Abstract
Two groups of unknown bacteria, which phenotypically resemble members of the Bacteroides fragilis group but phylogenetically display >5% 16S rRNA gene sequence divergence from their nearest validly described species, Bacteroides thetaiotaomicron, were characterized by phenotypic and molecular taxonomic methods. Phylogenetically and phenotypically, the unidentified bacteria displayed a relatively close association with each other. However, a 16S rRNA gene sequence divergence of approximately 4% between the two unknown bacteria, as well as distinguishable biochemical characteristics, demonstrates that these organisms are genotypically and phenotypically distinct, and each group may represent a previously unknown subline within the Bacteroides phylogenetic cluster. Subsequent DNA-DNA hybridization studies confirmed that the two novel organisms were indeed distinct from each other. The previously described species closest to both of them is B. thetaiotaomicron (approximately 94% sequence similarity), but they can be differentiated easily from B. thetaiotaomicron by virtue of not utilizing trehalose. DNA-DNA pairing studies also documented the separateness of the unknown species and B. thetaiotaomicron. Based on the phenotypic and phylogenetic findings, two new species, “Bacteroides nordii” sp. nov. and “Bacteroides salyersae” sp. nov, are proposed. The G+C content of the DNA is 41.4 mol% for Bacteroides nordii and 42.0 mol% for Bacteroides salyersae. The type strains of Bacteroides nordii and Bacteroides salyersae are WAL 11050 (ATCC BAA-998 or CCUG 48943) and WAL 10018 (ATCC BAA-997 or CCUG 48945), respectively.
The Bacteroides fragilis group is part of the commensal flora in humans and is commonly associated with a variety of human infections, such as intra-abdominal abscesses, wound infections, and bacteremia (5). B. fragilis group bacteremia contributes significantly to morbidity and mortality (12). The choice of antibiotics for therapy is limited because the species of the B. fragilis group are among the most resistant of all anaerobes to antimicrobial agents, and this resistance has increased recently (4, 14). Although B. fragilis group species have been found to be clinically very important, studies of the significance of isolates of the B. fragilis group might have been hindered by an inadequate taxonomy. The taxonomy of Bacteroides has undergone significant changes in the past few years (8). It has recently been proposed that the genus Bacteroides be restricted to highly fermentative species that phenotypically resemble B. fragilis, the type species of the genus (13), and related taxa. The taxonomic positions of some other species still included in the genus remain uncertain; all of these species will ultimately be transferred to other genera. Furthermore, several clinically important species still await formal description.
In this paper, we report on the characterization of two groups of clinical isolates which were isolated from clinical specimens of human intestinal origin. Phenotypically, the two novel species are very much like Bacteroides stercoris and Bacteroides uniformis, respectively; therefore, they were misidentified as B. stercoris and B. uniformis previously. However, 16S rRNA sequencing reveals approximately 7% sequence divergence between the novel species and B. stercoris or B. uniformis. Phylogenetically, the previously described species closest to both of them is Bacteroides thetaiotaomicron (approximate 94% similarity). Although these two novel species have 96% sequence similarity, they can be distinguished easily by phenotypic characteristics. DNA-DNA hybridization studies also confirmed that these two groups of unknown organisms were indeed distinct from each other and from their nearest valid species B. thetaiotaomicron. Based on the phenotypic and phylogenetic findings presented here, two new species, “Bacteroides nordii” sp. nov. and “Bacteroides salyersae” sp. nov, are proposed. In addition, we also describe the phenotypic tests useful in distinguishing between the organisms mentioned above.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The study included 12 B. stercoris-like strains (group I); 9 B. uniformis-like strains (group II); 8 strains each of B. fragilis, B. stercoris, B. thetaiotaomicron, B. uniformis; and 3 strains each of Prevotella nigrescens, Prevotella disiens, Prevotella corporis, Prevotella intermedia, Porphyromonas asaccharolytica, Porphyromonas endodontalis, and Porphyromonas gingivalis (Table 1). The novel isolates were recovered from clinical sources such as peritoneal fluid, appendix tissue, and intra-abdominal abscess; therefore they are likely of intestinal origin. All the clinical isolates of each species were identified by 16S DNA sequencing in our laboratory. All the strains were cultivated on brucella agar (Difco, Detroit, Mich.) supplemented with 5% sheep blood and incubated anaerobically at 37°C under N2 (86%), H2 (7%), and CO2 (7%) gas phase.
TABLE 1.
List of strains used in this study
| Species | Strain or isolates | No. of strains |
|---|---|---|
| Bacteroides nordii | ATCC BAA-998T | 1 |
| Clinical isolates | 11 | |
| Bacteroides salyersae | ATCC BAA-997T | 1 |
| Clinical isolates | 8 | |
| Bacteroides fragilis | ATCC 25285T | 1 |
| Clinical isolates | 7 | |
| Bacteroides thetaiotaomicron | ATCC 29741T | 1 |
| Clinical isolates | 7 | |
| Bacteroides stercoris | ATCC 43183T | 1 |
| Clinical isolates | 7 | |
| Bacteroides uniformis | ATCC 8492T | 1 |
| Clinical isolates | 7 | |
| Prevotella nigrescens | ATCC 33563T | 1 |
| Clinical isolates | 2 | |
| Prevotella disiens | ATCC 29426T | 1 |
| Clinical isolates | 2 | |
| Prevotella corporis | ATCC 33547T | 1 |
| Clinical isolates | 2 | |
| Prevotella intermedia | ATCC 25611T | 1 |
| Clinical isolates | 2 | |
| Porphyromonas asaccharolytica | ATCC 25260T | 1 |
| Clinical isolates | 2 | |
| Porphyromonas endodontalis | ATCC 35406T | 1 |
| Clinical isolates | 2 | |
| Porphyromonas gingivalis | ATCC 33277T | 1 |
| Clinical isolates | 2 |
Biochemical characterization.
The strains were characterized biochemically by using a combination of conventional tests described previously in the Wadsworth and VPI anaerobe manuals (7, 9), plus the API ZYM and Rapid ID 32A systems (API bioMérieux, Marcy l'Etoile, France) and the RapID ANA II system (Remel, Inc., Lenexa, Kans.), according to the respective manufacturers' instructions. All biochemical tests were performed in duplicate. Fermentation tests were performed using prereduced, anaerobically sterilized peptone-yeast-sugar broth tubes (Anaerobe Systems, Morgan Hill, Calif.). The strains were grown in peptone-yeast broth and peptone-yeast-glucose broth (Anaerobe Systems) for metabolic end product (short-chain volatile and nonvolatile fatty acids) analysis by gas-liquid chromatography (9). Antimicrobial susceptibility studies were done using various antimicrobial agents, which were selected either as representative of a class of compound or as drugs for which MICs for quality control strains were published, by means of the NCCLS-approved Wadsworth plate dilution method (11).
Cellular fatty acid composition.
Long-chain cellular fatty acids were analyzed as previously described (16).
DNA base composition.
The mol percent G+C content of DNA was determined by high-performance liquid chromatography as described by Mesbah et al. (10) except that the methanol content of the chromatographic buffer was decreased to 8% and the temperature was increased to 37°C.
16S rRNA sequencing and phylogenetic analysis.
The 16S rRNA genes were amplified by PCR with universal primers 8UA (positions 8 to 28, Escherichia coli numbering) and 1485B (positions 1485 to 1507) as described previously (2). The amplified product was purified by using the QIAamp PCR purification kit (QIAGEN, Inc., Chatsworth, Calif.) and directly sequenced with a Biotech Diagnostic (Laguna Niguel, Calif.) Big Dye sequencing kit on an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.). The closest known relatives of the new isolates were determined by performing database searches using the BLAST software (1). Almost the full lengths of the 16S rRNA gene sequences (>1,400 nucleotides) of the unidentified bacteria and of closely related bacteria were aligned with CLUSTAL-W (http://genome.kribb.re.kr). A phylogenetic tree was reconstructed with DNA analysis software PAUP*, version 4.0 (Sinauer Associates, Inc., Sunderland, Mass.). The stability of the groupings was estimated by bootstrap analysis (1,000 replications) using the same program.
DNA-DNA reassociation.
DNA-DNA reassociation experiments were carried out according to the spectrophotometric method of De Ley et al. (3), using a Gilford System model 2600 spectrophotometer equipped with a Gilford model 2527-R thermal programmer.
Nucleotide sequence accession numbers.
The 16S rRNA sequences of strains WAL 11050T and WAL 10018T have been deposited in GenBank under accession numbers AY608697 and AY608696, respectively.
RESULTS
The unusual bacteria recovered from clinical infections were always isolated together with other anaerobes (mostly other species of the B. fragilis group) and/or aerobes. The majority of isolates were recovered in heavy growth on primary isolation. They were found to be gram-negative, rod-shaped organisms. Typical cells of both groups were 0.8 to 1.5 μm by 0.5 to 5.0 μm. Colonies on brucella blood agar plates at 48 h were grey, circular, convex, entire, and opaque and attained a diameter of 1 to 2 mm. All of the isolates grew well (resistant to 20% bile) and blackened the Bacteroides bile esculin agar by hydrolyzing esculin. They all grew well anaerobically, but no growth occurred following subculture in air or in atmospheres of 2 or 6% O2. All of the strains were resistant to the kanamycin (1,000 μg), vancomycin (5 μg), and colistin sulfate (10 μg) special-potency disks. They were indole positive and lipase, catalase, urease, and nitrate negative. They were capable of hydrolyzing esculin and gelatin. They all produced acid from cellobiose, glucose, rhamnose, sucrose, and xylose but did not produce acid from salicin, trehalose, and xylan. The strains of group I (B. stercoris-like) did not utilize arabinose, in contrast to strains of group II (B. uniformis-like). Tests with the API ZYM, Rapid ID 32A, and RapID ANA II systems showed that all isolates of the same group produced the same profile. Positive reactions were obtained for β-galactosidase, α-glucosidase, β-glucosidase, alkaline phosphatase, β-N-acetyl-glucosaminidase, leucyl glycine arylamidase, alanine arylamidase, glutamyl glutamic acid arylamidase, acid phosphatase, naphthol-AS-Bl-phosphohydrolase, and p-nitrophenylphosphatase. Mannose and raffinose were fermented in tests with the Rapid ID 32A system. In tests with the RapID ANA II system, p-nitrophenyl-β,d-disaccharidase reactions were strongly positive for isolates of group II but were either negative or weakly positive for group I. All the other tests were negative. α-Fucosidase testing by all three systems was negative. In peptone-yeast broth and peptone-yeast-glucose broth, a major amount of acetic acid and minor amounts of isovaleric acid, propionic acid, and formic acid were produced by all isolates. The principal long-chain cellular fatty acid of the isolates was C15:0 ANTEISO FAME (20 to 23% of total). Significant amounts of C15:0 ISO FAME, C16:0 FAME,C16:0 3OH FAME, and C18:1 CIS 9 FAME (9 to 18% each of the total) were also present. Agar dilution tests showed that most of the strains were susceptible to metronidazole (MIC ≤ 4 μg/ml), imipenem (MIC ≤ 2 μg/ml), clindamycin (MIC ≤ 4 μg/ml), amoxicillin-clavulanate (MIC ≤ 8 μg/ml), and ertapenem (MIC ≤ 1 μg/ml). Resistance to penicillin G (MIC ≥ 32 μg/ml), ceftizoxime (MIC ≥ 32 μg/ml), cefotetan (MIC ≥ 64 μg/ml), and vancomycin (MIC ≥ 128 μg/ml) was shown by all strains. All strains were β-lactamase positive.
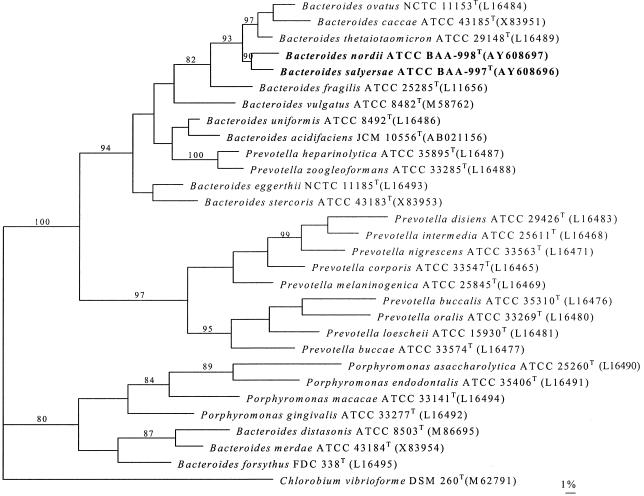
To assess the genealogical affinity between the unknown bacteria and their relationship with other taxa, their 16S rRNA gene sequences were determined. Pairwise analysis showed all of the isolates of the same group were phylogenetically closely related to each other (>99.5% sequence similarity). Sequence searches of GenBank and Ribosomal Database Project libraries for validly described species revealed that the unknown organisms were members of the Cytophaga-Flavobacter-Bacteroides (CFB) phylum, with a loose affinity with Prevotella and Porphyromonas phylogenetic clusters (>15% sequence divergence from species of the genera Prevotella and Porphyromonas sensu stricto) and a relatively close association with the Bacteroides cluster (>93% sequence similarity). A tree, constructed by the maximum-parsimony method, depicting the phylogenetic affinity of the two novel bacteria as exemplified by strains ATCC BAA-998T and ATCC BAA-997T is shown in Fig. 1; it confirmed the placement of the unknown bacteria in the Bacteroides phylogenetic cluster. It is evident from the branching pattern in the tree that the two novel species possess a significantly close relationship with each other (90% bootstrap resampling value). Chlorobium vibrioforme was used as the outgroup, since this species represents the phylum that branches closest to the CFB phylum (6). Pairwise comparison revealed approximately 4% sequence divergence between the two novel bacteria and approximately 5% sequence divergences between the unknown bacteria and the type strain of their closest valid species, B. thetaiotaomicron, based on almost the full lengths of the 16S rRNA gene sequences (>1,400 nucleotides). Although there is no precise correlation between percentages of 16S rRNA sequence divergence and species delineation, it is now generally accepted that organisms displaying divergence close to 3% or more do not belong to the same species (15). Furthermore, DNA-DNA reassociation values of 30.9, 13.5, and 16.8% for isolates WAL 11050T and WAL 10018T, WAL 11050T and ATCC 29741T (B. thetaiotaomicro type strain), and WAL 10018T and ATCC 29741T, respectively, were observed, thereby confirming that the unidentified bacteria represent two different species. The data clearly show that each group represents a previously unknown subline within the Bacteroides phylogenetic cluster.
FIG. 1.
Unrooted tree showing the phylogenetic position of B. nordii sp. nov. and B. salyersae sp. nov. within the Bacteroides phylogenetic cluster. The tree, constructed by the maximum-parsimony method, was based on a comparison of approximately 1,400 nucleotides. Bootstrap values, expressed as a percentage of 1,000 replications, are given at branching points. Scale bar = 1% sequence divergence.
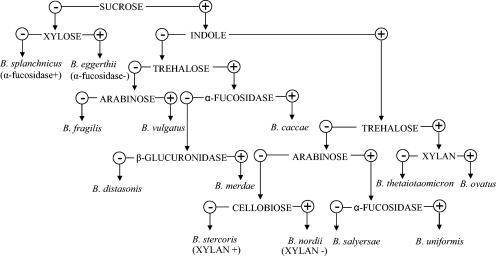
Support for the separation of the unknown bacteria from their related bacterial species also comes from the phenotypic characterizations. The two groups of unknown organisms can be readily distinguished from species of the genera Prevotella and Porphyromonas by their resistance to 20% bile. In addition, the unknown bacteria are highly fermentative, in contrast to asaccharolytic species in the genus Porphyromonas and moderately saccharolytic species in Prevotella. Within the Bacteroides phylogenetic cluster, although two groups of unknown bacteria are very much like each other in terms of biochemical characteristics, they can be differentiated from each other by virtue of arabinose fermentation: group II produces acid from arabinose, in contrast to group I. In addition, RapID ANA II testing indicated that group II showed a strongly positive reaction for p-nitrophenyl-β,d-disaccharidase, whereas group I was either negative or only weakly positive. The unknown organisms of groups I and II are also similar biochemically to B. stercoris and B. uniformis, respectively; therefore they were misidentified as B. stercoris and B. uniformis previously. However, organisms of group I can be differentiated from B. stercoris by several features, such as utilization of cellobiose but not xylan, production of glutamic acid decarboxylase but not arginine dihydrolase, and reaction to p-nitrophenyl-β,d-disaccharidase. Organisms of group II can also be differentiated from B. uniformis by not producing α-fucosidase, α-arabinosidase, or α-chymotrypsinase, whereas B. uniformis does. Although 16S rRNA gene sequence analysis showed the closest phylogenetic relative to both of the unknown bacteria is B. thetaiotaomicron, they can be distinguished easily from it by several biochemical characteristics. In particular, B. thetaiotaomicron produces acid from trehalose but the unknown organisms do not. The characteristics that distinguish the unknown bacteria from their phenotypically or phylogenetically closely related species are summarized in Table 2. Figure 2 presents a flow chart of key characteristics for identification and differentiation of the unknown organisms from the other species in the B. fragilis group.
TABLE 2.
Some properties by which B. nordii sp. nov. and B. salyersae sp. nov. can be differentiated from related Bacteroides spp.a
| Test | Resultb for:
|
|||||
|---|---|---|---|---|---|---|
| B. nordii (n = 12) | B. salyersae (n = 9) | B. thetaiotaomicron (n = 8) | B. stercoris (n = 8) | B. uniformis (n = 8) | B. fragilis (n = 8) | |
| PRASc tests | ||||||
| Arabinose | − | + | + | − | + | − |
| Trehalose | − | − | + | − | − | − |
| Cellobiose | + | + | + | − | W | + or − |
| Xylan | − | − | − | + | − | − |
| Biochemical tests | ||||||
| α-Fucosidase | − | − | + | + or − | + | + |
| α-Arabinosidase | − | − | + | − | + | − |
| Arginine dihydrolase | − | − | − | + | − | − |
| Glutamic acid | + | + | + | − | + | + |
| Arginine arylamidase | − | − | + | − | − | + |
| Leucine arylamidase | − | − | + | − | − | + |
| p-Nitrophenyl-β,d-disaccharidase | − | + | + | + | + | + |
| α-Chymotrypsinase | − | − | − | − | + | − |
Data are from the present study.
Symbols and abbreviations: +, positive; −, negative; W, weak reaction; + or −, variable.
PRAS, prereduced anaerobically sterilized.
FIG. 2.
Flow chart with key characteristics for identification and differentiation of the unknown organisms from the other species in the B. fragilis group and from Bacteroides splanchnicus.
DISCUSSION
In this study, we report on the characterization of two groups of unknown bacteria that are commonly isolated from clinical infections and are likely of intestinal origin. They appear to be of relatively low virulence since they were always found in mixed culture and were not recovered in blood cultures or in very serious infection. These two groups of clinical isolates were misidentified as B. stercoris and B. uniformis by routine biochemical tests. However, 16S rRNA sequencing revealed approximately 7% sequence divergence between the novel species and B. stercoris or B. uniformis, and the previously described species closest to both of them phylogenetically is B. thetaiotaomicron (approximate 94% sequence similarity). Although the two unknown bacteria display a number of similarities, 16S rRNA gene sequence divergence of approximately 4% between them, as well as distinguishable biochemical characteristics, demonstrated that these organisms are distinct. Subsequent DNA-DNA hybridization studies confirmed that the two novel organisms were indeed distinct from each other. A flow chart summarizes the key characteristics for identification and differentiation of the two organisms that we describe here from the other species in B. fragilis group. Based on both phenotypic and genotypic evidence, it is clear that the two groups of unknown isolates recovered from infections of intestinal origin in humans represent two novel species. We propose that the unknown isolates of group I and group II be classified as two new Bacteroides species, B. nordii and B. salyersae, respectively.
Description of B. nordii sp. nov.
B. nordii (to honor Carl Erik Nord, who has contributed so much to our knowledge of anaerobic bacteriology in general and intestinal bacteriology) cells are rod shaped and stain gram negative. Typical cells are 0.8 to 1.5 μm by 0.5 to 5.0 μm. Colonies on brucella blood agar plates at 24 h are grey, circular, convex, entire, and opaque and attain a diameter of 1 to 2 mm. The cells are obligately anaerobic, indole positive, and lipase, catalase, urease, and nitrate negative. Esculin and gelatin are hydrolyzed. Cells are resistant to 20% bile. Acid is produced from cellobiose, glucose, mannose, raffinose, rhamnose, sucrose, and xylose but not from arabinose, salicin, trehalose, and xylan. In peptone-yeast broth and peptone-yeast-glucose broth, a major amount of acetic acid and minor amounts of isovaleric acid, propionic acid, and formic acid are produced by all isolates. Positive reactions were obtained for β-galactosidase, α-glucosidase, β-glucosidase, alkaline phosphatase, β-N-acetyl-glucosaminidase, leucyl glycine arylamidase, alanine arylamidase, glutamyl glutamic acid arylamidase, acid phosphatase, naphthol-AS-Bl-phosphohydrolase, and p-nitrophenylphosphatase with the API ZYM, Rapid ID 32A, and RapID ANA II systems. Negative reactions for α-fucosidase were obtained with all three systems. The major long-chain cellular fatty acid is C15:0 ANTEISO FAME (20 to 23% of total). Significant amounts of C15:0 ISO FAME, C16:0 FAME, C16:0 3OH FAME, and C18:1 CIS 9 FAME (9 to 18% of the total each) are also present. Cells showed resistance to kanamycin (1,000 μg), vancomycin (5 μg), and colistin sulfate (10 μg) identification disks; agar dilution tests showed that most of the strains are susceptible or had intermediate susceptibility to metronidazole (MIC ≤ 4 μg/ml), imipenem (MIC ≤ 2 μg/ml), clindamycin (MIC ≤ 4 μg/ml), amoxicillin-clavulanate (MIC ≤ 8 μg/ml), and ertapenem (MIC ≤ 1 μg/ml). Resistance to penicillin G (MIC ≥ 32 μg/ml), ceftizoxime (MIC ≥ 32 μg/ml), cefotetan (MIC ≥ 64 μg/ml), and vancomycin (MIC ≥ 128 μg/ml) is shown by all strains. All strains were β-lactamase positive (Table 3).
TABLE 3.
Summary of characteristics of B. nordii sp. nov. and B. salyersae sp. nov.
| Parameter | Resulta for:
|
|
|---|---|---|
| B. nordii | B. salyersae | |
| Cell size (μm) | 0.8-1.5 by 0.5-5.0 | 0.8-1.5 by 0.5-5.0 |
| Growth in 20% bile | + | + |
| Indole production | + | + |
| Esculin hydrolysis | + | + |
| Nitrate reduction | − | − |
| Acid production | ||
| Arabinose | − | + |
| Cellobiose | + | + |
| Glucose | + | + |
| Mannose | + | + |
| Raffinose | + | + |
| Sucrose | + | + |
| Xylose | + | + |
| Production of: | ||
| α-Galactosidase | + | + |
| β-Galactosidase | + | + |
| α-Glucosidase | + | + |
| β-Glucosidase | ||
| Alkaline phosphatase | + | + |
| Acid phosphatase | + | + |
| α-Fucosidase | − | − |
| p-Nitrophenyl-β,d- disaccharidase | − | + |
| β-Lactamase | + | + |
| Susceptibility | ||
| Metronidazole | S | S |
| Imipenem | S | S |
| Clindamycin | S | S |
| Amoxicillin-clavulanate | S | S |
| Ertapenem | S | S |
| Penicillin G | R | R |
| Ceftizoxime | R | R |
| Cefotetan | R | R |
| Vancomycin | R | R |
| CFAb | C15:0 ANTEISO FAME | C15:0 ANTEISO FAME |
| G+C content (mol%) | 41.4% | 42.0% |
| Fermentation products | A, p, iv, f | A, p, iv, f |
Data were obtained with the API ZYM, Rapid ID 32A, and RapID ANA II systems. S, sensitive; R, resistant, A, acetic acid; p, propionic acid; iv, isovaleric acid; f, formic acid.
CFA, cellular fatty acid.
The cells were isolated from human clinical specimens of intestinal origin. The habitat is probably the human gut. The type strain is WAL 11050 (ATCC BAA-998 or CCUG 48943). The G+C content of the type strain is 41.4 mol%.
Description of B. salyersae sp. nov.
B. salyersae (to honor Abigail Salyers, who has contributed so much to our knowledge of intestinal bacteriology and anaerobic bacteriology in general) cells are rod shaped and stain gram negative. Typical cells are 0.8 to 1.5 μm by 0.5 to 5.0 μm. Colonies on brucella blood agar plates at 24 h are grey, circular, convex, entire, and opaque and attain a diameter of 1 to 2 mm. Cells are obligately anaerobic, indole positive, and lipase, catalase, urease, and nitrate negative. Esculin and gelatin are hydrolyzed. Cells are resistant to 20% bile. Acid is produced from arabinose, cellobiose, glucose, mannose, raffinose, rhamnose, sucrose, and xylose but not from salicin, trehalose, and xylan. In peptone-yeast broth and peptone-yeast-glucose broth, a major amount of acetic acid and minor amounts of isovaleric acid, propionic acid, and formic acid are produced by all isolates. Positive reactions are obtained for α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, alkaline phosphatase, β-N-acetyl-glucosaminidase, leucyl glycine arylamidase, alanine arylamidase, glutamyl glutamic acid arylamidase, acid phosphatase, naphthol-AS-Bl-phosphohydrolase, p-nitrophenylphosphatase and p-nitrophenyl-β,d-disaccharidase with the API ZYM, Rapid ID 32A, and RapID ANA II systems. Negative reactions for α-fucosidase were obtained with all three systems. The long-chain cellular fatty acid produced by most of the isolates is C15:0 ANTEISO FAME (20 to 23% of total). Significant amounts of C15:0 ISO FAME, C16:0 FAME, C16:0 3OH FAME,and C18:1 CIS 9 FAME (9 to 18% each of total) are also present. Cells showed resistance to kanamycin (1,000 μg), vancomycin (5 μg), and colistin sulfate (10 μg) identification disks; agar dilution tests showed that most of the strains are susceptible or had intermediate susceptibility to metronidazole (MIC ≤ 4 μg/ml), imipenem (MIC ≤ 2 μg/ml), clindamycin (MIC ≤ 4 μg/ml), amoxicillin-clavulanate (MIC ≤ 8 μg/ml), and ertapenem (MIC ≤ 1 μg/ml). Resistance to penicillin G (MIC ≥ 32 μg/ml), ceftizoxime (MIC ≥ 32 μg/ml), cefotetan (MIC ≥ 64 μg/ml), and vancomycin (MIC ≥ 128 μg/ml) is shown by all strains. All strains were β-lactamase positive (Table 3).
Cells were isolated from human clinical specimens of intestinal origin. The habitat is probably the human gut. The type strain is WAL 10018 (ATCC BAA-997 or CCUG 48945). The G+C content of the type strain is 42.0 mol%.
Acknowledgments
This work has been carried out, in part, with financial support from Veterans Administration Merit Review funds.
REFERENCES
- 1.Benson, D. A., M. S. Boguski, D. J. Lipman, and J. Ostell. 1997. GenBank. Nucleic Acids Res. 25:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Ley, J., H. Cattoir, and A. Reynaerts. 1970. The quantitative measurements of DNA hybridization from renaturation rates. Eur. J. Biochem. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 4.Finegold, S. M. 1995. Anaerobic infections in humans: an overview. Anaerobe 1:3-9. [DOI] [PubMed] [Google Scholar]
- 5.Finegold, S. M. 1977. Anaerobic bacteria in human disease. Academic Press, New York, N.Y.
- 6.Gherna, R., and C. R. Woese. 1992. A partial phylogenetic analysis of the “Flavobacter-Bacteroides” phylum: basis for taxonomic restructuring. Syst. Appl. Microbiol. 15:513-521. [DOI] [PubMed] [Google Scholar]
- 7.Holdeman, L. V., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg.
- 8.Jousimies-Somer, H. R. 1995. Update on the taxonomy and the clinical and laboratory characteristics of pigmented anaerobic gram-negative rods. Clin. Infect. Dis. 20(Suppl. 2):S187-S191. [DOI] [PubMed] [Google Scholar]
- 9.Jousimies-Somer, H. R., P. Summanen, D. M. Citron, E. J. Baron, H. M. Wexler, and S. M. Finegold. 2002. Wadsworth anaerobic bacteriology manual, 6th ed. Star Publishing Co., Belmont, Calif.
- 10.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2001. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 5th ed. Approved standard M11-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Redondo, M. C., M. D. Arbo, J. Grindlinger, and D. R. Snydman. 1995. Attributable mortality of bacteremia associated with the Bacteroides fragilis group. Clin. Infect. Dis. 20:1492-1496. [DOI] [PubMed] [Google Scholar]
- 13.Shah, H. N., and D. M. Collins. 1990. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int. J. Syst. Bacteriol. 40:205-208. [DOI] [PubMed] [Google Scholar]
- 14.Snydman, D. R., N. V. Jacobus, L. A. McDermott, R. Ruthazer, E. J. C. Goldstein, S. M. Finegold, L. J. Harrell, D. W. Hecht, S. G. Jenkins, C. Pierson, R. Venezia, J. Rihs, and S. L. Gorbach. 2002. National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends for 1997-2000. Clin. Infect. Dis. 35(Suppl. 1):S126-S134. [DOI] [PubMed] [Google Scholar]
- 15.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 16.Wexler, H. M., D. Reeves, P. H. Summanen, E. Molitoris, M. McTeague, J. Duncan, K. H. Wilson, and S. M. Finegold. 1996. Sutterella wadsworthensis gen. nov., sp. nov., bile-resistant microaerophilic Campylobacter gracilis-like clinical isolates. Int. J. Syst. Bacteriol. 46:252-258. [DOI] [PubMed] [Google Scholar]