Abstract
We studied the feasibility of genotyping human immunodeficiency virus (HIV) type 2 groups A and B by real-time PCR. Two group-specific PCRs were developed. Real-time genotyping of 22 samples of genotype A, 10 samples of genotype B, and the isolate of new group H were compared to genotyping by sequencing and phylogeny. The group-specific PCRs specifically identified 84.3% of group A or B samples; isolate H was not detected. This method allowed rapid and specific discrimination between HIV-2 groups A and B and could be a useful tool for molecular epidemiological studies.
Infection by human immunodeficiency virus type 2 (HIV-2) was discovered in Senegalese patients in 1985 (1) and was first isolated from a Cape Verdian patient suffering from AIDS in 1986 (4). In contrast to HIV-1, the HIV-2 epidemic has remained restricted mainly to west Africa (3, 10, 22), with some cases being reported occasionally in central Africa (Gabon, Cameroon, and Equatorial Guinea) (9, 14, 21) and east Africa (16). Its presence reported in Europe (France, Portugal, and Spain) (13, 15, 20) reflects historical links to the areas where the virus is endemic. A limited number of cases are present in Argentina, the Philippines, and the United States (18, 19, 23). However, there is no epidemic in these regions, in contrast to India, where HIV-2 seems to be spreading (17, 24). Analysis of genetic divergence of HIV-2 isolates has led to classification into subtypes. As the different subtypes of HIV-2 are analogous in term of genetic distance to the different groups of HIV-1, and as they are most likely to have arisen by independent cross-species transmission events (12), the HIV nomenclature has been recently revised (http://www.hiv.lanl.gov/content) and HIV-2 is presently defined by eight phylogenetic groups, designated A to H (3, 8, 10, 27).
Numerous data are available on strain diversity and molecular epidemiology for HIV-1, but little is known about variation and geographic distribution of HIV-2 groups. Only groups A and B are epidemic in west Africa. Group A, which contains most HIV-2 strains characterized so far, has been identified in Cape Verde Islands, Côte d'Ivoire, The Gambia, Ghana, Guinea-Bissau, Mali, and Senegal (16). Group B viruses, exhibiting greater geographic restriction, have been reported mainly in Côte d'Ivoire and Ghana (16). The groups C, D, E, and F, represented by only partial sequences of single virus genomes, have been identified in Liberia (groups C and D) (10) and Sierra Leone (groups E and F) (3, 10). Groups G and H are represented by the full-length genomic sequence of strains collected in Côte d'Ivoire from an asymptomatic blood donor (27) and a symptomatic patient (8), respectively. During recent years, extensive molecular epidemiological studies have been performed to survey the dynamics of HIV-1 groups and subtypes. Such molecular epidemiological studies are also needed to monitor dynamics and changes in the distribution of the HIV-2 groups in countries where the virus is endemic and to monitor emerging groups outside of west Africa. It might be also critical to discriminate between these groups to investigate if different clinical and biological properties exist.
It was previously shown that V3 serotyping, a rapid and simple laboratory method used for HIV-1 subtyping, was not relevant for HIV-2 (25). HIV-2 genotyping is essentially performed by nucleotidic sequencing of genomic fragments followed by phylogenetic analysis. Although this methodology is the gold standard and allows direct group classification, it is time-consuming, expensive, requires a qualified operator, and does not allow rapid analysis of numerous samples. Alternative methods are needed. Recently, a combination of long terminal repeat (LTR) sequence analysis and group-specific amplification of groups A and B with an ethidium bromide staining visualization has been developed (2); however, no group B samples were present in the evaluation panel. The aim of this study was to determine the feasibility of simple and rapid genotyping by real-time PCR based on LightCycler technology (Roche Diagnostics, Mannheim, Germany).
For the present work, real-time genotyping of 33 samples was performed and compared to data generated by sequencing and phylogenetic analysis. Samples were collected in the Bichat hospital, Paris, France (n = 21), and in the Infectious Diseases unit at the Rouen hospital (n = 12), France. Seventeen samples (including the new isolate defined as group H) were previously genotyped (5, 8). Genotyping of the 16 remaining samples was performed by sequencing and phylogenetic analysis of reverse transcriptase (RT; the pol gene).
Briefly, proviral DNA was extracted with a QIAamp DNA Blood Mini kit (Qiagen, Courtaboeuf, France). A genomic region of 1,054 bp encoding the 5′ end of the RT was amplified by nested PCR (amino acids 32 to 383) that used outer (RTC and RT2) and inner (RT3 and RT4) primers (10, 26). Cycling temperatures and times were slightly modified. The first round of PCR had an initial denaturation step at 95°C for 15 min. It was followed by 35 amplification cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min 30 s) and then by an elongation step at 72°C for 7 min. Nested PCR conditions were as follows: denaturation at 95°C for 15 min, followed by 30 amplification cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min) and then an elongation step at 72°C for 7 min. The PCR products were purified with the QIAquick PCR purification kit (Qiagen) and were sequenced on both strands with sense primers RT3b (5′-GAGGCACTAAAAGAGATCTG-3′) and RT5 and antisense primers RT4 and RT6, as previously described (10, 26). The sequence reaction was carried out with a CEQ Dye Teminator Cycle Sequencing with Quick Start kit (Beckman, Fullerton, Calif.) and was purified by alcoholic precipitation. Sequencing was processed on an automated capillary CEQ 8000 DNA sequencer (Beckman). Nucleotide sequences based on 949 sites were aligned with the CLUSTAL W program with minor manual adjustments to take into account the protein sequences. A phylogenetic tree was produced by the neighbor-joining method, and the reliability of the branching orders was assessed by the bootstrap approach (1,000 replicates) with the CLUSTAL W program. Genetic distances were calculated by Kimura's two-parameter method.
Real-time PCR genotyping was processed on templates including DNA from infected cells by coculture with patients' isolates (n = 10) and from uncultured peripheral blood mononuclear cells (PBMCs) from HIV-2-seropositive patients (n = 23). PBMCs, separated on a Ficoll gradient, were washed twice in phosphate-buffered saline (PBS), pelleted, and stored at −80°C. DNA was extracted by using the QIAamp DNA Blood Mini kit (Qiagen) according to the manufacturer's recommendations. Two group-specific real-time PCRs were developed for a region that spans the 3′ end of the LTR to the 5′ start of gag, amplifying a 286-bp group A-specific fragment and a 171-bp group B-specific fragment. Primers and hydrolysis probes were defined to be specific for each group. The probes were synthesized with a reporter fluorescein dye (6-carboxyfluorescein [FAM]) attached to the 5′ end and the quencher TAMRA (6-carboxytetramethylrhodamine) linked to the 3′ end. The PCR conditions with LightCycler-FastStart DNA Master Hybridization Probes (Roche Diagnostics) were defined as follows. The LightCycler master mix (2 μl) was mixed with 4 mM MgCl2, 0.5 μM group A-specific primers (U HIV2 A, 5′-TAGTCGCCGCCTGGTCA-3′; and L HIV2 A, 5′-GGTGTAGGTACTTACCTTCACCC-3′), and 0.25 μM group A-specific probe S HIV2A (FAM-AGTAACAAGACCCTGGTCTGTTAGGACC-TAMRA) in one capillary. A similar mixture was prepared in a second capillary with 0.25 μM group B-specific primers (U HIV2 B, 5′-AGTAAGGGCGGCAGGAAT-3′; and L HIV2 B, 5′-CGCCCATCTCCCACTTAT-3′) and 0.25 μM group B-specific probe S HIV2 B (FAM-AGGCCGGTACCAGGCAGCGT-TAMRA). A variable volume of extract, containing 500 ng of DNA (if possible), was added. The final volume of 20 μl was reached by supplementing the solution with water. Amplification was carried out as follows: 95°C for 10 min (one cycle), denaturation at 95°C for 10 s, and annealing from 70°C to 60°C for 30 s. The reduction in the annealing temperature was done for 10 cycles (Touchdown PCR); denaturation was at 95°C for 10 s, and annealing was at 60°C for 30 s (40 cycles).
Analytical sensitivity and intra- and interrun reproducibility of both group-specific PCRs were performed by using two reference strains representing groups A (ROD; GenBank accession number BD131284) and B (P1102; GenBank accession number AF170058). These HIV-2 DNA standards were cellular extracts after coculture, quantified by using primers defined specifically to amplify HIV-2 ROD and P1102 as well as HIV-1 BRU strains (GenBank accession number K02013). For the cell line 8E5, containing a single provirus of HIV-1 BRU per cell, we quantified the extracted DNA of HIV-2 ROD and P1102 strains with a known range of DNA extract from the 8E5 cell line (11). The sensitivity of the real-time PCR method was determined by testing serial dilutions of DNA from the two strains at 10,000, 1,000, 100, 50, 20, and 10 copies/capillary (five replicates). To assess intrarun reproducibility, dilutions containing 100 or 20 copies/capillary were tested 10 times each in the same experiment. To determine interrun variability, dilutions containing 100 or 20 copies/capillary were run in duplicate in four different experiments.
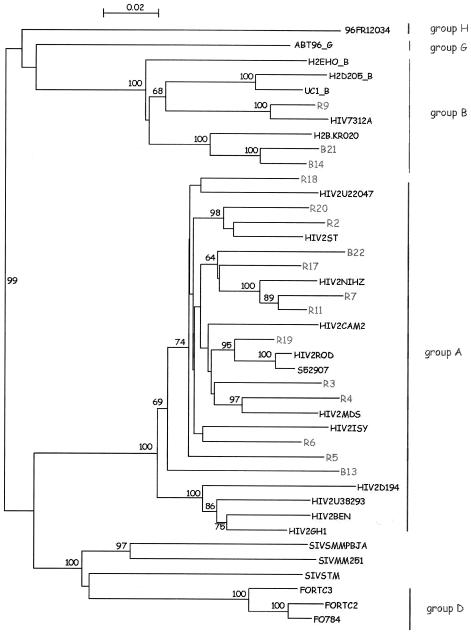
Genotyping by sequencing confirmed 22 genotype A samples and 10 genotype B samples. One more sample, corresponding to the new isolate fully sequenced and described recently as group H, was also analyzed (8). Results of the RT region sequencing are presented in Fig. 1.
FIG. 1.
Phylogenetic tree based on 949 unambiguously aligned nucleotides from the RT region. The 16 samples sequenced are included in the tree; samples from Bichat and Rouen hospitals are identified with a B or an R, respectively. The numbers near nodes indicate bootstrap values greater than 60%. The analysis was performed as described in the text.
All 33 samples, plus human DNA as negative control, were tested in both group-specific PCRs. Twenty-seven out of the 32 group A or B samples (84.3%) were amplified and correctly identified by the group-specific PCRs (Table 1). Analysis by electrophoresis on agarose gel and ethidium bromide staining indicated no amplification for the five undetectable samples of DNA (four from fresh cells and one from culture cells). The isolate H and DNA control were not detected by any group-specific PCR. Seventeen of the 22 group A samples (77.2%) were specifically detected by the group A-specific PCR. All of the group B samples were specifically detected by the group B-specific PCR.
TABLE 1.
Correlation between reference genotype and genotype by group-specific PCR
| Group-specific real-time PCR | Reference genotype
|
|||
|---|---|---|---|---|
| Culture cells
|
Fresh cells
|
|||
| A (n = 5) | B (n = 4) | A (n = 17) | B (n = 6) | |
| A | 4 | 0 | 13 | 0 |
| B | 0 | 4 | 0 | 6 |
To assess analytical performances of both PCRs, sensitivity as well as intrarun and interrun reproducibility studies were carried out with two representative strains of groups A and B. The detection rate of both PCRs was 100% in dilutions of 20 copies/capillary and were 20 and 40% at 10 copies/capillary for group A- and group B-specific PCR, respectively. Results of the intrarun and interrun reproducibility analyses are presented in Table 2. The coefficients of variation lie between 0.71 and 0.97 for the intrarun assay and between 0.8 and 1.22 for the interrun assay.
TABLE 2.
Intra- and interrun reproducibility of group A- and group B-specific PCRs
| Reproducibility criterion | Group A
|
Group B
|
||
|---|---|---|---|---|
| 100 copies/capillary | 20 copies/capillary | 100 copies/capillary | 20 copies/capillary | |
| Intrarun | ||||
| Crossing point mean | 25.97 | 28.01 | 22.69 | 24.87 |
| Crossing point SD | 0.25 | 0.27 | 0.16 | 0.21 |
| Coefficient of variation (%) | 0.95 | 0.97 | 0.71 | 0.84 |
| Interrun | ||||
| Crossing point mean | 26.15 | 28.50 | 22.58 | 24.74 |
| Crossing point SD | 0.96 | 0.33 | 0.28 | 0.20 |
| Coefficient of variation (%) | 1.13 | 1.15 | 1.22 | 0.8 |
The aim of the present study was to determine if rapid and simple genotyping of HIV-2 could be efficient and relevant. Our results indicate a strong correlation between the gold standard and our technique. A higher sensitivity for group B detection was observed (all samples were correctly identified). This result could be due to the analytical performance of each PCR rather than being linked to diversity. The analytical threshold was fixed at 20 copies/capillary because of 100% detection with both PCRs, but group B PCR seems to be more sensitive than group A at 10 copies/capillary (40% of detection versus 20% for group A). The hypothesis of a higher proviral load in group B leading to a better sensitivity cannot be rejected, but such a hypothesis would not be consistent with recent data (6). Among the five undetectable group A samples, four had amounts of DNA below 500 ng (from 25 ng to 275 ng). Small amounts of DNA associated with low proviral load and/or with a lesser sensitivity of the group A-specific PCR may explain these results, as may possible mismatch due to genetic variability in the amplification region. This latter hypothesis seems not to be relevant, because no mismatch was observed with the HIV-2 group A sequences published in the HIV sequence database (http://www.hiv.lanl.gov/content). However, variability of this region is difficult to assess, because only 15 group A and 4 group B sequences are available in this database.
In terms of analytical performance, the LightCycler system offers high intra- and interrun reproducibility for both PCRs. The real-time PCR allows a simplified protocol with fewer steps than classical PCR. This technology is presently available in some reference centers in developing countries, in particular in west Africa, where HIV-2 is endemic. This tool could be useful for studying HIV-2 infection in field conditions and for a rapid screening of non-A, non-B divergent HIV-2 strains. Actually, only HIV-2 groups A and B are prevalent, the epidemic being more restricted than that for HIV-1 group M. However, efficient tools could be useful to discriminate simply and rapidly between these two groups in order to investigate if different biological properties exist. The large nucleotidic distances between HIV-2 groups may lead to such differences. A direct consequence of HIV-2 diversity is observed in serological diagnosis: group B samples are involved in a specific pattern of intensive cross-reactivities on HIV-1 Western blotting (5), which can lead to the misidentification of a dual HIV-1/HIV-2 infection and, thus, a maladapted follow-up. Recently, it was observed that group B samples had lower viral loads by RNA real-time quantification (7). A population bias was considered, but further evaluation is needed to confirm or refute that point. It was also proposed that group A might be more pathogenic than group B (10), but this was not confirmed by a recent study based on in vivo data in France (5). Moreover, little is known about the consequences of HIV-2 diversity. Whatever the groups, HIV-2 is naturally resistant to nonnucleosidic reverse transcriptase inhibitors, but the consequences for viral load determination and for drug resistance profiles are not clearly known. An RNA version of this format could be suitable but will be extremely difficult to achieve, due to very low plasma RNA viral load in HIV-2 infection (7).
In conclusion, our method allows rapid and specific discrimination between HIV-2 groups A and B without sequencing. This PCR method is not a molecular screening tool but was developed as a complementary tool for grouping strains of HIV-2. HIV-2 infection must first be diagnosed. Larger evaluation of this technique is now needed. This technique could be a useful and simple tool for molecular epidemiological studies by allowing for discriminating rapidly between these two groups.
Acknowledgments
We thank the technicians of the Virology Unit from Rouen and Bichet hospitals, especially J. M. Dupré and S. Delarue.
This work was supported by the Ministère de la Santé and the Agence Nationale de Recherche sur le Sida (ANRS), Paris, France.
REFERENCES
- 1.Barin, F., S. M'Boup, F. Denis, P. Kanki, J. S. Allan, T. H. Lee, and M. Essex. 1985. Serological evidence for virus related to simian T-lymphotropic retrovirus III in residents of west Africa. Lancet ii:1387-1389. [DOI] [PubMed] [Google Scholar]
- 2.Berry, N., K. Ariyoshi, P. Balfe, R. Tedder, and H. Whittle. 2001. Sequence specificity of the human immunodeficiency virus type 2 (hiv-2) long terminal repeat u3 region in vivo allows subtyping of the principal hiv-2 viral subtypes a and b. AIDS Res. Hum. Retrovir. 17:263-267. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z., A. Luckay, D. L. Sodora, P. Telfer, P. Reed, A. Gettie, J. M. Kanu, R. F. Sadek, J. Yee, D. D. Ho, L. Zhang, and P. A. Marx. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol. 71:3953-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clavel, F., D. Guétard, F. Brun-Vézinet, S. Chamaret, M. A. Rey, M. O. Santos-Ferreira, A. G. Laurent, C. Dauguet, C. Katlama, C. Rouzioux, D. Klatzmann, J. L. Champalimaud, and L. Montagnier. 1986. Isolation of a new human retrovirus from west African patients with AIDS. Science 233:343-346. [DOI] [PubMed] [Google Scholar]
- 5.Damond, F., C. Apetrei, D. L. Robertson, S. Souquiere, A. Lepretre, S. Matheron, J. C. Plantier, F. Brun-Vezinet, and F. Simon. 2001. Variability of human immunodeficiency virus type 2 (HIV-2) infecting patients living in France. Virology 280:19-30. [DOI] [PubMed] [Google Scholar]
- 6.Damond, F., D. Descamps, I. Farfara, J. N. Telles, S. Puyeo, P. Campa, A. Lepretre, S. Matheron, F. Brun-Vezinet, and F. Simon. 2001. Quantification of proviral load of human immunodeficiency virus type 2 subtypes A and B using real-time PCR. J. Clin. Microbiol. 39:4264-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damond, F., M. Gueudin, S. Pueyo, I. Farfara, D. L. Robertson, D. Descamps, G. Chene, S. Matheron, P. Campa, F. Brun-Vezinet, and F. Simon. 2002. Plasma RNA viral load in human immunodeficiency virus type 2 subtype A and subtype B infections. J. Clin. Microbiol. 40:3654-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damond, F., M. Worobey, P. Campa, I. Farfara, G. Colin, S. Matheron, F. Brun-Vézinet, D. L. Robertson, and F. Simon. 2004. Identification of a highly divergent HIV-2 and proposal for a change in HIV-2 classification. AIDS Res. Hum. Retrovir. 20:666-672. [DOI] [PubMed] [Google Scholar]
- 9.Delaporte, E., W. Janssens, M. Peeters, A. Buve, G. Dibanga, J. L. Perret, V. Ditsambou, J. R. Mba, M. C. Courbot, A. Georges, A. Bourgeois, B. Samb, D. Henzel, L. Heyndrickx, K. Fransen, G. van der Groen, and B. Larouze. 1996. Epidemiological and molecular characteristics of HIV infection in Gabon, 1986-1994. AIDS 10:903-910. [DOI] [PubMed] [Google Scholar]
- 10.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, G. M. Shaw, et al. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gueudin, M., F. Damond, and F. Simon. Quantification of proviral DNA load of human immunodeficiency virus type 2 subtypes A and B using real time PCR. Human retrovirus protocols. In T. Zhu (ed.), Virology and Molecular Biology, vol. 1, in press. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 12.Hahn, B., G. M. Shaw, K. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 13.Heredia, A., A. Vallejo, V. Soriano, A. Aguilera, A. Mas, J. S. Epstein, and I. K. Hewlett. 1997. Genetic analysis of an HIV type 2 subtype B virus from a Spanish individual with AIDS. AIDS Res. Hum. Retrovir. 13:899-900. [DOI] [PubMed] [Google Scholar]
- 14.Heredia, A., A. Vallejo, V. Soriano, M. Gutierrez, S. Puente, J. S. Epstein, and I. K. Hewlett. 1997. Evidence of HIV-2 infection in Equatorial Guinea (central Africa): partial genetic analysis of a B subtype virus. AIDS Res. Hum. Retrovir. 13:439-440. [DOI] [PubMed] [Google Scholar]
- 15.Heredia, A., A. Vallejo, V. Soriano, A. Silva, K. Mansinho, S. Fevereiro, A. Mas, M. Gutierrez, J. S. Epstein, and I. K. Hewlett. 1998. Phylogenetic analysis of HIV type 2 strains from Portugal. AIDS Res. Hum. Retrovir. 14:471-473. [DOI] [PubMed] [Google Scholar]
- 16.Kanki, P. J., and K. M. De Cock. 1994. Epidemiology and natural history of HIV-2. AIDS 8:(Suppl.) 585-593. [Google Scholar]
- 17.Kannangai, R., R. V. Shaji, S. Ramalingam, M. V. Jesudason, O. C. Abraham, R. George, A. P. Shanmugam, D. H. Schwartz, and G. Sridharan. 2003. HIV-2 subtype circulating in India (south). J. Acquir. Immun. Defic. Syndr. 33:219-222. [DOI] [PubMed] [Google Scholar]
- 18.Leano, P. S., S. Kageyama, A. Espantaleon, J. Maniar, M. Iwasaki, D. Saple, N. Yoshihara, T. Kurimura, and D. M. Agdamag. 2003. Introduction of human immunodeficiency virus type 2 infection in the Philippines. J. Clin. Microbiol. 41:516-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libonatti, O., M. Avila, R. Zlatkes, S. Pampuro, L. M. Peralta, E. Rud, M. Wainberg, and H. Salomon. 1998. First report of HIV-2 infection in Argentina. J. Acquir. Immun. Defic. Syndr. Hum. Retrovirol. 18:190-191. [DOI] [PubMed] [Google Scholar]
- 20.Loussert-Ajaka, I., D. Descamps, F. Simon, F. Brun-Vezinet, M. Ekwalanga, and S. Saragosti. 1995. Genetic diversity and HIV detection by polymerase chain reaction. Lancet 346:912-913. [DOI] [PubMed] [Google Scholar]
- 21.Mauclere, P., I. Loussert-Ajaka, F. Damond, P. Fagot, S. Souquieres, M. Monny Lobe, F. X. Mbopi Keou, F. Barre-Sinoussi, S. Saragosti, F. Brun-Vezinet, and F. Simon. 1997. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS 11:445-453. [DOI] [PubMed] [Google Scholar]
- 22.Norrgren, H., S. Marquina, T. Leitner, P. Aaby, M. Melbye, A. G. Poulsen, O. Larsen, F. Dias, D. Escanilla, S. Andersson, J. Albert, and A. Naucler. 1997. HIV-2 genetic variation and DNA load in asymptomatic carriers and AIDS cases in Guinea-Bissau. J. Acquir. Immun. Defic. Syndr. Hum. Retrovirol. 16:31-38. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien, T. R., J. R. George, and S. D. Holmberg. 1992. Human immunodeficiency virus type 2 infection in the United States. Epidemiology, diagnosis, and public health implications. JAMA 267:2775-2779. [PubMed] [Google Scholar]
- 24.Pfutzner, A., U. Dietrich, U. von Eichel, H. von Briesen, H. D. Brede, J. K. Maniar, and H. Rubsamen-Waigmann. 1992. HIV-1 and HIV-2 infections in a high-risk population in Bombay, India: evidence for the spread of HIV-2 and presence of a divergent HIV-1 subtype. J. Acquir. Immun. Defic. Syndr. 5:972-977. [PubMed] [Google Scholar]
- 25.Plantier, J. C., F. Damond, S. Souquieres, F. Brun-Vezinet, F. Simon, and F. Barin. 2001. V3 serological subtyping of human immunodeficiency virus type 2 infection is not relevant. J. Clin. Microbiol. 39:3803-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodes, B., A. Holguin, V. Soriano, M. Dourana, K. Mansinho, F. Antunes, and J. Gonzalez-Lahoz. 2000. Emergence of drug resistance mutations in human immunodeficiency virus type 2-infected subjects undergoing antiretroviral therapy. J. Clin. Microbiol. 38:1370-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi, J., S. G. Devare, and C. A. Brennan. 2000. Identification of a new HIV-2 subtype based on phylogenetic analysis of full-length genomic sequence. AIDS Res. Hum. Retrovir. 16:925-930. [DOI] [PubMed] [Google Scholar]