGCAGAACCC | 1,159 | 54 |
| exoTR | TGTCGTAGAGGATCTCCTG | ||
| phzHF | GGGTTGGGTGGATTACAC | 1,752 | 51 |
| phzHR | CTCACCTGGGTGTTGAAG | ||
| aprF | TGTCCAGCAATTCTCTTGC | 1,017 | 51 |
| aprR | CGTTTTCCACGGTGACC | ||
| lasAF | GCAGCACAAAAGATCCC | 1,075 | 47 |
| lasAR | GAAATGCAGGTGCGGTC | ||
| phzIIF | GCCAAGGTTTGTTGTCGG | 1,036 | 51 |
| phzIIR | CGCATTGACGATATGGAAC | ||
| pvdAF | GACTCAGGCAACTGCAAC | 1,281 | 59 |
| pvdAR | TTCAGGTGCTGGTACAGG | ||
| lasBF | ACAGGTAGAACGCACGGTTG | 1,220 | 57 |
| lasBR | GATCGACGTGTCCAAACTCC | ||
| exoYF | TATCGACGGTCATCGTCAGGT | 1,035 | 61.8 |
| exoYR | TTGATGCACTCGACCAGCAAG | ||
| exoSF | ATCCTCAGGCGTACATCC | 328 | 53 |
| exoSR | ACGACGGCTATCTCTCCAC | ||
| exoUF | GATTCCATCACAGGCTCG | 3,308 | 61.6 |
| exoUR | CTAGCAATGGCACTAATCG | ||
| phzMF | ATGGAGAGCGGGATCGACAG | 875 | 54 |
| phzMR | ATGCGGGTTTCCATCGGCAG | ||
| phzIF | CATCAGCTTAGCAATCCC | 392 | 49 |
| phzIR | CGGAGAAACTTTTCCCTC | ||
| phzSF | TCGCCATGACCGATACGCTC | 1,752 | 63 |
| phzSR | ACAACCTGAGCCAGCCTTCC | ||
| pilAF | ACAGCATCCAACTGAGCG | 1,675 | 59 |
| pilAR | TTGACTTCCTCCAGGCTG | ||
| pilBF | TCGAACTGATGATCGTGG | 408 | 54 |
| pilBR | CTTTCGGAGTGAACATCG | ||
| Prophage primers | |||
| Bacto 1F | CCTGGCTCATCTTAAACAGTC | 2,103 | 59.8 |
| Bacto 1R | TGTTCACCCGAATGGTCTGG | ||
| Pf1F | GGATCAGTGGACATCCTGGTCAAC | 834 | 62 |
| Pf1R | GCGTAGTCGGAGATACAGAGGGAG | ||
| Genomic island primers | |||
| Flagellum | |||
| FlagF | ACAACGGATAAAGGGCAG | 1,689 | 59.5 |
| FlagR | AACGAGCGATTCTCGATG | ||
| ORF FF | CAGTTCTTCACCCGCAAGATCGCC | 946 | 64 |
| ORF FR | CCTCGTAGTCGCCGTGCATGAACG | ||
| PAGI-1 | |||
| ORF 3F | TGGTGCTGACCAGCGACAAG | 958 | 59 |
| ORF 3R | TCCATCGACTCGGTGCGTAG | ||
| ORF 18F | ATTCCTCCACTGCCGTTCACAACG | 1,039 | 58 |
| ORF 18R | CCTTGCTCATCTGGAACAGGTAGC | ||
| ORF 42F | CGGAGAACCATCTCTCGCACAC | 675 | 58 |
| ORF 42R | GGCTAAGACGTTCGACTGATTCC | ||
| PAGI-2 | |||
| C22F | CCTTCGTCCATTACCTGTGGAAC | 943 | 62.4 |
| C22R | AACTTGCGAGCCAACTCACG | ||
| C105F | GATTGATGCTCAACGACGATGG | 681 | 59 |
| C105R | GCTGTTCCGCCTTCAGTTCC | ||
| PAGI-3 | |||
| SG8F | TACAGAGTGCCCGAGCTGATG | 732 | 61 |
| SG8R | GTGCTTCCCTGAGAGACAGACG | ||
| SG100F | GCAATCTGTACGTCCTGCACG | 553 | 60.4 |
| SG100R | AGCACGGCTTGTCGCTGTTC |
All primers used were designed in this study.
Southern blot analysis.
To confirm negative PCR results, Southern hybridization analysis was carried out. DNA from each strain of interest was digested with the restriction enzyme EcoRI (Roche Molecular Biochemicals, East Sussex, United Kingdom) and separated by electrophoresis in 0.6% (wt/vol) 1× Tris-borate-EDTA agarose. Separated DNA fragments were transferred to a nitrocellulose membrane for Southern hybridization. A single DNA probe was generated for each of the regions examined by PCR amplification by using P. aeruginosa strain PAO1 as a template and was radiolabeled with 32P to verify the absence of a particular gene. Southern hybridization was carried out by using a radioactive nucleic acid labeling and detection system according to the work of Sambrook and Russell (43).
RESULTS
Genetic variation at the mdh loci among P. aeruginosa isolates.
To determine the evolutionary genetic relationships among our collection of P. aeruginosa isolates, a 714-bp region of the housekeeping gene mdh (encoding malate dehydrogenase) was analyzed. Previous studies have shown that comparative nucleotide sequence analysis of the mdh locus is a reliable indicator of overall genetic relationships between strains (6). Within the 714-bp region from the 17 P. aeruginosa strains examined, there was a total of 22 polymorphic sites, which included 2 amino acid replacement (nonsynonymous) sites (Table 3). Of the 22 polymorphic sites, all were phylogenetically informative (at least two or more sequences contained the polymorphism). The average pairwise difference for the 17 P. aeruginosa mdh sequences was 1.24%, with a maximum pairwise difference of 2.85% observed between strains CF194, CF208, and CF242, on the one hand, and strains CF95, CF139, CF175, CF177, and CF195, on the other (Table 3). The synonymous (kS) and nonsynonymous (kN) substitution rates at the mdh locus are similar to those for other species (Table 3).
TABLE 3.
Sequence variation at the mdh locus among P. aeruginosa isolates
| Grouping | No. of strains | Fragment size (bp) | No. of polymorphic sites
|
ks ± SE | kN ± SE | ||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Synonymous | Nonsynonymous | Informative | Singleton | |||||
| All P. aeruginosa isolates | 17 | 714 | 22 | 20 | 2 | 22 | 0 | 0.044 ± 0.0099 | 0.0015 ± 0.001 |
| Clinical isolates | 12 | 714 | 21 | 19 | 2 | 21 | 0 | 0.051 ± 0.0116 | 0.0019 ± 0.001 |
| Hospital isolates | 5 | 714 | 4 | 4 | 0 | 1 | 3 | 0.0115 ± 0.005 | 0.000 ± 0.000 |
P. aeruginosa strains CF95, CF139, CF175, CF177, and CF195, all recovered from CF patients, had mdh sequences that were identical to each other. P. aeruginosa strains CF194, CF208, and CF242, recovered from CF patients, also had mdh sequences that were identical to each other. In addition, P. aeruginosa strains BR220 and BR227 (environmental isolates) have mdh sequences identical to that of the sequenced strain PAO1. P. aeruginosa strains CF93 and CF198, both recovered from CF patients, had mdh sequences identical to that of BR764, an environmental isolate. Among the 12 P. aeruginosa clinical isolates for which the mdh locus was examined, there were a total of 21 polymorphic sites, of which 19 were synonymous and 2 were nonsynonymous (Table 3). The average pairwise difference for the mdh sequences of the 12 P. aeruginosa clinical isolates was 1.44%. Among the five P. aeruginosa environmental isolates examined, there was a total of four synonymous polymorphic sites (Table 3). The average pairwise difference for the four P. aeruginosa environmental isolates was 0.30%. From the P. aeruginosa mdh sequences, we constructed a neighbor-joining tree based on synonymous polymorphic sites, that is, sites in a codon predicted not to result in amino acid replacements and therefore not under selective pressure (Fig. 1). Figure 1 represents the overall genetic diversity of the isolates examined. Interestingly, the mdh sequences from clinical and environmental isolates clustered together on the mdh gene tree.
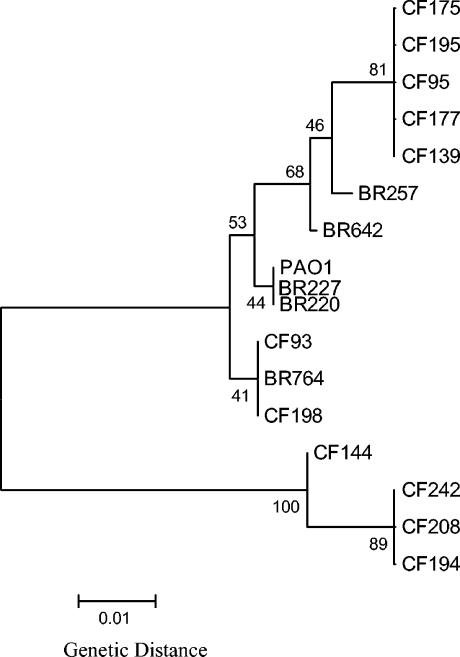
FIG. 1.
Neighbor-joining tree constructed by the Jukes-Cantor method with the nucleotide sequences of mdh gene fragments of P. aeruginosa strains. Construction and bootstrapping of the tree were carried out with the MEGA suite of programs. One thousand bootstrap replicates were performed for each analysis. Bootstrap values are given at the nodes.
Of the 17 P. aeruginosa isolates examined, 8 were serial isolates from single patients. Strains CF177 and CF195, collected on the same day from CF patient 26, had identical mdh sequences and therefore clustered on the same branch of the mdh tree. Similarly, strains CF194 and CF242, collected 39 days apart from CF patient 39, also had mdh sequences identical to each other and clustered together on the most divergent branch of the gene tree with strain CF208. Strains CF208 and CF95 were collected 208 days apart from CF patient 35; based on their mdh sequences, strain CF95 clustered with CF195 and CF177. P. aeruginosa strains CF175 and CF198 were collected 24 days apart from patient 50; strain CF175 clustered with CF195, CF177, CF95, and CF139, whereas strain CF198 clustered with CF93 and an environmental strain, BR764.
Analysis of an additional housekeeping gene, groEL, encoding a chaperonin, from the 17 P. aeruginosa isolates also demonstrated a close genetic relationship among the P. aeruginosa isolates examined. Among the 17 P. aeruginosa groEL sequences, only 2 polymorphic sites were observed within the 337-bp region examined, which does not have the power to distinguish between isolates (data not shown). The comparative sequence data from both the mdh and groEL loci, however, do emphasize the conserved nature of the core P. aeruginosa genome among these isolates.
Phenotype of P. aeruginosa isolates.
To determine if there were phenotypic differences between our P. aeruginosa clinical and environmental isolates, a number of phenotypes associated with virulence were examined (Table 4). First we examined our collection of P. aeruginosa isolates for a range of motility phenotypes and found that all isolates examined were positive in swimming, swarming, and twitching assays. All P. aeruginosa isolates were also positive for protease, HCN, and pyoverdine production, but all isolates were negative for DNase activity. All P. aeruginosa clinical isolates, but only two of the five environmental isolates, BR220 and BR257, were positive for elastase activity. Most isolates, with the exception of CF208 and two environmental isolates (BR642 and BR227), showed lipase activity. In addition, all clinical isolates tested positive for phenazine production; however, all environmental isolates were negative by this assay. Six clinical isolates (CF93, CF144, CF177, CF195, CF198, and CF208) and one environmental isolate (BR227) were negative for gelatinase activity. Four clinical isolates (CF93, CF139, CF144, and CF242) and one environmental isolate (BR642) were negative for rhamnolipid production.
TABLE 4.
Phenotypic analysis of P. aeruginosa clinical and environmental strains
| P. aeruginosa strain | Resulta by the following testb:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| PAO1 | + | + | + | + | − | + | + | + | + | + | + | + |
| CF93 | + | + | + | + | − | + | + | + | − | − | + | + |
| CF95 | + | + | + | + | − | + | + | + | + | + | + | + |
| CF139 | + | + | + | + | − | + | + | + | + | − | + | + |
| CF144 | + | + | + | + | − | + | + | + | − | − | + | + |
| CF175 | + | + | + | + | − | + | + | + | + | + | + | + |
| CF177 | + | + | + | + | − | + | + | + | − | + | + | + |
| CF194 | + | + | + | + | − | + | + | + | + | + | + | + |
| CF195 | + | + | + | + | − | + | + | + | − | + | + | + |
| CF198 | + | + | + | + | − | + | + | + | − | + | + | + |
| CF208 | + | + | + | + | − | + | + | − | − | + | + | + |
| CF242 | + | + | + | + | − | + | + | + | + | − | + | + |
| BR220 | + | + | + | + | − | + | + | + | + | + | − | + |
| BR227 | + | + | + | + | − | + | + | − | − | + | − | − |
| BR257 | + | + | + | + | − | + | + | + | + | + | − | + |
| BR642 | + | + | + | + | − | + | + | − | + | − | − | − |
| BR764 | + | + | + | + | − | + | + | + | + | + | − | − |
+, positive; −, negative.
1, swimming; 2, swarming; 3, twitching; 4, protease; 5, DNase; 6, HCN; 7, pyoverdin; 8, lipase; 9, gelatinase; 10, rhamnolipid; 11, phenazine; 12, elastase.
Molecular analysis of regions associated with virulence.
To examine our P. aeruginosa collection for genotypic differences, we next carried out PCR and Southern hybridization analyses for a range of pathogenicity-associated regions. A total of 15 regions encoding genes required for virulence were examined: 4 genes (exoT, exoU, exoS, and exoY) encoded type III effectors involved in lung injury; 3 genes (phzH, phzM, and phzS) and 2 operons (phzI and phzII) encoded proteins required for phenazine production, which causes cell death; 3 genes (apr, lasA, and lasB) encoded proteases; pvdA encoded l-ornithine hydroxylase, an enzyme in the pyoverdine biosynthetic pathway; and pilA and pilB encoded a type IV fimbrial precursor and a biogenesis protein, respectively. The distribution of the 15 virulence regions did not give any unique profile associated with clinical isolates. Among the clinical isolates, all gave the expected PCR bands with the primer pairs for phzH, phzII, exoS, and exoU, respectively, with the exception of PAO1, which was negative for exoU (Tables 2 and 5). For the six regions exoY, pvdA, lasB, phzM, phzI, and phzS, most clinical isolates were positive, giving the expected PCR bands. Strains CF144, CF194, CF208, CF242, and BR227 were all negative for exoY. Strain CF198 was negative for pvdA; strains CF93, CF95, CF194, and CF212 were negative for lasB; strain CF139 was negative for phzM; strains CF194 and CF189 were negative for phzI; and strains CF194 and CF198 were negative for phzS.
TABLE 5.
Distribution of virulence genes in P. aeruginosa isolates
| P. aeruginosa strain | Presence or absencea of the following virulence gene or prophage (locus or region):
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| exoT (PA0044)b | exoU (PA3831) | exoS (PA3841) | exoY (PA2191) | phzH (PA0051) | phzII (PA1899-1905) | phzM (PA4209) | phzI (PA4210-4216) | phzS (PA4217) | apr (PA1249) | lasA (PA1871) | lasB (PA3724) | pvdA (PA2386) | pilA (PA4525) | pilB (PA4526) | Bacto1 (PA617-620) | Pf1 (PA722-724) | |
| PAO1 | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| CF93 | − | + | + | + | + | + | + | + | + | − | − | − | + | + | − | + | − |
| CF95 | − | + | + | + | + | + | + | + | + | − | − | − | + | − | − | + | + |
| CF139 | − | + | + | + | + | + | − | − | + | + | − | − | + | + | − | − | + |
| CF144 | − | + | + | − | + | + | + | + | + | − | − | + | + | + | − | − | + |
| CF175 | − | + | + | + | + | + | + | + | + | + | − | + | + | − | − | + | + |
| CF177 | − | + | + | + | + | + | + | + | + | + | − | + | + | − | − | + | + |
| CF194 | − | + | + | − | + | + | + | − | − | + | + | − | + | + | − | − | − |
| CF195 | − | + | + | + | + | + | + | + | + | + | − | + | + | − | − | + | + |
| CF198 | − | + | + | + | + | + | + | − | − | − | − | + | − | − | − | + | + |
| CF208 | − | + | + | − | + | + | + | + | + | − | − | + | + | + | − | + | + |
| CF242 | − | + | + | − | + | + | + | + | − | − | + | − | + | + | − | + | − |
| BR220 | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | + |
| BR227 | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | + |
| BR257 | − | + | + | + | + | + | + | + | + | − | − | + | − | − | − | + | + |
| BR642 | − | + | − | + | + | + | + | + | + | − | + | + | + | − | − | + | + |
| BR764 | − | + | − | + | + | + | + | + | + | − | + | + | + | − | − | + | + |
+, presence; −, absence.
PA numbers indicate ORFs on sequenced gene.
For the three regions apr, lasA, and pilA, most isolates were negative by PCR assays and Southern blot analysis. However, strains CF175, CF177, and CF195 were positive for apr, giving the expected PCR band, and strains CF93, CF144, and CF208 were positive for pilA, giving the expected PCR band (see Fig. 4). CF242 was positive for lasA and pilA, giving the expected PCR bands.
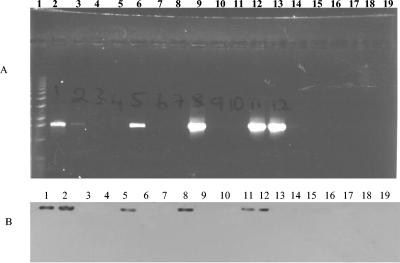
FIG. 4.
PCR and Southern blot analysis of the pilA genes of our P. aeruginosa isolates. (A) PCR analysis. Lane 1, 1-kb marker; lane 2, PAO1 (positive control); lanes 3 to 18, P. aeruginosa isolates CF93, CF95, CF139, CF144, CF175, CF177, CF194, CF195, CF198, CF208, CF242, BR220, BR227, BR257, BR642, and BR764, respectively; lane 19, negative control. (B) Southern blot analysis. Lanes 1 to 17, P. aeruginosa PAO1 (positive control) and isolates CF93, CF95, CF139, CF144, CF175, CF177, CF194, CF195, CF198, CF208, CF242, BR220, BR227, BR257, BR642, and BR764, respectively; lane 19, negative control.
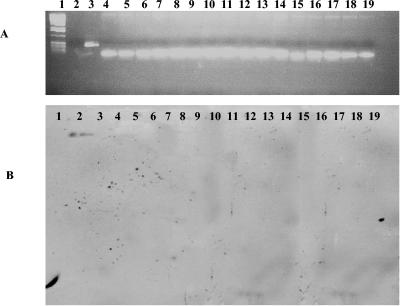
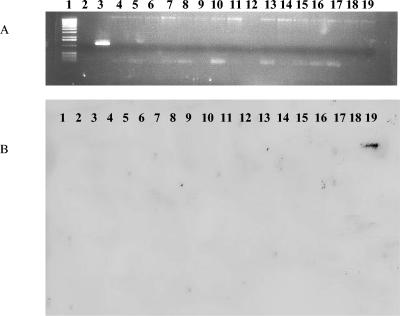
Among some of the environmental isolates, unique profile patterns could be distinguished. For example, strains BR220 and BR227 were negative for 13 of the 15 regions examined. Strain BR220 gave the expected PCR product band with primers for the exoY gene, and the phzH primer pair gave expected PCR product bands for both BR220 and BR227 (Table 2). Strains BR642 and BR764 were also negative by PCR and Southern blot analysis for 5 (exoT, apr, exoS, pilA, and pilB) of the 15 regions examined (Fig. 2, 3, and 4). BR257, a rhizosphere isolate, was negative by PCR assays and Southern blot analysis for six of the regions of interest, exoT, apr, lasA, pvdA, pilA, and pilB. All five P. aeruginosa environmental isolates were negative by PCR assay for the presence of exoT, apr, pilA, and pilB (Fig. 2, 3, and 4).
FIG. 2.
PCR and Southern blot analysis of the pilB genes from our P. aeruginosa isolates. (A) PCR analysis. Lane 1, 1-kb marker; lane 2, negative control; lanes 3 to 19, PAO1 (positive control), CF93, CF95, CF139, CF144, CF175, CF177, CF194, CF195, CF198, CF208, CF242, BR220, BR227, BR257, BR642, and BR764, respectively. (B) Southern blot analysis. Lane 1, negative control; lane 2, PAO1 (positive control); lanes 3 to 19, P. aeruginosa isolates CF93, CF95, CF139, CF144, CF175, CF177, CF194, CF195, CF198, CF208, CF242, BR220, BR227, BR257, BR642, and BR764.
FIG. 3.
PCR and Southern blot analysis of the exoT genes of our P. aeruginosa isolates. (A) PCR analysis. Lane 1, 1-kb marker; lane 2, negative control; lanes 3 to 19, PAO1 (positive control), CF93, CF95, CF139, CF144, CF175, CF177, CF194, CF195, CF198, CF208, CF242, BR220, BR227, BR257, BR642, and BR764, respectively. (B) Southern blot analysis. Lanes 1 to 17, P. aeruginosa isolates CF93, CF95, CF139, CF144, CF175, CF177, CF194, CF195, CF198, CF208, CF242, BR220, BR227, BR257, BR642, and BR764; lane 18, negative control; lane 19, PAO1 (positive control).
The presence or absence of the two prophages (Bacto1 and Pf1) and four P. aeruginosa GEIs (the flagellum island, PAGI-1, PAGI-2, and PAGI-3) (3, 28, 29) among our collection of isolates was examined. With primers for phage Bacto1, most isolates gave the expected PCR band, with the exception of strains CF144, CF194, BR220, and BR227. Most isolates gave the expected PCR band when the primers for phage Pf1 were used, with the exception of strains CF93, CF194, and CF242. None of the P. aeruginosa isolates examined contained PAGI-1, PAGI-2, or PAGI-3. Two P. aeruginosa strains, CF93 and BR642, did contain the flagellum island (data not shown).
DISCUSSION
Genotypic and phenotypic variation within a collection of 17 P. aeruginosa isolates from the airways of CF patients, from a hospital environment, and from a plant rhizosphere was investigated. The evolutionary genetic relationships of P. aeruginosa were examined by comparative sequence analysis of a 714-bp region of the housekeeping gene mdh from each of our isolates. Phylogenetic analysis of the mdh sequences did not reveal distinct and separate branching of clinical and environmental isolates. Clinical and environmental isolates clustered with each other on the mdh tree. In addition, analysis of a second chromosomal locus, groEL, also demonstrated a close genetic relationship among strains and underscored the conserved nature of the core genome. These findings correlate with those of previous studies showing that clinical and environmental isolates do not form distinct lineages. In our study, of the four pairs of serial isolates from CF patients examined, we found that two P. aeruginosa isolates (CF177 and CF195) had identical mdh sequences and virulence profiles as determined by PCR analysis even though they showed different colony morphologies when collected, indicating that these two isolates are morphological variants of the same strain. In contrast, strains CF194 and CF242, which were collected 39 days apart, had identical mdh sequences and phenotypic profiles but different genotypic profiles. Strain CF242 differed from CF194 in that it was positive for phzI and prophage Bacto1 but negative for apr, likely indicating the presence of two independent strains with an identical core genome. Four additional serial P. aeruginosa isolates taken from two CF patients, patient 35 (CF95 and CF208) and patient 50 (CF175 and CF198), showed differing mdh sequences and PCR profiles of virulence regions, suggesting two different strains colonizing the same patient. Whether this is cocolonization or succession of P. aeruginosa isolates was not determined. Individual CF patients colonized by different P. aeruginosa strains have been reported previously (20, 21, 52).
Previously, restriction mapping of the P. aeruginosa genome indicated that large rearrangements, and the acquisition and loss of large blocks of DNA, were responsible for most of the genetic diversity among strains (39, 45). In general, the results of our phylogenetic, phenotypic, and genotypic analyses were consistent with these observations. Much of the P. aeruginosa genome was relatively well conserved based on mdh and groEL sequence analysis. We selected mdh and groEL because both these loci encode basic metabolic housekeeping genes present in all strains of the species and therefore are not under selective pressure, evolve in a neutral manner, and are reliable indicators of genetic relationships. Previously, the mdh locus has been used in several evolutionary studies of a number of enteric bacteria including Escherichia coli, Salmonella enterica, and Vibrio cholerae. These studies demonstrated that phylogenetic trees based on mdh are congruent with analyses of evolutionary relatedness based on other methods and that mdh is a reliable estimate of overall genetic diversity (5, 6, 10). The conserved core P. aeruginosa genome was interrupted in some isolates by the sporadic distribution of a number of virulence-associated regions. Analysis of the 17 P. aeruginosa strains revealed the presence, absence, or divergence of 15 regions associated with virulence, but no specific pattern was associated with strains isolated from the airways of CF patients. However, two of the environmental isolates lacked the majority of the 15 regions examined. In a recent study of the genetic diversity of clinical and environmental P. aeruginosa isolates from a Spanish hospital, Ruiz and colleagues concluded, based on analyses of whole-cell proteins, outer membrane proteins, antibiotic susceptibility patterns, pulsed-field gel electrophoresis, and randomly amplified polymorphic DNA, that there was not a close relationship between clinical and environmental isolates (41). Their findings are not unexpected given that the methods they used to determine genetic diversity were developed to differentiate between isolates and are not good indicators of overall evolutionary genetic relationships among strains. Indeed, none of the five dendrograms presented on the basis of randomly amplified polymorphic DNA analyses was congruent (41).
Of the four type III effector proteins (ExoT, ExoS, ExoU, and ExoY) analyzed, most isolates contained at least two. Feltman and colleagues (14) have suggested that all P. aeruginosa isolates harbor at least some of the genes encoding the type III secretion apparatus but that three of the four type III effector proteins are variable and that the vast majority of isolates contain either the exoS or the exoU gene but not both. Subsequently, Wolfgang et al. (54) showed that 13 out of 19 strains examined contained both the exoU and exoS genes. In agreement with this finding, our study showed that the vast majority of our P. aeruginosa isolates contain both genes. Among the environmental isolates, only BR257, a plant rhizosphere isolate, contained both exoU and exoS; this finding is of interest, because both these genes are involved in phagocytosis and lung injury in the human host.
The majority of the clinical isolates carried all the genes involved in phenazine production, with the exception of CF194, which lacked both phzI and phzS, and CF242, which lacked phzS. Two environmental isolates, BR220 and BR227, were positive for phzH but lacked all the remaining phenazine genes examined. Phenotypic analysis indicated that all the clinical isolates, but none of the environmental isolates, produced phenazines. The presence of phenazine genes does not necessarily correlate with expression. There are more than 50 known phenazine compounds with the same basic structure, differing only in the heterocyclic core. These modifications largely determine the physical properties of phenazines and influence their biological activity against plant and animal pathogens (34). It was found that all P. aeruginosa isolates in our collection were positive for a range of motility phenotypes (swimming, swarming, and twitching). However, PCR analysis of the pilA and pilB genes showed that pilA is present only in six isolates and pilB is present only in the sequenced strain PAO1. Kiewitz and Tummler (25) suggested that pilA was localized in chromosomal regions of extended interclonal variability and did not have consistently high codon adaptation indices, which correlates with low levels of gene expression. In addition, Wolfgang and colleagues (54) identified 24 variable segments, one of which contained pilA and pilB, that were polymorphic in P. aeruginosa strains. The pilB gene product has been shown to be essential for the synthesis of type IV pili, which confer twitching motility (7). Our phenotypic analysis shows that all strains have twitching motility, suggesting the presence of an additional locus involved in twitching motility in P. aeruginosa. Our study also looked at the presence of the two prophages Bacto1 and Pf1 and of four GEIs (the flagellum island, PAGI-1, PAGI-2, and PAGI-3) among isolates. A high proportion of our P. aeruginosa isolates contained both prophages, without any differentiation between clinical and environmental isolates. None of our isolates contained PAGI-1, -2, or -3, but two isolates, CF93 and BR764, contained the flagellum island. These data were consistent with several other studies which showed that the prophages and GEIs were localized to regions of the chromosome that show a high degree of variability (28, 29, 54).
From our analysis, it appears that P. aeruginosa strains recovered from clinical or environmental sources have a highly conserved core genome but are highly variable with regard to the presence of regions involved in the virulence phenotype. Furthermore, as others have suggested, a key component of P. aeruginosa virulence appears to be the regulation of gene expression rather than the presence or absence of genes, since no distinct phenotypic or genotypic profile was found for clinical isolates. In addition, our data demonstrate that the P. aeruginosa genome is a dynamic entity in which horizontal gene transfer and recombination, together with gene loss, are a driving force in genetic diversity and play an important role in the evolution of this species.
Acknowledgments
We are grateful to Claire Adams for supplying the P. aeruginosa isolates from CF patients and to P. Cornelis for the P. aeruginosa environmental isolates.
Research in E.F.B.'s laboratory is funded by a PRTLI-3 grant from the Higher Education Authority of Ireland and by an Enterprise Ireland basic research grant. Research in J.P.M. and F.O.G.'s groups is supported by grants from the European Union (QLK3-2000-31759, QLK3-2001-00101, and QLK5-CT-2002-00914), the HEA PRTLI programs, Enterprise Ireland (SC/02/0520 and SC/02/517), and the Science Foundation of Ireland (02/IN.1/B1261).
REFERENCES
- 1.Adams, C., M. Morris-Quinn, F. McConnell, J. West, B. Lucey, C. Shortt, B. Cryan, J. B. Watson, and F. O'Gara. 1998. Epidemiology and clinical impact of Pseudomonas aeruginosa infection in cystic fibrosis using AP-PCR fingerprinting. J. Infect. 37:151-158. [DOI] [PubMed] [Google Scholar]
- 2.Allewelt, M., F. T. Coleman, M. Grout, G. P. Priebe, and G. B. Pier. 2000. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect. Immun. 68:3998-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. USA 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, E. F., F.-S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, E. F., K. Nelson, F. S. Wang, T. S. Whittam, and R. K. Selander. 1994. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 91:1280-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 8.Brown, M. R., and J. H. Foster. 1970. A simple diagnostic milk medium for Pseudomonas aeruginosa. J. Clin. Pathol. 23:172-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns, J. L., R. L. Gibson, S. McNamara, D. Yim, J. Emerson, M. Rosenfeld, P. Hiatt, K. McCoy, R. Castile, A. L. Smith, and B. W. Ramsey. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444-452. [DOI] [PubMed] [Google Scholar]
- 10.Byun, R., L. D. Elbourne, R. Lan, and P. R. Reeves. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castric, P. A. 1975. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can. J. Microbiol. 21:613-618. [DOI] [PubMed] [Google Scholar]
- 12.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2000. Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochim. Biophys. Acta 1485:145-152. [DOI] [PubMed] [Google Scholar]
- 13.Ernst, R. K., D. A. D'Argenio, J. K. Ichikawa, M. G. Bangera, S. Selgrade, J. L. Burns, P. Hiatt, K. McCoy, M. Brittnacher, A. Kas, D. H. Spencer, M. V. Olson, B. W. Ramsey, S. Lory, and S. I. Miller. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5:1341-1349. [DOI] [PubMed] [Google Scholar]
- 14.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 15.Frank, L. H., and R. D. DeMoss. 1959. On the biosynthesis of pyocyanine. J. Bacteriol. 77:776-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goehring, U. M., G. Schmidt, K. J. Pederson, K. Aktories, and J. T. Barbieri. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 274:36369-36372. [DOI] [PubMed] [Google Scholar]
- 17.Hacker, J., U. Hentschel, and U. Dobrindt. 2003. Prokaryotic chromosomes and disease. Science 301:790-793. [DOI] [PubMed] [Google Scholar]
- 18.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 19.Hamood, A. N., J. Griswold, and J. Colmer. 1996. Characterization of elastase-deficient clinical isolates of Pseudomonas aeruginosa. Infect. Immun. 64:3154-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haussler, S., B. Tummler, H. Weissbrodt, M. Rohde, and I. Steinmetz. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621-625. [DOI] [PubMed] [Google Scholar]
- 21.Haussler, S., I. Ziegler, A. Lottel, F. von Gotz, M. Rohde, D. Wehmhohner, S. Saravanamuthu, B. Tummler, and I. Steinmetz. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J. Med. Microbiol. 52:295-301. [DOI] [PubMed] [Google Scholar]
- 22.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 23.Hofte, M., S. Buysens, N. Koedam, and P. Cornelis. 1993. Zinc affects siderophore-mediated high affinity iron uptake systems in the rhizosphere Pseudomonas aeruginosa 7NSK2. Biometals 6:85-91. [DOI] [PubMed] [Google Scholar]
- 24.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism III. Academic Press, New York, N.Y.
- 25.Kiewitz, C., and B. Tummler. 2000. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182:3125-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 27.Kurahashi, K., O. Kajikawa, T. Sawa, M. Ohara, M. A. Gropper, D. W. Frank, T. R. Martin, and J. P. Wiener-Kronish. 1999. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Investig. 104:743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tummler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, X., X. Q. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, C., E. F. Boyd, R. Quentin, P. Massicot, and R. K. Selander. 1999. Enzyme polymorphism in Pseudomonas aeruginosa strains recovered from cystic fibrosis patients in France. Microbiology 145:2587-2594. [DOI] [PubMed] [Google Scholar]
- 31.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIver, K. S., J. C. Olson, and D. E. Ohman. 1993. Pseudomonas aeruginosa lasB1 mutants produce an elastase, substituted at active-site His-223, that is defective in activity, processing, and secretion. J. Bacteriol. 175:4008-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morihara, K. 1964. Production of elastase and proteinase by Pseudomonas aeruginosa. J. Bacteriol. 88:745-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrissey, J. P., C. Cullinane, A. Abbas, L. Mark, and F. O'Gara. 2004. Biosynthesis and regulation of antifungal metabolites by pseudomonads, p. 635-670. In J. L. Ramos (ed.), The Pseudomonads, vol. III. Biosynthesis of macromolecules and molecular metabolism. Kluyver Press, Dordrecht, The Netherlands.
- 35.Nakayama, K., S. Kanaya, M. Ohnishi, Y. Terawaki, and T. Hayashi. 1999. The complete nucleotide sequence of φCTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol. Microbiol. 31:399-419. [DOI] [PubMed] [Google Scholar]
- 36.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirnay, J. P., D. De Vos, C. Cochez, F. Bilocq, J. Pirson, M. Struelens, L. Duinslaeger, P. Cornelis, M. Zizi, and A. Vanderkelen. 2003. Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug-resistant clone and a silver sulfadiazine-resistant clone. J. Clin. Microbiol. 41:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pirnay, J. P., D. De Vos, C. Cochez, F. Bilocq, A. Vanderkelen, M. Zizi, B. Ghysels, and P. Cornelis. 2002. Pseudomonas aeruginosa displays an epidemic population structure. Environ. Microbiol. 4:898-911. [DOI] [PubMed] [Google Scholar]
- 39.Romling, U., K. D. Schmidt, and B. Tummler. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386-404. [DOI] [PubMed] [Google Scholar]
- 40.Romling, U., J. Wingender, H. Muller, and B. Tummler. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz, L., M. A. Dominguez, N. Ruiz, and M. Vinas. 2004. Relationship between clinical and environmental isolates of Pseudomonas aeruginosa in a hospital setting. Arch. Med. Res. 35:251-257. [DOI] [PubMed] [Google Scholar]
- 42.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Schmidt, K. D., B. Tummler, and U. Romling. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegmund, L., and F. Wagner. 1991. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 5:265-268. [Google Scholar]
- 46.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 47.Smirnov, V. V., E. A. Kiprianova, A. D. Garagulia, T. A. Dodatko, and I. I. Piliashenko. 1990. Antibiotic activity and siderophores of Pseudomonas cepacia. Prikl. Biokhim. Mikrobiol. 26:75-80. [PubMed] [Google Scholar]
- 48.Sokol, P. A., D. E. Ohman, and B. H. Iglewski. 1979. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J. Clin. Microbiol. 9:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wehmhoner, D., S. Haussler, B. Tummler, L. Jansch, F. Bredenbruch, J. Wehland, and I. Steinmetz. 2003. Inter- and intraclonal diversity of the Pseudomonas aeruginosa proteome manifests within the secretome. J. Bacteriol. 185:5807-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 54.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]