Abstract
We investigated the emergence and spread of community-associated strains of methicillin-resistant Staphylococcus aureus (CA-MRSA) in central and northern Wisconsin by determining the temporal and clonal relationships and geographic expansion among 581 of 956 clinical isolates of MRSA collected between 1989 and 1999. Based on EcoRI plasmid profiles (PP), two types, PP-11 and PP-13, were highly stable over time and were consistently associated with multidrug-sensitive strains recovered from outpatients treated at Native American community clinics. Pulsed-field gel electrophoresis (PFGE) yielded six major clonal groups (MCGs) and 19 minor clonal groups. The six MCGs represented 82% of the isolates. All strains with either PP-11 or -13 were present in MCG-2. Eighty-nine percent of the isolates in MCG-2 originated from Native American clinics, and 90% belonged to two PFGE types (19 and 20), the types associated with an outbreak of MRSA in a Native American community in 1992. MCG-2 isolates were multidrug sensitive, harbored type IVa staphylococcal cassette chromosome mec, and were very closely related by PFGE to the Midwestern CA-MRSA strain MW2. MCG-2 strains were mostly obtained from skin infections and affected patients with a mean age of 24 (±18.0) years. MCG-2 strains spread to four additional Native American communities and 20 other communities. Our findings suggest that CA-MRSA in Wisconsin likely originated in Native American communities in the early 1990s and since has become widespread throughout the state. Two early CA-MRSA strains (WI-33 and WI-34) in Wisconsin represent progenitors of the MW2 strain, based on their almost indistinguishable genotypic characteristics.
For many years, methicillin-resistant Staphylococcus aureus (MRSA) was considered a multidrug-resistant pathogen that was strongly associated with infections in individuals with established risk factors, the most important of which was recent hospitalization (7, 9). Surveillance in U.S. hospitals indicated that staphylococcal infections due to MRSA increased from 2.4% in 1975 to 54.5% in 1999 (31, 35). More recently, there have been reports of community-associated MRSA (CA-MRSA) strains affecting individuals who have few risk factors for infection. As CA-MRSA strains have become more thoroughly characterized, it has been found that CA-MRSA strains are less likely to be resistant to multiple antibiotic classes and are more often associated with superficial skin and soft tissue infections than hospital-associated MRSA (HA-MRSA) strains (17, 19, 27). Furthermore, strains of MRSA that originate in the community appear to have molecular features that are quite distinct from those of HA-MRSA. These include unique pulsed-field gel electrophoresis (PFGE) clonal types (1, 2, 15, 21, 28), the presence of type IV staphylococcal cassette chromosome mec (SCCmec) (12, 34), and the presence of the Panton-Valentine leukocidin (pvl) genes (22, 47). Recently, our group showed that almost all CA-MRSA strains in rural Wisconsin contain both SCCmec IV and pvl genes (41).
Certain ethnic groups are reported to have a propensity for the acquisition of CA-MRSA. These include Native American populations in the Midwest United States (15, 18) and aboriginals in Canada (14, 44) and Australia (24, 46). CA-MRSA infections have also been reported for other patient populations across many regions of the United States, including Minnesota and North Dakota (8, 28), Illinois (16, 19, 43), Texas (2, 27), Hawaii (17), Nebraska (15), and California (10). In states adjacent to Wisconsin, studies have documented CA-MRSA infections in pediatric populations (16, 19), including four deaths (8) and an increased incidence among young individuals (median age range, 16 to 20 years) (28). The CA-MRSA strain MW2 is a hypervirulent strain that was responsible for the death of a Native American child in North Dakota in 1998 (4, 8) and has been linked to several other reports (15, 18, 28). The MW2 strain has been classified as part of the USA400 clonal group in a proposed MRSA database in the United States (26).
Although MRSA was detected in the United States as early as 1968 (6), clinical isolates of MRSA were not identified at Marshfield Laboratories until 1989. Marshfield Laboratories is a large reference laboratory that serves a significant portion of rural central and northern Wisconsin. Since 1989, the number of MRSA cases has increased dramatically, and currently about 20% of S. aureus isolates in our laboratory are resistant to methicillin. In the spring of 1992, we detected an outbreak of multidrug-sensitive MRSA among outpatients in a Native American community in rural Wisconsin (42). At that time, we had begun using EcoRI restriction analysis of plasmid DNA as a genetic typing method and determined that the outbreak isolates shared a common restriction profile. Although only a limited epidemiologic investigation was undertaken, the index cases were believed to be children with impetigo who attended a day care center. Subsequent phenotypic and genetic characterizations of these early isolates were consistent with CA-MRSA (M. E. Stemper, S. K. Shukla, and K. D. Reed, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. C-296, 2002). The purpose of this study was to determine the temporal and geographic spread of CA-MRSA strains in central and northern Wisconsin for the period of 1989 to 1999.
MATERIALS AND METHODS
MRSA isolates and their identification.
A total of 956 clinical strains of MRSA were collected between October 1989 and December 1999. Of the 956 isolates, 581 were stored at −70°C and were available for plasmid and PFGE typing. The remaining 375 isolates were recovered exclusively from patients who were treated at five Native American clinics between 1994 and 1999, and these samples were not saved in frozen storage. All strains (n = 956) were recovered from clinical specimens and occasional surveillance cultures submitted to our reference laboratory for routine microbiological testing. The isolates were identified as S. aureus by standard laboratory methods, including colony morphology determination, Gram staining, and tests for catalase and coagulase.
This investigation was approved by the Institutional Review Board of Marshfield Clinic Research Foundation.
Demographic data.
Data collected for each isolate included the date of collection, the specimen source, the patient's age and sex, and the submitting healthcare facility. Duplicate isolates from the same patients were excluded. The healthcare facilities were categorized as private clinics, hospitals, or long-term-care facilities. The type of infection associated with each isolate was defined as invasive or noninvasive based upon the source of the clinical specimen. For the purposes of this study, isolates recovered from blood, spinal fluid, sputum, tracheal or bronchial aspirates, and urine sources were considered invasive. Isolates from all other sources, such as skin and soft tissue infections, were considered noninvasive.
Susceptibility testing.
Antimicrobial susceptibilities were determined for all 956 isolates by broth microdilution with the Vitek system (bioMèrieux Inc., Durham, N.C.) according to the protocols of the National Committee for Clinical Laboratory Standards (30). The antimicrobial agents tested included penicillin, oxacillin (OXA), ampicillin, cephalothin, erythromycin (ERY), ciprofloxacin (CIP), gentamicin, clindamycin (CLI), tetracycline (TET), trimethoprim-sulfamethoxazole (SXT), and rifampin. Isolates were considered resistant to methicillin if the OXA MIC was ≥4 μg/ml. OXA resistance was confirmed by the use of OXA screen agar (Becton Dickinson, Franklin Lakes, N.J.).
Molecular methods. (i) Plasmid analysis.
The extraction of plasmid DNAs and the separation of EcoRI restriction fragments were performed as described previously (36, 37).
(ii) PFGE.
Preparations of chromosomal DNAs were performed by modifications of previously described methods (5, 25). The modifications included making cell suspensions from overnight growth on blood agar plates. Plugs were made by adding 4 μl of lysostaphin (1 mg/ml) to 250 μl of cell suspension and combining this mixture with 250 μl of 1.8% SeaPlaque agarose. The plugs were incubated for 4 h at 37°C in lysis buffer followed by an overnight incubation at 55°C in proteinase K buffer. DNAs were digested with 35 U of SmaI at 25°C for 3 h (without agitation). Fragments were resolved in 1% SeaKem agarose in 0.5× Tris-borate-EDTA buffer at 14°C with a CHEF-DRIII system (Bio-Rad Laboratories, Hercules, Calif.) for 20 h at 6 V/cm, with pulse times ramping from 5 to 40 s. The gels were poststained with ethidium bromide and digitally imaged with a Gel Doc 2000 system (Bio-Rad Laboratories). A SmaI digest of DNA from S. aureus NCTC 8325 was applied to multiple lanes of each gel as a reference standard.
(iii) SCCmec typing.
Representative isolates from each major clonal group (MCG) were characterized by a PCR assay for SCCmec. The method was based on a modification (41) of the multiplexed PCR assay of Oliveira and de Lencastre (34).
Analysis of genetic relatedness.
PFGE patterns were analyzed with Molecular Analyst Fingerprinting Plus, version 1.1, software (Bio-Rad Laboratories). The patterns were also analyzed visually to resolve occasional ambiguities in band assignments. Dendrograms were created by means of the Dice coefficient and the unweighted pair group method using averages. The position tolerance between two bands for them to be considered a match was set to 1.5%. The evaluation of genetic relatedness among the strains was based on the following definitions: (i) clonal groups were defined as isolates having PFGE patterns with six or fewer band differences based on the criteria of Tenover et al. (45) and having a similarity index of ≥80%, (ii) MCGs were defined as groups that contained ≥5% of the study isolates, and (iii) minor clonal groups (mCGs) were MRSA groups that accounted for <5% of the study isolates. Isolates were assigned PFGE patterns and were placed into clonal groups based on the order in which they had been collected. The CA-MRSA strain MW2 was used for comparison (a kind gift from Timothy Naimi of the Minnesota Department of Health).
Statistical analysis.
Associations between MCGs, patient healthcare facilities, and isolate invasiveness were analyzed by the two-tailed Fisher exact test (StatXact-3; Cytel Software Corporation, Cambridge, Mass.). P values of ≤0.05 were considered statistically significant. The age distribution for the MCGs was analyzed by the use of SAS software (version 8.2; SAS Institute, Inc., Cary, N.C.).
RESULTS
MRSA isolates and demographic data.
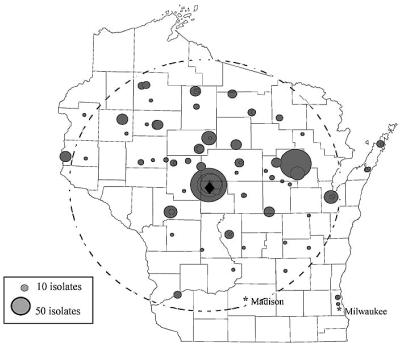
A total of 956 MRSA isolates were collected between 1989 and 1999. These isolates were submitted from 77 Wisconsin healthcare facilities located in 33 counties within an approximately 160-mile radius of Marshfield, Wis. (Fig. 1). The submitting facilities included private clinics (64%), hospitals (23%), and long-term-care facilities (13%). The isolates came from a patient population consisting of 51% males and 49% females. Of the 956 isolates, 581 were available for molecular subtyping by PFGE and plasmid analysis.
FIG. 1.
Map of Wisconsin showing the 77 patient care facilities that submitted clinical specimens containing MRSA between 1989 and 1999. Each circle represents a patient care facility. The sizes of the circles are proportional to the numbers of isolates collected from the sites. The dashed circle indicates a 160-mile radius from Marshfield (♦).
Description of an MRSA outbreak in a single Native American clinic.
Between April and December 1992, 20 cases of MRSA infections were identified among patients seen at a small Native American clinic designated NAC-A. All of the isolates had the same antimicrobial susceptibility pattern, showing resistance only to ERY in addition to the β-lactam antibiotics. The initial genetic typing of these isolates in 1992 was done by plasmid analysis, the only molecular typing method available in our laboratory at that time. Remarkably, all 20 isolates had the same plasmid profile (PP), PP-11. The first two MRSA index strains, WI-33 and WI-34, were recovered on 21 April and 6 May 1992 from skin wounds on the arm of a 16-year-old girl and the knee of a 7-year-old boy, respectively. The two children were not related and neither had known risk factors for MRSA infection. Laboratory records showed that there were no previously documented reports of MRSA in this community prior to these index cases. Shortly after the onset of the outbreak, MRSA strains with a similar multidrug-sensitive antibiogram were isolated from additional patients in this community. These strains had PP-13, which had a similar band pattern to PP-11 but lacked two fragments. During the course of the decade, both PP-11 and PP-13 strains were observed in other clinics serving primarily Native American communities.
Molecular analysis.
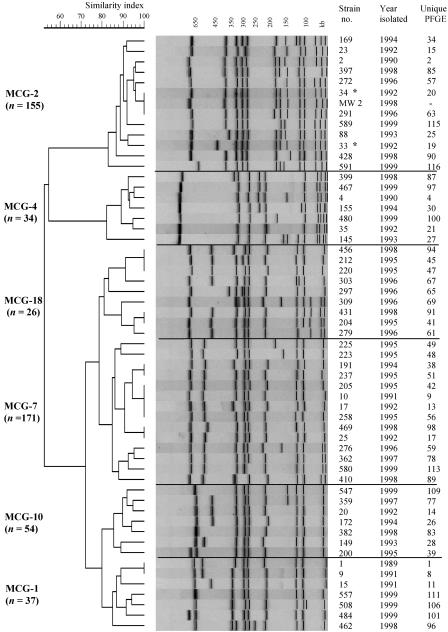
The clonal relationship of 581 MRSA isolates was determined by SmaI PFGE analysis. PFGE identified 109 unique macrorestriction patterns based on one or more band differences. A total of 25 clonal groups were established based on related PFGE patterns with six or fewer fragment differences. Eighty-two percent of the isolates clustered into six MCGs, designated MCG-1 (6.4%), MCG-2 (26.6%), MCG-4 (5.8%), MCG-7 (29.4%), MCG-10 (9.3%), and MCG-18 (5.0%) (Fig. 2). The number of PFGE types present in the different MCGs ranged from 7 to 14. The PFGE patterns within each MCG had a genetic similarity index of ≥80.0% and were represented by ≥5.0% of the study isolates. The remaining 18% of the isolates were classified into 19 mCGs.
FIG. 2.
Dendrogram of representative MRSA isolates from six major clonal groups (MCGs) based on the unweighted pair group method using averages and the Dice similarity coefficient. The number of isolates (n) within each MCG is indicated along the left margin. The first column on the right identifies MRSA strain numbers. The second column indicates the years of isolation. The third column describes the unique PFGE designations present within the MCGs. MW2 is a CA-MRSA control strain. The two asterisks (*) identify the index cases of CA-MRSA from the Native American community outbreak in 1992. The horizontal lines across the dendrogram were drawn to separate each clonal group.
The resulting dendrogram revealed that MCG-2 was genetically divergent from the other Wisconsin MCGs, as it had a similarity index of <60% (Fig. 2). MCG-2 was comprised of 12 closely related PFGE patterns (similarity index, >83%) representing 155 isolates, including the outbreak index isolates WI-33 and WI-34. Even though the outbreak strains had the same PP, their PFGE types differed by two bands, consistent with a single genetic event (45). Eighty-nine percent of the isolates in MCG-2 (n = 155) were either PFGE type 19 (PFGE 19) (16%) or PFGE 20 (73%). All of the MCG-2 isolates were closely related (three or fewer band differences) to PFGE 20. In addition, isolate WI-34 (PFGE 20) was indistinguishable by PFGE from the MW2 strain, but it had a different PP. However, strain WI-99, isolated in 1993 from a patient in the outbreak community, was the earliest strain in MCG-2 to have an indistinguishable PFGE pattern in addition to the same PP (PP-13) as the MW2 strain.
Stability of PPs among MCG-2 strains.
PPs were determined for all of the isolates in this investigation (data not shown) and were compared to the clonality of strains based on PFGE. We observed a strong correlation between MCG-2 and PP-11 or PP-13. In fact, 96% of the isolates in MCG-2 (n = 155) had PP-11 (74%) or PP-13 (22%), while the remaining 4% represented six isolates with unique PPs. PP-11 and -13 remained stable within MCG-2 during the course of the decade. Furthermore, neither of these plasmid types was identified in strains belonging to other clonal groups. The presence of PP-11 also consistently correlated with resistance to ERY, while strains with PP-13 were sensitive to all antibiotic classes other than the β-lactams.
Antimicrobial susceptibilities.
There were 24 antibiogram patterns among the 956 study isolates. Sixty-nine percent of the isolates were sensitive to all but one or two antimicrobial agents (typically ERY) in addition to the β-lactam antibiotics. Strains were rarely resistant to gentamicin (1.7%) or rifampin (1.3%). The antibiograms of MRSA isolates within the MCGs are shown in Table 1. MCG-2 isolates were susceptible to multiple antibiotic classes, including CIP, CLI, TET, and SXT. ERY resistance was present in 73% of the isolates in this group. Isolates belonging to MCG-2 did not show an increasing trend in the resistance pattern but rather remained multidrug sensitive throughout the study period. In contrast, >65% of the MRSA isolates within the other five MCGs, MCG-1, -4, -7, -10, and -18, were resistant to three or more antibiotic classes, including CIP, CLI, and ERY. The average levels of resistance to CIP, CLI, and ERY among these five MCGs were 93, 82, and 94%, respectively.
TABLE 1.
Antimicrobial resistance patterns and clinical data for MRSA isolates within the six major clonal groups
| MCG | n | Mean age of patients | % Antimicrobial resistance
|
No. (%) of MRSA isolates by healthcare site
|
% MRSA infections
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | CLI | ERY | TET | SXT | Clinic | Hospital | LTCFa | P | Invasiveb | Noninvasive | P | |||
| 2c | 155 | 24.3 | 0.0 | 0.6 | 73.1 | 2.6 | 0.6 | 121 (78.0) | 34 (21.9) | 0 (0) | 3.2 | 96.8 | ||
| 1 | 37 | 70.6 | 78.4 | 81.1 | 91.9 | 0.0 | 5.4 | 17 (45.9) | 20 (54.0) | 0 (0) | 0.0002 | 32.4 | 67.6 | <0.0001 |
| 4 | 34 | 71.0 | 91.7 | 88.9 | 97.2 | 0.0 | 8.3 | 9 (26.5) | 23 (67.6) | 2 (5.8) | <0.0001 | 44.4 | 55.6 | <0.0001 |
| 7 | 171 | 73.6 | 98.8 | 66.3 | 95.9 | 1.7 | 3.5 | 51 (29.8) | 56 (32.7) | 64 (37.4) | <0.0001 | 36.0 | 64.0 | <0.0001 |
| 10 | 54 | 74.6 | 98.2 | 88.9 | 94.4 | 0.0 | 5.6 | 12 (22.2) | 21 (38.9) | 21 (38.9) | <0.0001 | 50.0 | 50.0 | <0.0001 |
| 18 | 26 | 69.2 | 100.0 | 84.6 | 88.5 | 0.0 | 3.9 | 11 (42.3) | 12 (46.2) | 3 (1.2) | 0.0005 | 34.6 | 65.4 | <0.0001 |
LTCF, long-term-care facility.
Based on source (blood, spinal fluid, sputum, endotracheal sample, urine).
Reference group.
MCG-2 strains are CA-MRSA.
Isolates belonging to MCG-2 were designated as CA-MRSA based on multiple established phenotypic and genotypic features associated with CA-MRSA strains. All of the isolates had multidrug-sensitive antibiograms and PFGE patterns that were indistinguishable from or within one to three band differences of the MW2 strain. In addition, the SCCmec types were determined for 49 isolates within MCG-2. Ninety-eight percent of the isolates harbored type IVa SCCmec, including the index strains. In contrast, 48 isolates representing the other five MCGs harbored type II SCCmec (41). Type II SCCmec has been shown to be more commonly associated with HA-MRSA (29). Further evidence that MCG-2 is CA-MRSA is based on the following demographic data and clinical presentations of infections associated with this group of MRSA.
(i) Age distribution.
The mean age of patients with isolates belonging to MCG-2 was 24.3 ± 18.0 years (Table 1). This was compared to an average mean age of 72.0 ± 21.0 years for patients with isolates in MCG-1, -4, -7, -10, and -18.
(ii) Healthcare setting.
A correlation was observed between outpatient healthcare settings and MCG-2 isolates compared to all other clonal groups (Table 1). Seventy-eight percent of the isolates in MCG-2 were recovered from patients who were seen in outpatient clinic settings. These data were statistically significant compared to the number of isolates in the other five MCGs that came from outpatient clinic settings (P < 0.001).
(iii) Isolate invasiveness.
The type of infection (i.e., invasive or noninvasive [see Materials and Methods]) caused by each MRSA strain was correlated with the MCG by PFGE (Table 1). Ninety-seven percent of infections caused by MCG-2 isolates were skin related. Only 3.2% (n = 155) of the isolates in MCG-2 were considered invasive, compared to 32.4 to 50.0% invasive isolates within the other clonal groups (P < 0.001). A seasonal trend in infections caused by MCG-2 isolates was noticed, with the incidence of infections increasing during the summer and fall (data not shown). This was possibly due to more skin exposure during the summer and to vulnerability to secondary infection of wounds such as insect bites.
Earliest CA-MRSA strains in our investigation.
We recognized two isolates in MCG-2, WI-2 and WI-23, that occurred prior to the 1992 outbreak. The first isolate, WI-2, was from the nares of a 46-year-old male from Wood County in central Wisconsin in November 1990. This strain was sensitive to all non-β-lactam antibiotics except TET. The isolate had PFGE 2 and PP-5 and harbored SCCmec IVa. The second strain, WI-23, was identified in January 1992 in a tracheal culture of a 1-year-old male from Forest County in northeast Wisconsin. This isolate was sensitive to all non-β-lactam antibiotics and had PP-9 and PFGE 15, respectively. It harbored SCCmec IV, although not the IVa subtype. Both isolates had PFGE patterns that were closely related to PFGE 20, but they possessed unique genotypic and phenotypic features compared to the other CA-MRSA strains in our collection. No definitive epidemiologic link could be identified between these patients and the outbreak cases.
Emergence and geographic spread of CA-MRSA from a Native American community.
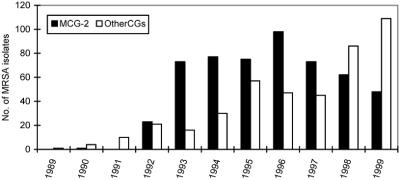
The yearly distribution of the 956 isolates during this study period is shown in Fig. 3. The data are divided into isolates in MCG-2 and those in all other MCGs and mCGs. The emergence of MCG-2 isolates corresponded to the recognition of the MRSA outbreak in 1992. Since 98.2% (n = 110) of multidrug-sensitive MRSA isolates that were submitted from Native American clinics clustered in MCG-2, it is very likely that the 375 MRSA isolates with multidrug-sensitive antibiograms that were recovered from patients at the same five clinics between 1994 and 1999 but were unavailable for PFGE were also members of MCG-2. Altogether, the incidence of MCG-2 peaked in 1996 and then declined between 1997 and 1999, as a steady increase in MRSA groups other than MCG-2 was observed (Fig. 3). MCG-2 represented the most predominant clonal group during the study period, with a total isolation rate of 55.4% (n = 956). Eighty-nine percent (n = 530) of the patients with MCG-2 MRSA were seen at Native American facilities. Only 10.7% of patients with MCG-2 strains were identified in other rural Wisconsin communities. These patients had a similar average age of 26 years, and they had no apparent differences in the epidemiology of their CA-MRSA.
FIG. 3.
Frequency distribution of MRSA isolates in Wisconsin between 1989 and 1999 at Marshfield Laboratories. The solid bars represent MCG-2 isolates. The open bars specify isolates belonging to all other MCGs and mCGs.
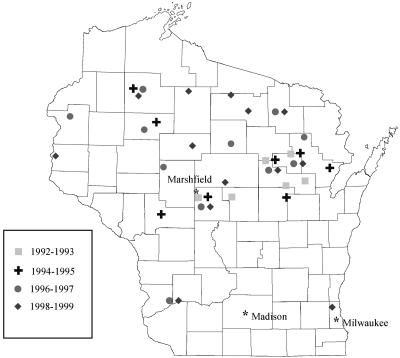
The geographic distribution of MCG-2 MRSA isolates within Wisconsin expanded over the course of the study period (Fig. 4). New locations were identified each year after the onset of the outbreak. Altogether, strains belonging to MCG-2 were also identified in 24 new Wisconsin communities. Four of these locations were Native American communities identified between 1993 and 1995. The remaining 20 Wisconsin communities were identified during the following 2-year periods, with an incidence of 4, 3, 6, and 7 communities, respectively: 1992-1993, 1994-1995, 1996-1997, and 1998-1999. Most of these communities were rural locations within an approximately 45-mile distance from a Native American community. Collectively, the spread of MCG-2 encompassed 21 of the 33 counties that submitted cultures that were positive for MRSA. All but 2 of the 20 counties lie in the northern half of Wisconsin (Fig. 4).
FIG. 4.
Map of Wisconsin showing the temporal and geographic distribution of MCG-2 isolates throughout the period 1992-1999. Each symbol represents the time period in which the isolates were identified.
DISCUSSION
This retrospective investigation of 956 isolates from 1989 to 1999 provides the first comprehensive epidemiological analysis of MRSA strains from patients in the largely rural area of central and northern Wisconsin. During the 11-year study period, an outbreak of CA-MRSA occurred in a Native American community in 1992, and over the course of the decade, the outbreak strain spread geographically to other locations in Wisconsin. Our retrospective analysis provided a unique opportunity to study the clonal relationship of MRSA strains at a time when CA-MRSA was being recognized as an emerging health concern. Furthermore, this study provides evidence that the outbreak of CA-MRSA in Wisconsin in 1992 predated reports from the late 1990s of the presence of CA-MRSA in neighboring states of Wisconsin (8, 19, 28).
Genetic relationships based on plasmid analysis led to the earliest evidence of the MRSA outbreak in 1992. Even though the utility of plasmid analysis for MRSA outbreak investigations has been described previously (23, 38, 48), we recognize that plasmid typing is no longer a widely used tool for molecular epidemiology. However, in our case, EcoRI PP data showed the presence of two distinct PPs that were consistently associated with CA-MRSA in MCG-2 throughout the decade. The fact that these plasmid types remained stable in MCG-2 suggests that they may provide additional fitness factors that give strains a selective advantage. The correlation between one of the PPs and ERY resistance also suggests that there are plasmid-mediated determinants involved in the expression of ERY resistance. Recently, O'Brien et al. (32) described inducible ERY resistance associated with a 2-kb plasmid in CA-MRSA in Australia.
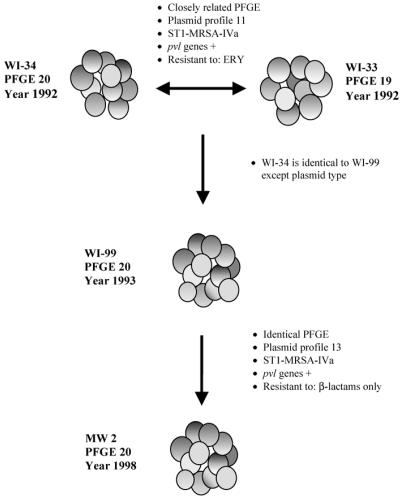
Our SmaI PFGE data for 581 isolates showed the clustering of 82% of the isolates into six MCGs. MCG-2 was quite divergent from the other MCGs in Wisconsin based on its similarity index of <60% (Fig. 2). Several of the other MCGs contained multidrug-resistant strains of MRSA that are known to be endemic to a nursing home setting (13), supporting the likelihood that these MCGs represent HA-MRSA. Indeed, we recently showed that representative isolates within these five MCGs had molecular features of HA-MRSA (41). The two most prevalent PFGE patterns within MCG-2, PFGE 19 and 20, not only characterized the community outbreak isolates, but were also indistinguishable from or closely related by PFGE to the well-characterized CA-MRSA strain MW2. Since MW2 has been described as part of the lineage of the USA400 clonal group established by the Centers for Disease Control (26), MCG-2 in Wisconsin corresponds to this clonal group. In addition to related PFGE patterns, the outbreak index strains WI-33 and WI-34 in MCG-2 shared multiple common features, including the SCCmec IVa type. Specifically, they also possessed a multilocus-sequence-type allelic profile 1-1-1-1-1-1-1 (ST1) and the pvl genes (41). In an evolutionary context, the clonal expansion within MCG-2 appeared to have favored the genetic background of strain WI-34 over that of the related WI-33 strain given the fact that all of the isolates in the group were closely related to PFGE 20. This suggests that strain WI-34 was ecologically more successful. Since strains WI-34 and WI-99 had an identical PFGE 20 pattern and SCCmec IVa type, we suggest that WI-34 was probably the precursor strain of WI-99 identified in 1993 (Fig. 5). Furthermore, WI-99 has identical features, including its antibiogram, PFGE type, SCCmec type, and PP, as the MW2 strain (Fig. 5). Analogous deletions were also detected in the mecI and mecR1 genes of strain WI-99 compared to the MW2 strain (40). Taken together, the events described in Fig. 5 show that strain WI-34 was likely the progenitor of the MW2 strain and that WI-99, which has been in Wisconsin since 1993, appears identical to MW2.
FIG. 5.
Schematic diagram showing suggested precursor and predecessor strains of the MW2 strain. Strains WI-33 and WI-34 share the same common phenotypic and genotypic features, except that they have different PFGE profiles due to a single genetic event. “ST1-MRSA-IVa” represents strains with the multilocus-sequence-type allelic profile 1-1-1-1-1-1-1 and SCCmec type IVa. The bidirectional arrow between WI-33 and WI-34 suggests that either strain could have been a precursor to the other. The straight arrows suggest definitive events. Strain WI-99 is genetically identical to WI-34, except that it has a different plasmid type. The MW2 strain has the same phenotypic and genotypic features as strain WI-99.
The geographic expansion of MCG-2 during the study period was remarkable. After the 1992 outbreak, MCG-2 isolates spread to 24 additional communities in the state. The temporal pattern of dissemination over time coincided with the geography of five Native American communities and their surrounding vicinities in central and northern Wisconsin. This geographic relationship suggests that residents in Native American communities were one of the reservoirs for CA-MRSA within Wisconsin. The fact that nearly 90% of the patients infected with MCG-2 isolates were treated at one of these Native American clinics also suggests that this population is at a higher risk of CA-MRSA infection. Whether Native Americans are essentially predisposed to acquiring S. aureus is not known at this time. However, some authors have speculated about risk factors among Native Americans, including depressed socioeconomic levels, overcrowded living conditions, sanitation concerns, limited access to health care, and empirical use of antibiotics (3, 11, 14, 18, 28). The majority of the individuals seeking medical care at a Native American clinic are likely of Native American ethnicity. However, the true ethnic prevalence within this clonal group could not be ascertained, as patient medical records were unavailable for review. A study by Groom et al. (18), which included medical record reviews, similarly showed that CA-MRSA strains in an American Indian population in the Midwest were clonally related to MW2. Fey et al. (15) further described the CA-MRSA in a Native American community in Nebraska as indistinguishable from or highly related to the CA-MRSA reported by Groom et al. (18) and Naimi et al. (28).
This is the first large-scale investigation of temporal MRSA isolates from Wisconsin. Our data indicate that CA-MRSA strains were present as early as 1990 and accounted for more than half (55%) of the 956 isolates in our 11-year collection. Not only were MCG-2 isolates divergent from other Wisconsin clonal groups by PFGE, but they also harbored SCCmec IVa, in contrast to the SCCmec II types found in the other MCGs. Other reports of CA- and HA-MRSA strains preferentially harboring SCCmec type IV and other types, respectively, have been described previously (1, 15, 20, 29, 33). Oftentimes, accurate information about the timeline of exposure to MRSA is difficult to obtain. Molecular analysis is likely to play a critical role in identifying CA-MRSA strains as they are transmitted (39) or become endemic to healthcare facilities. In the same way, molecular distinctions would also allow basic comparisons of isolates across the globe. Nevertheless, the distinctions associated with MCG-2 were not limited to molecular features. One of the more remarkable observations for this group was the younger mean age than the average of 72 years for patients infected with the other MCGs. Naimi et al. (28) made a similar observation in Minnesota, where CA-MRSA was prevalent among individuals with a mean age of 16 years. In addition, we identified a statistical difference in the number of patients with CA-MRSA presenting with skin and soft tissue infections at outpatient clinic facilities. This was in spite of the fact that cultures submitted from hospital facilities included those collected in emergency rooms, urgent care units, obstetric departments, and other outpatient settings and may have allowed an overrepresentation of CA-MRSA from hospitals.
In conclusion, we have demonstrated that the CA-MRSA strains in MCG-2 in Wisconsin are clonally related to the MW2 strain. More specifically, we showed that the CA-MRSA strain WI-34 that emerged in Wisconsin in 1992 was likely a progenitor of the MW2 strain. The plasmid types associated with CA-MRSA were stable over time and appeared to play a role in ERY expression. The largest proportion of CA-MRSA isolates were associated with young individuals from Native American communities. Given the prevalence of CA-MRSA strains circulating within rural areas of Wisconsin, the treatment of outpatient skin infections, particularly in young individuals, should be guided by susceptibility testing. More detailed epidemiological studies are warranted to determine the specific risk factors associated with acquisition and to establish preventive measures within the community.
Acknowledgments
This work was supported by the Marshfield Clinic Research Foundation.
We acknowledge Carla Schofield for creating the study database and Linda Weis and Alice Stargardt for manuscript preparation. We also thank the staff of the microbiology section of Marshfield Laboratories for their assistance with the collection of isolates.
REFERENCES
- 1.Abi-Hanna, P., A. L. Frank, J. P. Quinn, S. Kelkar, P. C. Schreckenberger, M. K. Hayden, and J. F. Marcinak. 2000. Clonal features of community-acquired methicillin-resistant Staphylococcus aureus in children. Clin. Infect. Dis. 30:630-631. [DOI] [PubMed] [Google Scholar]
- 2.Adcock, P. M., P. Pastor, F. Medley, J. E. Patterson, and T. V. Murphy. 1998. Methicillin-resistant Staphylococcus aureus in two child care centers. J. Infect. Dis. 178:577-580. [DOI] [PubMed] [Google Scholar]
- 3.Anthony, B. F., L. V. Perlman, and L. W. Wannamaker. 1967. Skin infections and acute nephritis in American Indian children. Pediatrics 39:263-279. [PubMed] [Google Scholar]
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimin, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett, F. F., R. F. McGehee, and M. Finland, Jr. 1968. Methicillin-resistant Staphylococcus aureus at Boston City Hospital. Bacteriologic and epidemiologic observations. N. Engl. J. Med. 279:441-448. [DOI] [PubMed] [Google Scholar]
- 7.Boyce, J. M. 1998. Are the epidemiology and microbiology of methicillin-resistant Staphylococcus aureus changing? JAMA 279:623-624. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. [PubMed] [Google Scholar]
- 9.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlebois, E. D., D. R. Bangsberg, N. J. Moss, M. R. Moore, A. R. Moss, H. F. Chambers, and F. Perdreau-Remington. 2002. Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin. Infect. Dis. 34:425-433. [DOI] [PubMed] [Google Scholar]
- 11.Dajani, A. S., P. Ferrieri, and L. Wannamaker. 1973. Endemic superficial pyoderma in children. Arch. Dermatol. 108:517-522. [PubMed] [Google Scholar]
- 12.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 13.Drinka, P., J. T. Faulks, C. Gauerke, B. Goodman, M. Stemper, and K. Reed. 2001. Adverse events associated with methicillin-resistant Staphylococcus aureus in a nursing home. Arch. Intern. Med. 161:2371-2377. [DOI] [PubMed] [Google Scholar]
- 14.Embil, J., K. Ramotar, L. Romance, M. Alfa, J. Conly, S. Cronk, G. Taylor, B. Sutherland, T. Louie, E. Henderson, and L. E. Nicolle. 1994. Methicillin-resistant Staphylococcus aureus in tertiary care institutions on the Canadian prairies 1990-1992. Infect. Control Hosp. Epidemiol. 15:646-651. [DOI] [PubMed] [Google Scholar]
- 15.Fey, P. D., B. Said-Salim, M. E. Rupp, S. H. Hinrichs, D. J. Boxrud, C. C. Davis, B. N. Kreiswirth, and P. M. Schlievert. 2003. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank, A. L., J. F. Marcinak, P. D. Mangat, and P. C. Schreckenberger. 1999. Community-acquired and clindamycin-susceptible methicillin-resistant Staphylococcus aureus in children. Pediatr. Infect. Dis. J. 18:993-1000. [DOI] [PubMed] [Google Scholar]
- 17.Gorak, E. J., S. M. Yamada, and J. D. Brown. 1999. Community-acquired methicillin-resistant Staphylococcus aureus in hospitalized adults and children without known risk factors. Clin. Infect. Dis. 29:797-800. [DOI] [PubMed] [Google Scholar]
- 18.Groom, A. V., D. H. Wolsey, T. S. Naimi, K. Smith, S. Johnson, D. Boxrud, K. A. Moore, and J. E. Cheek. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 10:1201-1205. [DOI] [PubMed] [Google Scholar]
- 19.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 20.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Layton, M. C., W. J. Hierholzer, and J. E. Patterson. 1995. The evolving epidemiology of methicillin-resistant Staphylococcus aureus at a university hospital. Infect. Control Hosp. Epidemiol. 16:12-17. [DOI] [PubMed] [Google Scholar]
- 22.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 23.Liu, P. Y., Z. Y. Shi, Y. J. Lau, B. S. Hu, J. M. Shyr, W. S. Tsai, Y. H. Lin, and C. Y. Tseng. 1996. Use of restriction endonuclease analysis of plasmids and pulsed-field gel electrophoresis to investigate outbreaks of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 22:86-90. [DOI] [PubMed] [Google Scholar]
- 24.Maguire, G. P., A. D. Arthur, P. J. Boustead, B. Dwyer, and B. J. Currie. 1996. Emerging epidemic of community-acquired methicillin-resistant Staphylococcus aureus infection in the Northern Territory. Med. J. Aust. 164:721-723. [DOI] [PubMed] [Google Scholar]
- 25.Matushek, M. G., M. J. Bonten, and M. K. Hayden. 1996. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J. Clin. Microbiol. 34:2598-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno, F., C. Crisp, J. H. Jorgensen, and J. E. Patterson. 1995. Methicillin-resistant Staphylococcus aureus as a community organism. Clin. Infect. Dis. 21:1308-1312. [DOI] [PubMed] [Google Scholar]
- 28.Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O'Boyle, R. N. Danila, J. E. Cheek, M. T. Osterholm, K. A. Moore, and K. E. Smith. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996-1998. Clin. Infect. Dis. 33:990-996. [DOI] [PubMed] [Google Scholar]
- 29.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. 1999. Approved standard M100-S9: performance standards for antimicrobial susceptibility testing. Ninth supplement, vol. 19. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 31.National Nosocomial Infections Surveillance System. 1999. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien, F. G., T. T. Lim, F. N. Chong, G. W. Coombs, M. C. Enright, D. A. Robinson, A. Monk, B. Said-Salim, B. N. Kreiswirth, and W. B. Grubb. 2004. Diversity among community isolates of methicillin-resistant Staphylococcus aureus in Australia. J. Clin. Microbiol. 42:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panlilio, A. L., D. H. Culver, R. P. Gaynes, S. Banerjee, T. S. Henderson, J. S. Tolson, and W. J. Martone. 1992. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975-1991. Infect. Control Hosp. Epidemiol. 13:582-586. [DOI] [PubMed] [Google Scholar]
- 36.Reed, K. D., M. F. Vandermause, and M. E. Stemper. 1992. Magic minipreps: rapid isolation of plasmid DNA from clinical isolates of staphylococci. Promega Notes 37:9-10. [Google Scholar]
- 37.Reed, K. D., M. E. Stemper, M. F. Vandermause, and P. D. Mitchell. 1993. Evaluation of a commercial DNA purification system for plasmid analysis of nosocomial bacterial pathogens. Am. J. Clin. Pathol. 100:304-307. [DOI] [PubMed] [Google Scholar]
- 38.Rhinehart, E., D. M. Shlaes, T. F. Keys, J. Serkey, B. Kirkley, C. Kim, C. A. Currie-McCumber, and G. Hall. 1987. Nosocomial clonal dissemination of methicillin-resistant Staphylococcus aureus. Elucidation by plasmid analysis. Arch. Intern. Med. 147:521-524. [PubMed] [Google Scholar]
- 39.Saiman, L., M. O'Keefe, P. L. Graham III, F. Wu, B. Said-Salim, B. Kreiswirth, A. LaSala, P. M. Schlievert, and P. Della-Latta. 2003. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin. Infect. Dis. 37:1313-1319. [DOI] [PubMed] [Google Scholar]
- 40.Shukla, S. K., S. V. Ramaswamy, J. Conradt, M. E. Stemper, R. Reich, K. D. Reed, and E. A. Graviss. 2004. Identification of novel polymorphisms in mec genes and a new mec complex type in methicillin-resistant Staphylococcus aureus from rural Wisconsin. Antimicrob. Agents Chemother. 48:3080-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla, S. K., M. E. Stemper, S. V. Ramaswamy, J. M. Conradt, R. Reich, E. A. Graviss, and K. D. Reed. 2004. Molecular characteristics of nosocomial and Native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J. Clin. Microbiol. 42:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stemper, M. E. 2002. Molecular epidemiology of a community-acquired strain of methicillin-resistant Staphylococcus aureus in Wisconsin. M.S. thesis. University of Wisconsin, La Crosse.
- 43.Suggs, A. H., M. C. Maranan, S. Boyle-Vavra, and R. S. Daum. 1999. Methicillin-resistant and borderline methicillin-resistant asymptomatic Staphylococcus aureus colonization in children without identifiable risk factors. Pediatr. Infect. Dis. J. 18:410-414. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, G., T. Kirkland, K. Kowalewska-Grochowska, and Y. Wang. 1990. A multistrain cluster of methicillin-resistant Staphylococcus aureus based in a native community. Can. J. Infect. Dis. 1:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turnidge, J. D., and J. M. Bell. 2000. Methicillin-resistant Staphylococcus aureus evolution in Australia over 35 years. Microb. Drug Resist. 6:223-229. [DOI] [PubMed] [Google Scholar]
- 47.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuccarelli, A. J., I. Roy, G. P. Harding, and J. J. Couperus. 1990. Diversity and stability of restriction enzyme profiles of plasmid DNA from methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 28:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]