Abstract
During 2002 to 2003, eight Salmonella enterica serotype Virchow poultry and poultry product isolates from various sources (chicken farms, poultry slaughterhouse, or retail store) and one S. enterica rough strain isolated from human feces were found to produce extended-spectrum β-lactamase CTX-M-9. Poultry and poultry product isolates were recovered from different locations in the southwest of France. The human rough isolate had sequences of flagellin genes (fliC and fljB) typical of serotype Virchow and ribotyping and pulsed-field gel electrophoresis (PFGE) patterns closely similar to those of serotype Virchow strains. PFGE confirmed the clonal relationship between the poultry isolates, while the human isolate displayed a pattern with 94% homology. The blaCTX-M-9 gene was located on a conjugative plasmid and was shown to be linked to orf513. Plasmid profiling found a very similar EcoRI restriction pattern in six transconjugants studied, including transconjugants obtained from the human isolate. A single hatchery, supplying chicks to the six farms, was identified. Emergence of extended-spectrum β-lactamase-producing S. enterica strains in food animals is a major concern, as such strains could disseminate on a large scale and lead to antibiotic therapy difficulties.
Salmonella enterica is a frequent pathogen of animals and humans. Food-borne diseases caused by this species represent an important public health problem worldwide. An increase in the β-lactam resistance in Salmonella spp. has been observed during the 1990s with the emergence of a multiresistant epidemic strain of S. enterica serotype Typhimurium of phage type DT 104, with chromosomal integration of the genes coding for resistance to ampicillin, streptomycin (and spectinomycin), chloramphenicol, sulfonamides, and tetracycline (7, 10). The emergence of Salmonella strains that are resistant to extended-spectrum cephalosporins have been reported throughout the world since 1991 (1, 3, 4, 9, 15, 18, 19, 23, 27, 30). These strains, isolated mostly in hospitalized patients, produced plasmid-mediated class A extended-spectrum β-lactamases belonging to the TEM, SHV, CTX-M, or PER families. More recently, the emergence of strains of S. enterica producing plasmidic class C cephamycinase CMY-2 has been reported in the United States (14). These strains, belonging mostly to serotype Newport (known as Newport-MDRAmpC), have been isolated in food animals, in retail ground meat, and in humans.
We report here the characterization of nine CTX-M-9 extended-spectrum β-lactamase-producing multiresistant Salmonella strains isolated in France from 2002 to 2003. The human isolate was recovered from stools of a 4-year-old girl with gastroenteritis in February 2002. The eight poultry and poultry product isolates were recovered in 2003 from various sources (chicken farms, poultry slaughterhouse, or retail store) located in the southwest of France. Molecular characterization of these isolates was done by pulsed-field gel electrophoresis (PFGE) and plasmid fingerprinting. The β-lactamases were characterized by isoelectric focusing and molecular methods.
MATERIALS AND METHODS
Bacterial strains.
The human S. enterica isolate ROU was received for serotyping at the French National Reference Center for Salmonella in February 2002. The eight poultry and poultry product isolates were received at the Agence Française de Sécurité Sanitaire des Aliments (French Agency for Food Safety) laboratory between April and June 2003. Table 2 shows details of the nine isolates studied. Escherichia coli J5 (rif) was used for conjugation experiments. E. coli ATCC 25922 was used as a susceptible control in disk diffusion method and in MIC determinations. Four non-β-lactamase-producing strains of S. enterica serotype Virchow from the Agence Française de Sécurité Sanitaire des Aliments collection isolated in poultry in 2003 and S. enterica serotype Braenderup H9812 were used as controls and molecular size markers, respectively, in the PFGE experiment. The reference strains used for automated ribotyping, 41K of serotype Virchow (6,7:r:1,2), 16K of serotype Heidelberg (1,4,5,12:r:1,2), 7404/90 of serotype Bsilla (6,8:r:1,2), 1741/76 of serotype Lode (17:r:1,2), 3363/81 of serotype Grandhaven (30:r:1,2), and 1136/72 of serotype Crossness (67:r:1,2), were from the World Health Organization Collaborative Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
TABLE 2.
S. enterica isolates
| Isolate | Serotype | Source | Date of isolation | Area of isolation | Additional antimicrobial resistance phenotypesa | β-Lactamase pI(s) | bla gene(s) as determined by PCR or PCR and sequencing | PFGE type |
|---|---|---|---|---|---|---|---|---|
| ROU | Rough | Human feces | Feb 2002 | Haut-Rhin | S, Sp, K, Nal, Su, Tmp, Te | 5.4, 7.9 | blaTEM-1, blaCTX-M-9 | Vir.Xb.0015 |
| 2437 | Virchow | Chicken feces/farm no. 1 | Apr 2003 | Lot | S, Sp, K, Nal, Su, Tmp, Te | 5.4, 7.9 | blaTEM, blaCTX-M-9-like | Vir.Xb.0006 |
| 2731 | Virchow | Retail chicken meat | Apr 2003 | Landes | S, Sp, K, Nal, Su, Tmp, Te | 5.4, 7.9 | blaTEM, blaCTX-M-9 | Vir.Xb.0003 |
| 2983 | Virchow | Chicken environment/farm no.2 | Apr 2003 | Dordogne | S, Sp, K, Nal, Su, Tmp, Te | 5.4, 7.9 | blaTEM, blaCTX-M-9-like | Vir.Xb.0003 |
| 3279 | Virchow | Chicken feces/farm no. 3 | May 2003 | Tarn-et-Garonne | S, Sp, K, Nal, Su, Tmp, Te | 5.4, 7.9 | blaTEM, blaCTX-M-9-like | Vir.Xb.0003 |
| 3292 | Virchow | Environmental/poultry slaughterhouse | May 2003 | Landes | S, Sp, K, Nal, Su, Tmp, Te | 5.4, 7.9 | blaTEM, blaCTX-M-9-like | Vir.Xb.0003 |
| 3464b | Virchow | Chicken environment/farm no. 4 | May 2003 | Dordogne | S, Sp, Nal, Su, Tmp, Te | 7.9 | blaCTX-M-9 | Vir.Xb.0011 |
| 3670 | Virchow | Chicken feces/farm no. 5 | May 2003 | Tarn-et-Garonne | S, Sp, K, Nal, Su, Tmp, Te | 5.4, 7.9 | blaTEM, blaCTX-M-9-like | Vir.Xb.0003 |
| 4300 | Virchow | Chicken environment/farm no. 6 | June 2003 | Dordogne | S, Sp, Nal, Su, Tmp, Te | 5.4, 7.9 | blaTEM, blaCTX-M-9 | Vir.Xb.0002 |
Abbreviations: S, streptomycin; Sp, spectinomycin, K, kanamycin; Nal, nalidixic acid; Te, tetracycline; Su, sulfonamides; Tmp, trimethoprim.
Serotyping.
Isolates were serotyped on the basis of somatic O and phase 1 and phase 2 flagellar antigens by agglutination tests with antisera (Bio-Rad, Marnes la Coquette, France, and World Health Organization Collaborative Centre for Reference and Research on Salmonella) as specified by the White-Kauffman-Le Minor scheme (21).
Automated ribotyping.
Automated ribotyping was performed with the RiboPrinter microbial characterization system (Qualicon, Wilmington, Del.) according to the manufacturer's instructions. Digestion by restriction endonuclease PstI, gel separation, transfer, and hybridization with a chemiluminescent-labeled DNA probe containing the E. coli rRNA operon were carried out in 8 h.
Pulsed-field gel electrophoresis.
PFGE with XbaI (Amersham Biosciences, Freiburg, Germany) was carried out with a CHEF-DRIII system (Bio-Rad) as described previously (20). The running conditions were 6 V/cm at 14°C for 20 h with pulse times ramped from 2.2 to 63.8 s. BioNumerics software (Applied Maths, Kortrijk, Belgium) was used to compare the PFGE profiles. The bands generated were analyzed by the Dice coefficient and the unweighted pair group method with arithmetic averages.
Antimicrobial susceptibility testing.
Antibiotic susceptibility was determined by the disk diffusion method on Mueller-Hinton (MH) agar according to the guidelines of the Antibiogram Committee of the French Society for Microbiology (28). The following antimicrobials (Bio-Rad) were tested: amoxicillin, amoxicillin-clavulanic acid, ticarcillin, ticarcillin-clavulanic acid, piperacillin, piperacillin-tazobactam, cephalothin, cefamandole, cefoperazone, cefoxitin, ceftriaxone, ceftazidime, cefepime, aztreonam, moxalactam, imipenem, streptomycin, spectinomycin, kanamycin, tobramycin, netilmicin, gentamicin, amikacin, isepamicin, nalidixic acid, pefloxacin, ciprofloxacin, sulfonamides, trimethoprim, chloramphenicol, and tetracycline.
The MICs of the β-lactams and ciprofloxacin were determined by Etest (AB Biodisk, Solna, Sweden). The extended-spectrum β-lactamase phenotype was detected with the extended-spectrum β-lactamase detection Etest strips and the double disk diffusion test (16).
Preparation of crude extracts of β-lactamase and isoelectric focusing.
Crude extracts of β-lactamases were obtained by sonication followed by 45 min of ultracentrifugation at 100,000 x g with a Beckman L8-55 M ultracentrifuge. Isoelectric focusing was performed with a PhastSystem apparatus (Amersham-Pharmacia Biotech, Freiburg, Germany) as described previously (31).
PCR amplification and DNA sequencing.
Total DNA was extracted with the InstaGene matrix kit (Bio-Rad) in accordance with the manufacturer's recommendations. The sequences of the primers used in the PCR amplifications are given in Table 1. PCR amplifications of fliC and fljB genes coding the two flagellar phases were performed with the FLIC-F and FLIC-R or FLJB-F and FLJB-R primers (13).
TABLE 1.
Primers
| PCR target | Primer | Oligonucleotide sequencea (5′→3′) | Reference | PCR product size (bp) |
|---|---|---|---|---|
| fliC | FLIC-F | CAAGTCATTAATACAAACAGC | 13 | 1,500 |
| FLIC-R | TTAACGCAGTAAAGAGAGGAC | |||
| fljB | FLJB-F | CAAGTAATCAACACTAACAGT | 13 | 1,500 |
| FLJB-R | TTAACGTAACAGAGACAGCAC | |||
| blaTEM | TEM-F | ATAAAATTCTTGAAGACGAAA | 31 | 1,080 |
| TEM-R | GACAGTTACCAATGCTTAATC | |||
| blaCTX-M consensus | CTX-M-F | CRATGTGCAGYACCAGTAA | This study | 540 |
| CTX-M-R | CGCRATATCRTTGGTGGTG | |||
| blaCTX-M-9 | CTX-M-9-F | GTGACAAAGAGAGTGCAACGG | 27 | 856 |
| CTX-M-9-R | ATGATTCTCGCCGCTGAAGCC | |||
| gyrA | STGYRA1 | TGTCCGAGATGGCCTGAAGC | 2 | 470 |
| STGYRA12 | CGTTGATGACTTCCGTCAG | |||
| Class I integron | 5′-CS | GGCATCCAAGCAGCAAGC | 17 | Variable |
| 3′-CS | AAGCAGACTTGACCTGAT | |||
| Region upstream of blaCTX-M-9 | ORF513-F | GCAGCACTACCCAGCCTTCA | This study | 2,700 |
| MA-2-R | CCGCRATATGRTTGGTGGTG |
Y, C or T; R, A or G.
CTX-M-specific PCR analysis was performed with in-house primers designed on the basis of consensus sequences of various CTX-M genes; primer forward CTX-M-F (located from positions 209 to 227 with respect to the CTX-M translational starting point), and primer reverse CTX-M-R (located from positions 750 to 732). A CTX-M-9 group-specific PCR assay was carried out with primers CTX-M-9-F and CTX-M-9-R amplifying an 856-bp fragment of blaCTX-M-9 like (27). Primers TEM-F and TEM-R were used to amplify a 1,080-bp fragment of the blaTEM gene. All amplifications except the quinolone resistance-determining region of gyrA and upstream region of blaCTX-M-9 were performed on 50-μl samples containing DNA (2.5 μl), primers (50 pmol each), deoxynucleoside triphosphate (200 μM), Taq DNA polymerase (1.25 U; Ampli Taq Gold; Roche) and its buffer, MgCl2 (2 mM), and dimethyl sulfoxide (10%). The cycling conditions included 10 min of denaturation at 94°C (one cycle); 30 s (1 min for fliC and fljB) of denaturation at 94°C, 30 s (1 min for fliC and fljB) of annealing at 50°C (53°C for blaCTX-M consensus, fliC, and fljB and 54°C for blaCTX-M-9) and 1 min (1 min 30 s for fliC and fljB) of polymerization at 72°C (35 cycles), followed by 10 min of extension at 72°C.
Amplification of class I integrons with primers 5′-CS (17) and 3′-CS was performed as described previously (31). PCR amplification of the quinolone resistance-determining region of gyrA was carried out with primers STGYRA1 and STGYRA12 as described previously (2). PCR amplification of the upstream region of blaCTX-M-9 was performed with primers ORF513-F and MA-2-R. Amplification was performed on 25-μl samples containing DNA (2.5 μl), primers (20 pmol each), deoxynucleoside triphosphate (200 μM), and Taq DNA polymerase (2.5 U; Taq DNA polymerase, Promega, Charbonnières, France) and its buffer. The cycling conditions included 5 min of denaturation at 94°C (one cycle); 1 min of denaturation at 94°C, 1 min of annealing at 58°C, and 4 min of polymerization at 72°C (35 cycles), followed by 7 min of extension at 72°C.
The purified PCR fragments were sequenced on both strands by Genome Express (Meylan, France) with an ABI 100 DNA sequencer (Applied Biosystems, Foster City, Calif.).
The nucleotide sequence was analyzed with the Lasergene software (Dnastar, Madison, Wis.). The BLASTN program of NCBI (http://www.ncbi.nlm.nih.gov) was used for database searches.
Resistance transfer determination.
A resistance transfer experiment was carried out on liquid media with four selected isolates as described previously (12). E. coli J5 resistant to rifampin was used as the recipient strain. Transconjugants were selected on Drigalski agar supplemented with cefotaxime (2 mg/liter) and rifampin (250 mg/liter).
Plasmid analysis.
Plasmid DNA was purified from E. coli transconjugants by an alkaline lysis procedure (29) and subjected to 0.8% agarose gel electrophoresis. The molecular sizes of plasmids were determined with Taxotron software (Institut Pasteur, Paris, France) by reference to plasmids of known sizes (RP4, 54 kb, and pIP173, 126 kb) mixed with a supercoiled DNA ladder (Invitrogen, Groningen, The Netherlands). DNA restriction fragment length polymorphisms were analyzed by agarose gel electrophoresis of plasmid DNA cleaved with EcoRI and PstI (Roche, Mannheim, Germany).
RESULTS
Serotyping of Salmonella isolates.
The eight poultry and poultry product isolates belonged to serotype Virchow (6,7:r:1,2), while the human isolate ROU could not be serotyped due to autoagglutination (rough strain). PCR amplification of the fliC and fljB genes encoding the two flagellar phases from isolate ROU and subsequent sequencing identified sequences 99% identical to the H-1(r) gene (EMBL accession no. X04505) and the H-2(1, 2) gene (GenBank accession no. U17177), respectively, indicating that isolate ROU possesses at least the same flagellar phases as the Virchow isolates.
Automated ribotyping.
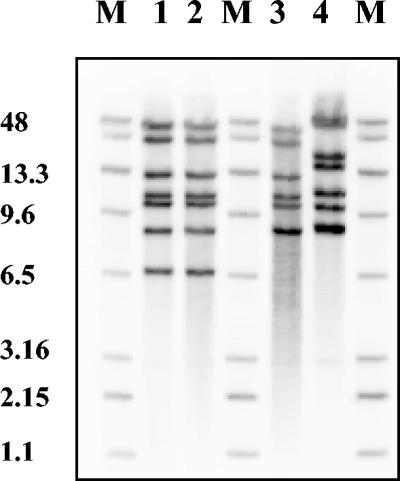
To determine if human isolate ROU, untypeable by conventional serotyping (rough), could derive from serotype Virchow, automated ribotyping was performed on isolate ROU, on poultry isolate 2731, on the reference strain of serotype Virchow, and on reference strains of serotypes Heidelberg, Bsilla, Lode, Grandhaven, and Crossness, which possess the same flagellar antigens (r:1,2) but different O antigens. Isolates ROU and 2731 displayed the same pattern, which differed from that of the Virchow reference strain by a single band (Fig. 1). The patterns obtained with the other reference strains were quite different (Fig. 1 and data not shown).
FIG. 1.
Automated ribotyping of PstI-digested genomic DNA from isolate ROU, isolate 2731, and reference strains of serotypes Virchow and Grandhaven. Lane M, RiboPrinter molecular size markers (band sizes in kilobase pairs); lane 1, isolate ROU (human rough isolate); lane 2, isolate 2731 (poultry Virchow isolate); lane 3, serotype Virchow reference strain 41K; lane 4, serotype Grandhaven reference strain 3363/81.
PFGE.
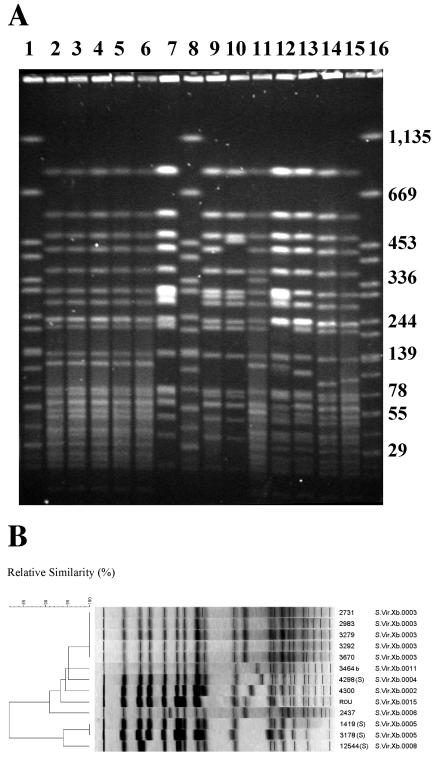
The clonal relatedness of the nine CTX-M-9-producing S. enterica isolates and four non-β-lactamase-producing S. enterica serotype Virchow isolates was assessed by PFGE with XbaI (Fig. 2). Eight distinct profiles were observed among the 12 strains and five among the nine CTX-M-9-producing isolates (Table 2 and Fig. 2). Two separate clusters (similarity < 82%) were found after cluster analysis of these fingerprints (Fig. 2). All the CTX-M-9-producing isolates and non-β-lactamase-producing isolate 4298 belonged to the main cluster, with a genetic similarity of about 91%.
FIG. 2.
(A) PFGE of XbaI-digested genomic DNA from the nine CTX-M-9-producing S. enterica isolates under study and four non-extended-spectrum β-lactamase-producing S. enterica serotype Virchow poultry isolates (S). Lanes 1, 8, and 16, S. enterica serotype Braenderup H9812 used as molecular size markers (band sizes in kilobase pairs); lane 2, isolate 3292; lane 3, isolate 3670; lane 4, isolate 3279; lane 5, isolate 2983; lane 6, isolate 2731; lane 7, isolate 3178 (S); lane 9, isolate 1419 (S); lane 10, isolate 12544 (S); lane 11, isolate 2437; lane 12, isolate ROU; lane 13, isolate 4300; lane 14, isolate 4298 (S); lane 15, isolate 3464. (B) Dendrogram generated by BioNumerics showing the results of cluster analysis on the basis of PFGE fingerprinting. Similarity analysis was performed with the Dice coefficient, and clustering was by unweighted pair group method with arithmetic averages.
Antimicrobial susceptibility.
All nine isolates were resistant to amoxicillin, ticarcillin, piperacillin, cephalothin, cefamandole, and ceftriaxone and remained apparently in vitro susceptible to piperacillin-tazobactam, cefoxitin, ceftazidime, and imipenem by the disk diffusion method. The isolates were also resistant to aminoglycosides (streptomycin, spectinomycin, and, for most isolates, kanamycin), sulfamethoxazole-trimethoprim, nalidixic acid, and tetracycline (Table 2). An extended-spectrum β-lactamase-producing phenotype was detected with the double disk diffusion test and Etest strips (data not shown). The MICs of the β-lactams determined by Etest in five selected isolates are shown in Table 3. The selected isolates showed decreased susceptibilities to ceftriaxone (MICs of 8 to 16 mg/liter) and slightly decreased susceptibilities to ceftazidime (MICs of 1 to 2 mg/liter). The five isolates showed decreased susceptibilities to ciprofloxacin (MIC of 0.25 mg/liter) (Table 3).
TABLE 3.
MICs of β-lactams and ciprofloxacina
| Antibiotics | MIC (mg/liter) for strain:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ROU | pROU-1 | 2437 | p2437-1 | 2731 | 3279 | p3279-1 | 4300 | p4300-1 | E. coli J5 | E. coli ATCC 25922 | |
| Ampicillin | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | 2 | 4 |
| Amoxicillin-CLAb | 8 | 8 | 8 | 4 | 8 | 8 | 4 | 4 | 4 | 4 | 4 |
| Ticarcillin | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | 2 | 4 |
| Ticarcillin-CLAc | 16 | 16 | 8 | 4 | 8 | 16 | 4 | 16 | 4 | 2 | 4 |
| Piperacillin | >256 | 128 | 256 | 128 | 256 | 128 | 128 | >256 | 64 | 1 | 2 |
| Piperacillin-TZBd | 4 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Cefoxitin | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ceftazidime | 2 | 0.125 | 1 | 0.125 | 1 | 1 | 0.125 | 1 | 0.125 | 0.125 | 0.25 |
| Ceftazidime-CLAc | 0.25 | 0.125 | 0.25 | <0.06 | 0.25 | 0.25 | 0.06 | 0.25 | <0.06 | NT | NT |
| Ceftriaxone | 16 | 2 | 8 | 2 | 16 | 16 | 2 | 16 | 2 | <0.06 | 0.06 |
| Cefotaxime-CLAe | 0.125 | 0.06 | 0.06 | <0.06 | 0.125 | 0.125 | <0.06 | 0.125 | <0.06 | NT | NT |
| Cefepime | 2 | 0.25 | 1 | 0.25 | 2 | 2 | 0.25 | 2 | 0.25 | <0.06 | 0.06 |
| Aztreonam | 2 | 0.25 | 1 | 0.25 | 1 | 2 | 0.125 | 1 | 0.25 | <0.06 | 0.06 |
| Imipenem | 0.25 | 0.125 | 0.25 | 0.25 | 0.125 | 0.125 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Ciprofloxacin | 0.25 | NT | 0.25 | NT | 0.25 | 0.25 | NT | 0.25 | NT | NT | 0.015 |
The MICs of β-lactams and ciprofloxacin (Etest) for CTX-M-9-producing Salmonella enterica isolates ROU, 2437, 2731, 3279, and 4300, E. coli J5 transconjugants pROU-1, p2437-1, p3279-1, and p4300-1, and E. coli strains J5 and ATCC 25922 are presented. CLA, clavulanic acid; TZB, tazobactam; NT, not tested.
Amoxicillin-CLA (2:1).
CLA, 2 mg/liter.
TZB, 4 mg/liter.
CLA, 4 mg/liter.
Isoelectric focusing.
Seven isolates produced two β-lactamases with isoelectric points of 5.4 and 7.9, while isolate 3464 b produced only pI 7.9 β-lactamase (Table 2).
Characterization of β-lactamase genes, quinolone resistance-determining region of gyrA, and class I integrons.
CTX-M consensus PCR performed on all the isolates gave the expected PCR product of 540 bp. Sequence analysis of the PCR product from isolate ROU revealed 100% homology with the blaCTX-M-9a sequence (GenBank accession number AF252621). To confirm the presence of blaCTX-M-9 in all the isolates, a CTX-M-9 group-specific PCR assay was carried out, and all the isolates gave the expected PCR product of 856 bp (Table 2). Sequencing of PCR products and deduced amino acid sequence analysis from isolates ROU, 2731, 3464b, and 4300 confirmed that the β-lactamase was CTX-M-9 (Table 2). It was consistent with the identification of a pI 7.9 β-lactamase by isoelectric focusing. A TEM-specific PCR assay gave the expected 1,080-bp fragment in all but one isolate, 3464b (Table 2). DNA sequencing of the PCR product from isolate ROU confirmed that the β-lactamase was TEM-1 (Table 2). It was consistent with the production of a pI 5.4 β-lactamase.
The PCR amplifications of the quinolone resistance-determining region of gyrA in two isolates tested (ROU and 2437) revealed identical single mutations leading to the amino acid substitution Ser83Phe, reported to be involved in quinolone resistance.
PCR amplification of class I integrons was performed in all the isolates. They all gave a PCR product of 1.5 kb (data not shown). The DNA sequences of 1.5 kb of the PCR products from isolates ROU and 2731 were 100% identical to the corresponding sequence of In36 (GenBank accession no. AY259085) and 99% identical to the corresponding sequence of In60 (25). Two different gene cassettes were detected. The first one contained the dfrA16 gene cassette, encoding resistance to trimethoprim. The second gene cassette contained an aadA2 gene, encoding resistance to streptomycin and spectinomycin. Analysis of the upstream region of blaCTX-M-9 by PCR with primers ORF513-F and MA-2-R showed that the nine isolates had the expected 2,700-bp regions between orf513 and blaCTX-M-9, as described for In60, a complex integron carrying blaCTX-M-9, identified in E. coli and S. enterica serotype Virchow in Spain (data not shown).
Resistance transfer determination and plasmid analysis.
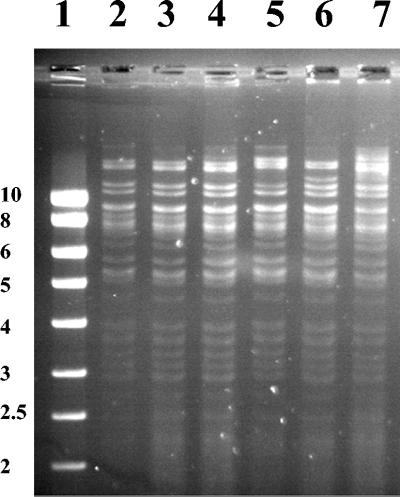
A resistance transfer experiment was carried out on liquid media with E. coli J5 (rif) as the recipient strain. E. coli transconjugants pROU-1, p2437-1, p2731-1, p3279-1, p3464b-1, and p4300-1 were obtained for all the isolates tested, ROU, 2437, 2731, 3279, 3464b, and 4300, respectively. They expressed β-lactamases with pIs of 5.4 and 7.9, except p4300-1, which expressed only a pI 7.9 β-lactamase. The MICs of ceftazidime and ceftriaxone were lower for the transconjugants than for the parental strains (MICs of ceftazidime were 0.125 versus 1 to 2 mg/liter and the MICs of ceftriaxone were 2 versus 8 to 16 mg/liter) (Table 3). Other resistance determinants, including streptomycin, spectinomycin, kanamycin (except p4300-1), tetracycline, and sulfamethoxazole-trimethoprim, were cotransferred to transconjugants with cefotaxime resistance. Extraction of plasmids isolated from all the transconjugants revealed single plasmids larger than 126 kb (data not shown). Plasmid DNAs extracted from all the transconjugants were compared by restriction endonuclease analysis (EcoRI and PstI). All the plasmids had a very similar fingerprint (Fig. 3 and data not shown).
FIG. 3.
Restriction analysis (EcoRI) of plasmids isolated from E. coli transconjugants. Lane 1, markers (SmartLadder, Eurogentec, Seraing, Belgium) (band sizes in kilobase pairs); lane 2, E. coli transconjugant p2437-1; lane 3, E. coli transconjugant p2731-1; lane 4, E. coli transconjugant p3279-1; lane 5, E. coli transconjugant p3464b-1; lane 6, E. coli transconjugant p4300-1; lane 7, E. coli transconjugant pROU-1.
DISCUSSION
CTX-Ms are extended-spectrum β-lactamases that are generally more active against cefotaxime (and ceftriaxone) than ceftazidime but are inhibited by clavulanic acid. CTX-Ms form a growing family that comprises at least 40 enzymes belonging to six groups (4). The CTX-M-9 group includes nine plasmid-mediated enzymes (CTX-M-9, CTX-M-13, CTX-M-14, CTX-M-16, CTX-M-17, CTX-M-19, CTX-M-21, CTX-M-27, and Toho-2). CTX-M-9 was first reported in a human isolate of E. coli in Spain in 1996 (24). Since then, this extended-spectrum β-lactamase has been sporadically identified in Spain (22, 24), France (26), Turkey (U. G. Bahar, T. Demiray, E. Kandirali, N. Apaydin, and A. Mert, Abstr. 13th Eur. Congress Clin. Microbiol. Infect. Dis., abstr. P580, 2003), Brazil (5), and China (11), mostly from E. coli isolated in hospitalized patients. In the genus Salmonella, CTX-M-9 has only been reported in Spain within serotype Virchow (27). Simarro et al. (27) described four cases of gastroenteritis caused by CTX-M-9-producing isolates of S. enterica serotype Virchow from 1997 to 1998. The source of these infections was not investigated.
In our study, we report a dual emergence in France of CTX-M-9-producing multiresistant strains of S. enterica serotype Virchow in poultry (and in a poultry-derived retail meat) and of an S. enterica rough strain in a human. This rough strain could derive from serotype Virchow because it possessed the same flagellar genes and close ribotyping and PFGE patterns. The reports of extended-spectrum β-lactamases in the genus Salmonella mostly described nosocomial outbreaks. Exchange of mobile genetic elements carrying extended-spectrum β-lactamase genes, such as plasmids and transposons, between enteric bacteria frequently encountered in hospitals and selected by traces of extended-spectrum cephalosporins used in humans has been suggested to explain the hospital outbreaks.
Another source of contamination with extended-spectrum β-lactamase-producing strains of S. enterica could be through the food chain, as seen with ceftriaxone-resistant Newport-MDRAmpC strains (14). Very few studies evoked this way of infection with extended-spectrum β-lactamase-producing Salmonella strains. Cardinale et al. (9) described the dual emergence of SHV-12-producing S. enterica serotype Keurmassar in humans and in a poultry product in Senegal. Extensive use of antimicrobial agents as feed additives for farm animals (especially in the poultry industry) has been suggested as an essential factor for the emergence of multiresistant Salmonella strains (9). In a second report, the authors also suspected transmission through the food chain for TEM-52-producing strains of S. enterica of various serotypes isolated in France during the period 2002 to 2003 (32). This is a major concern because food animals represent a large reservoir for dissemination of such resistant strains to humans, leading to important therapeutical difficulties, especially in children, for whom extended-spectrum cephalosporins are the treatment of choice.
CTX-M-14, which belongs to the CTX-M-9 group, but also SHV-12 and CMY-2 have been separately identified in three fecal isolates of healthy chickens in Spain in 2000 to 2001 among 120 recovered at the slaughterhouse level (8). The authors could not explain the emergence of such strains, as the use of extended-spectrum cephalosporins is very unusual in chickens. In 2001, dissemination of cephalosporin-resistant E. coli (carrying blaCTX-M-9 or blaCMY-2) among summer camp attendees in Spain was attributed to food or water (22). During a survey in Spain in 2001, CTX-M-14 has been identified in E. coli from 17 patients, of whom five (among 14 documented) never had contact with the hospital environment before isolation of the strain (6).
In our study, CTX-M-9 was found in only eight isolates among 247 isolates of S. enterica serotype Virchow of nonhuman origin collected by the Agence Française de Sécurité Sanitaire des Aliments network in 2003. CTX-M-9-producing human isolates are also extremely rare, as only one isolate has been identified among 1,200 Salmonella sp human isolates tested in 2002 and among 140 S. enterica serotype Virchow human isolates tested from 1997 to 2002. A single hatchery, located in the southwest of France, supplying chicks to the six farms (Table 1) was identified in our study. It is now important to conduct a more detailed investigation to determine the step of the supply chain where the contamination occurred (hatchery or breeding company). It is also important to know the nature of the antimicrobial agents used. Extended-spectrum cephalosporins are not adequate for use as growth promoters in chicken farms but could be used for therapeutical purposes in reproductive chickens.
We can speculate that the selective pressure may lead to the emergence of plasmid-mediated extended-spectrum β-lactamases harbored by enteric bacteria which could subsequently be transferred to Salmonella spp. The finding that tetracycline, streptomycin, or sulfamethoxazole-trimethoprim resistance genes reside on the same plasmid as blaCTX-M-9 raises the possibility that the use of these common antibiotics could coselect this extended-spectrum β-lactamase phenotype.
In conclusion, our investigation documented that Salmonella spp. producing the CTX-M-9 extended-spectrum β-lactamase have been identified in poultry and poultry product in France. Active surveillance of antimicrobial use in animal husbandry is important to reduce selective pressure and subsequent dissemination of multiresistant Salmonella spp. to humans.
Acknowledgments
We thank all the corresponding laboratories of the French National Reference Center Salmonella network and the French Food Safety Agency Salmonella Network, I. Castellanos (Laboratoire Vétérinaire Départemental de la Dordogne, France), I. Thiese (Laboratoire Vétérinaire Départemental du Tarn-et-Garonne, France), and M. Kretz (Service de Pédiatrie, Hôpitaux Civils de Colmar, France) for their collaboration.
REFERENCES
- 1.AitMhand, R., A. Soukri, N. Moustaoui, H. Amarouch, N. ElMaghri, D. Sirot, and M. Benbachir. 2002. Plasmid-mediated TEM-3 extended-spectrum β-lactamase production in Salmonella typhimurium in Casablanca. J. Antimicrob. Chemother. 49:169-172. [DOI] [PubMed] [Google Scholar]
- 2.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in High-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type 204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, O. Ang, C. Bal, and J. M. Casellas. 1996. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob. Agents Chemother. 40:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, R., C. Dutour, J. L. M. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp240Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou, G., M. Cartelle, M. Tomas, D. Canle, F. Molina, R. Moure, J. M. Eiros, and A. Guerrero. 2002. Identification and broad dissemination of the CTX-M-14 β-lactamase in different Escherichia coli strains in the northwest area of Spain. J. Clin. Microbiol. 40:4030-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, D., G. A. Peters, A. Cloeckaert, K. Sidi Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briñas, L., M. A. Moreno, M. Zarazaga, C. Porrero, Y. Sáenz, M. García, L. Dominguez, and C. Torres. 2003. Detection of CMY-2, CTX-M-14, and SHV-12 β-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob. agents Chemother. 47:2056-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardinale, E., P. Colbachini, J. D. Perrier-Gros-Claude, A. Gassama, and A. Aidara-Kane. 2000. Dual emergence in food and humans of a novel multiresistant serotype of Salmonella in Senegal: Salmonella enterica subsp. enterica serotype 35:c:1,2. J. Clin. Microbiol. 39:2373-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casin, I., J. Breuil, A. Brisabois, F. Moury, F. Grimont, and E. Collatz. 1999. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded β-lactamase PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 11.Chanawong, A., F. Hannachi M'Zali, J. Heritage, J. H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courvalin, P., F. Goldstein, A. Philippon, and J. Sirot (ed.). 1985. L'antibiogramme. Mpc-videom, Paris, France.
- 13.Dauga, C., A. Zabrovskaia, and P. A. D Grimont. 1998. Restriction fragment length polymorphism analysis of some flagellin genes of Salmonella enterica. J. Clin. Microbiol. 36:2835-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, A., J. Fontana, C. Crowe, B. Bolstorff, A. Stout, S. Van Duyne, M. P. Hoekstra, J. M. Whichard, T. J. Barrett, and F. J. Angulo. 2003. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188:1707-1716. [DOI] [PubMed] [Google Scholar]
- 15.Hammami, A., G. Arlet, S. Ben Redjeb, F. Grimont, A. Ben Hassen, A. Rekik, and A. Philippon. 1991. Nosocomial outbreak of acute gastroenteritis in a neonatal intensive care unit in Tunisia caused by multiply drug resistant Salmonella wien producing SHV-2 beta-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 10:641-646. [DOI] [PubMed] [Google Scholar]
- 16.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 17.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makanera, A., G. Arlet, V. Gautier, and M. Manai. 2003. Molecular epidemiology and characterization of plasmid-encoded β-lactamases produced by Tunisian clinical isolates of Salmonella enterica serotype Mbandaka resistant to broad-spectrum cephalosporins. J. Clin. Microbiol. 41:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morosini, M. I., J. Blazquez, M. C. Negri, R. Canton, E. Loza, and F. Baquero. 1996. Characterization of a nosocomial outbreak involving an epidemic plasmid encoding for TEM-27 in Salmonella enterica subspecies enterica serotype Othmarschen. J. Infect. Dis. 174:1015-1020. [DOI] [PubMed] [Google Scholar]
- 20.Peters, T. M., C. Maguire, E. J. Threlfall, I. S. T. Fisher, N. Gill, and A. J. Gatto. 2003. The Salm-gene project-a European collaboration for DNA fingerprinting for food-related salmonellosis. Euro Surveill. 8:46-50. [DOI] [PubMed] [Google Scholar]
- 21.Popoff, M. Y. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. W.H.O. Collaborating Center for Reference and Research on Salmonella. Institut Pasteur, Paris, France.
- 22.Prats, G., B. Mirelis, E. Miró, F. Navarro, T. Llovet, J. R. Johnson, N. Camps, A. Domínguez, and L. Salleras. 2003. Cephalosporin-resistant Escherichia coli among summer camp attendees with salmonellosis. Emerg. Infect. Dis. 9:1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revathi, G., K. P. Shannon, P. D. Stapleton, B. K. Jain, and G. L. French. 1998. An outbreak of extended-spectrum β-lactamase-producing Salmonella senftenberg in a burns ward. J. Hosp. Infect. 40:295-302. [DOI] [PubMed] [Google Scholar]
- 24.Sabaté, M., R. Tarragó, F. Navarro, E. Miró, C. Vergés, J. Barbé, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabaté, M., F. Navarro, E. Miró, S. Campoy, B. Mirelis, J. Barbé, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saladin, M., V. T. Cao, T. Lambert, J. L. Donay, J. L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 27.Simarro, E., F. Navarro, J. Ruiz, E. Miró, J. Gómez, and B. Mirelis. 2000. Salmonella enterica serovar Virchow with CTX-M-like β-lactamase in Spain. J. Clin. Microbiol. 38:4676-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soussy, C. J., G. Carret, J. D. Cavallo, H. Chardon, C. Chidiac, P. Choutet, P. Courvalin, H. Dabernat, H. Drugeon, L. Dubreuil, F. Goldstein, V. Jarlier, R. Leclercq, M. H. Nicolas-Chanoine, A. Philippon, C. Quentin, B. Rouveix, and J. Sirot. 2000. Comité de l'Antibiogramme de la Société Française de Microbiologie. Communiqué 2000-2001. Pathol. Biol. 48:832-871. [PubMed] [Google Scholar]
- 29.Takahashi, S., and Y. Nagano. 1984. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J. Clin. Microbiol. 20:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vahaboglu, H., S. Dodanli, C. Eroglu, R. Ozturk, G. Soyletir, I. Yildirim, and V. Avkan. 1996. Characterizations of multiple-antibiotic-resistant Salmonella typhimurium strains: molecular epidemiology of PER-1 producing isolates and evidence for nosocomial plasmid exchange by a clone. J. Clin. Microbiol. 34:2942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weill, F. X., M. Demartin, D. Tandé, E. Espié, I. Rakotoarivony, and P. A. D Grimont. 2004. Extended-spectrum-β-lactamase (SHV-12 like)-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 42:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weill, F. X., M. Demartin, L. Fabre, and P. A. D Grimont. 2004. Extended-spectrum-β-lactamase (TEM-52)-producing strains of Salmonella enterica of various serotypes isolated in France. J. Clin. Microbiol. 42:3359-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]