Abstract
It is becoming increasingly clear that the evolution of infectious disease is influenced by host population structure. Theory predicts that parasites should be more ‘prudent’—less transmissible—in spatially structured host populations. However, here we (i) highlight how low transmission, the phenotype being selected for in this in context, may also be achieved by rapacious host exploitation, if fast host exploitation confers a local, within-host competitive advantage and (ii) test this novel concept in a bacteria–virus system. We found that limited host availability and, to a lesser extent, low relatedness favour faster-killing parasites with reduced transmission. By contrast, high host availability and high relatedness favour slower-killing, more transmissible parasites. Our results suggest high, rather than low, virulence may be selected in spatially structured host–parasite communities where local competition and hence selection for a within-host fitness advantage is high.
This article is part of the themed issue ‘Opening the black box: re-examining the ecology and evolution of parasite transmission’.
Keywords: transmission, parasites, growth, virulence, population structure
1. Introduction
Owing to the importance of infectious diseases to human health, agriculture and wildlife populations [1–7], it is critical that we better understand how ecology shapes the evolution of life histories of infectious organisms. In particular, host spatial population structure is ubiquitous in natural and managed populations. A growing body of theoretical models, observational studies and experiments suggests that host population structure will select for ‘prudent’ parasites with lower rates of parasite transmission, because this strategy leads to more-efficient exploitation of the susceptible local host population and higher parasite densities. A reason for evolution towards lower transmission rate in structured populations is due to local extinctions of infected hosts: a higher transmission rate is selected until a critical point is reached beyond which any further increase in transmission will cause the local cluster of hosts to be wiped out very rapidly [8]. In addition to the direct fitness benefit of low transmission in structured host populations, there is also an indirect fitness benefit: host population structure increases parasite relatedness [9,10], hence low transmission reduces competition between kin [9,11–19].
The above models [9,11–15], and many others [20–25], assume that transmission is a positive function of within-host growth rate, which also affects pathogen virulence (reductions in host fitness caused by the parasite). Natural selection will favour an optimal level of virulence that tends to maximize parasite transmission rate and minimize infected-host death rate because death of the host ceases transmission [9,21,26]. Low transmission can therefore result from both high and low within-host growth rate and hence virulence. However, while mild parasites do not regulate the host population, high virulence may have negative consequences on host availability if infected hosts die and/or recovered hosts cannot reproduce; hence models typically predict that population structure will select for low transmission and low virulence [8].
Many parasites, however, must kill their hosts to achieve transmission, and in these contexts selection by definition favours maximal virulence. These obligately killing parasites are found among many groups of organisms, including bacteriophages, nuclear polyhedroviruses, bacteria, nematodes, fungi and microsporidia [27]. They grow inside their host and convert host biomass into parasite transmission stages that are released into the environment when the host ruptures [28,29]. However, obligate killing parasites face an analogous virulence-transmission trade-off to parasites that need to transmit from live hosts, in terms of the rate they kill their hosts. Specifically, some intermediate rate of kill will maximize transmission; too short, and there will be fewer propagules developed inside the host before host death, and too long inevitably reduces the rate of transmission [27]. Crucially, in the case of obligate killing parasites, infected hosts are unlikely to have opportunities to reproduce before they are killed, hence in principle, selection for low transmission could be achieved by fast and slow killing.
The optimal levels of parasite virulence and time to kill will be affected by a range of ecological factors, including extrinsic host mortality, host immune status and competition with other parasite genotypes [27,30,31]. This latter selection pressure is likely to be particularly relevant in structured populations, because limited host availability may influence the likelihood of coinfection of competing parasite genotypes. Assuming competition between coinfecting parasites is mediated purely by resource competition (as opposed to, e.g. interference competition or intraspecific exploitation of public goods [32–34]), then coinfection tends to favour higher growth rates (and hence higher virulence or faster time to kill) to maximize within-host competitive ability [18,20,35,36]. If fast and slow killing have the same consequences for disease transmission, the added advantage of increased within-host competitive ability may, in some contexts, favour faster time to kill in structured host populations.
Here, we investigate if (i) faster killing can be favoured over slow killing under conditions where there is limited local host availability, and (ii) whether selection for time to kill is altered by within-host competition between parasite genotypes, using an obligately killing virus and its bacterial host. We selected on standing variation in time to kill from viruses evolved in a previous study, in which we showed that faster-killing viruses are more competitive within hosts but have lower transmission [36]. To allow us to independently manipulate local host availability and relatedness, both of which will be affected by host spatial structure, we carried out selection in experimental metapopulations, where hosts were limited or not and single or multiple viral clones were inoculated into each subpopulation (high and low relatedness).
2. Material and methods
(a). Experimental viral strains
We previously evolved 12 populations of an obligately killing virus (bacteriophage ϕ2) on its host bacterium Pseudomonas fluorescens for 300 generations, under conditions of low or variable multiplicity of infection (MOI), six lines evolved under each treatment. Virus evolved under variable MOI killed host cells faster in multiple infections, at the cost of reduced transmission to new hosts. Virus evolved under low MOI had a slower time to kill in multiple infections, resulting in lower within-host competitiveness but increased transmission [36]. We isolated a single clone from each experimental population to be used in subsequent experiments.
(b). Selection experiment
We evolved 24 metapopulations of ϕ2 virus on P. fluorescens for three transfers, approximately 20 phage generations. This short time scale was sufficient for the experiment as we were selecting on large amounts of standing variation from our previously evolved viral populations, rather than relying on de novo mutation. Each metapopulation consisted of 12 subpopulations (wells in microtitre plates) containing 200 µl Kings Media B (KB) and 102 exponentially growing ancestral bacteria. It was necessary to establish high MOI in all treatments because our experimental viruses only kill host cells rapidly during multiple infections [36]. Thus, we inoculated each patch with 108 viruses. We previously demonstrated that coinfections readily occur in this system under these conditions by using marked phages [36,37].
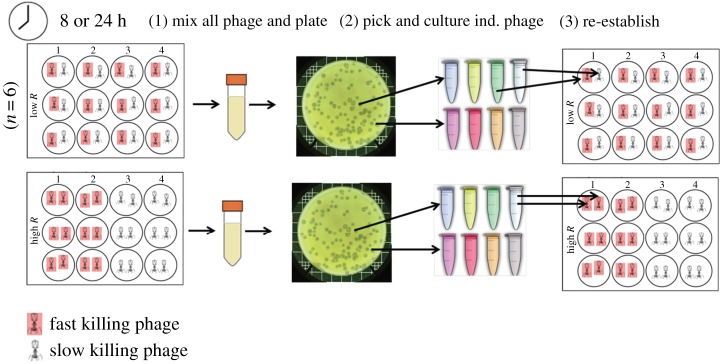
Each metapopulation was established with 12 virus subpopulations that varied in their time to kill (described above, fast: 28.69 ± 0.61 min, or slow: 36.54 ± 0.32 min). Six replicate metapopulations were founded with single (and unique) viral clones in each subpopulation (high relatedness (genetically identical) treatments), while six metapopulations had 12 individual subpopulations founded with one fast-killing clone and one slow-killing clone, with each of the 12 viral clones used twice per replicate metapopulation (low relatedness (genetically non-identical) treatments). To manipulate host availability, half of the metapopulations were transferred every 8 h and the other half of the metapopulations were transferred every 24 h. In both cases, hosts were a limiting resource, given the high MOI, but lower growth rates over 24 h than 8 h during preliminary work suggests that the former results in greater host limitation. The experimental design is shown in figure 1.
Figure 1.
Experimental set-up. We manipulated viral relatedness and host availability in a fully factorial design. We started the experiment with diverse metapopulations of fast- and slow-killing phage. Half the metapopulations were relatively high relatedness treatments—subpopulations (wells in microtitre plates) founded with single phage clones, while half were relatively low relatedness—founded with two clones. We manipulated host availability by varying the amount of time host-virus populations were cultured: 24 versus 8 h; hosts were a more or less limited resource, meaning that parasite growth was relatively constrained and unconstrained, respectively. (1) At each transfer, we mixed together all subpopulations within a metapopulation and (2) randomly selected virus clones from the plated mixture to (3) re-establish new subpopulations: the output of a subpopulation was consequently dependent on its productivity (global competition).
In each transfer, we combined all the subpopulations within a metapopulation to simulate global competition within spatially structured populations, by thoroughly mixing the entire metapopulation in a 10 ml falcon tube, treating with 10% chloroform and centrifuging at 14 000 r.p.m. for 2 min to isolate the phage, then isolated individual phage clones by plating dilutions of phage populations onto KB agar plates with a semisolid overlay P. fluorescens lawn. We then picked individual clones (phage plaques) with sterile pipette tips, amplified them overnight in liquid KB media plus 106 ancestral P. fluorescens so they reached the same densities, then redistributed 108 phage into a fresh set of wells containing 102 bacteria to reestablish the starting treatment conditions. There were four treatments in total: (i) low relatedness and high host availability, (ii) low relatedness and low host availability, (iii) high relatedness and high host availability and (iv) high relatedness and low host availability (figure 1). At the end of the selection, we assessed the mean lysis time (t50) of each of the populations of virus from each of the four treatments.
(c). Measuring time to kill
We measured the population-level time taken for viruses to lyse bacteria cells in each of our 24 metapopulations in a ‘one-step growth experiment’ [38]. We added 108 phage to 102 exponentially growing bacteria in 20 µl KB media and measured phage density by plating onto bacterial lawns at time zero and then at 5-min intervals from 25 min (we never observed increases in phage density prior to 30 min in our previous studies) [36]. Our specific measure of lysis time was t50, the number of minutes taken to reach 50% of maximal phage density during a single synchronized growth cycle.
(d). Statistical analysis
We analysed variation in phage growth as a factorial generalized linear model (GLM) with explanatory variable ‘relatedness’ (high or low) and ‘time’ (8 or 24) and their interaction. We analysed variation in phage population time to kill (t50) as a factorial GLM with explanatory variables ‘relatedness’ (high or low) and ‘host availability’ (unlimited or limited) and their interaction. We conducted pairwise comparisons of t50 with Tukey's pairwise comparison tests using function ‘glht’ in package ‘multcomp’ [39] in R. We carried out all analyses and drew all figures using R v. 2.15.3.
3. Results and discussion
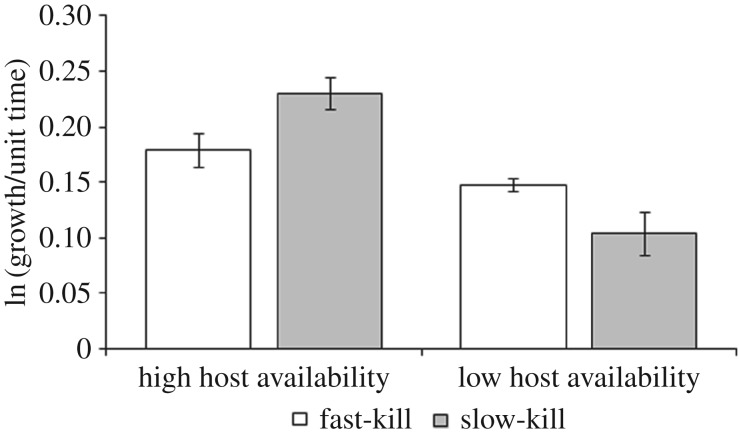
We started the experiment with viruses we had previously evolved to vary in their time to kill. Faster-killing viruses have greater within-host competitive ability, but lower between-host transmission [36]. We first wanted to determine whether the lower transmission of the faster-killing viruses resulted in a higher net growth rate than that of the more transmissible, slower-killing virus when there is limited host availability, and vice versa when hosts are abundant. To do this, we assayed the relative growth rate of populations of the experimental viruses over 8 and 24 h when host availability is relatively unlimited and limited, respectively. As predicted, (i) at 8 h when parasite growth is relatively unconstrained by host availability, the slower-killing viruses had a higher growth rate (figure 2) and (ii) at 24 h when parasite growth is constrained by limited host availability, the faster-killing populations had a higher growth rate (figure 2, significant effect of time × strain interaction F1,20 = 7.84, p = 0.012).
Figure 2.
Mean population growth (multiplication rate, logged) per unit time (hour), of fast- and slow-killing virus strains. Over 8 h, host availability was less limited, resulting in higher relative population growth rate of the higher transmitting and less virulent (slow-killing) populations, whereas hosts were more limiting over 24 h, resulting in higher population growth of the lower transmitting and virulent (fast-killing) populations. Data are means (n = 6) ± s.e.m.
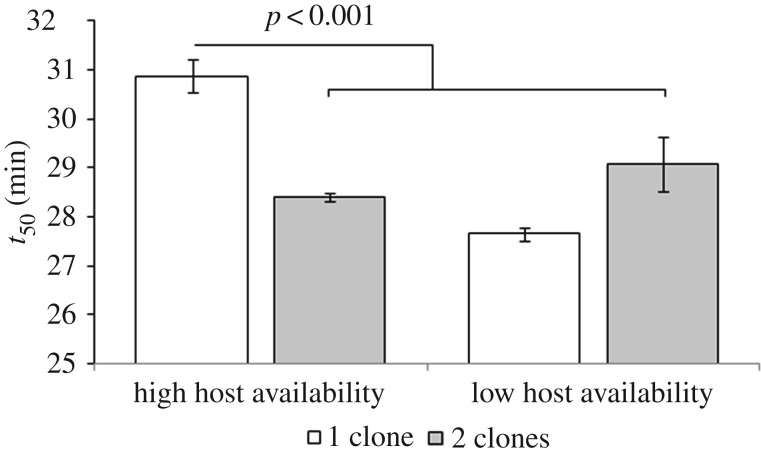
We next wanted to determine how competition between genotypes and host availability might interact to determine the relative success of faster-killing versus slower-killing viruses, given that both relatedness and host availability might be affected by spatial structure. To allow us to independently manipulate local host availability and competition between genotypes, we carried out selection in experimental metapopulations, where hosts were more or less limited and single or two viral clones (one fast and slow killing) were inoculated into each patch (relatedness is relatively high or low; r = 1 or r < 1). Note that given the high ratio of viruses to hosts, high and low relatedness refers to both within-subpopulation and within-host scales. We found an interaction between host availability and relatedness, such that when host availability was limited, fast time to kill (approx. 28 min) is favoured regardless of relatedness; whereas when host availability was unlimited and relatedness was high, time to kill was significantly slower (approx. 31 min; figure 3, significant relatedness × host availability: F1,14 = 37.82, p < 0.001, Tukey's pairwise comparisons, p < 0.001).
Figure 3.
Time to kill (t50) of evolved virus populations. Viruses were evolved in subpopulations where relatedness was either relatively high (1 clone) or low (2 clones), and where their growth was relatively unconstrained or constrained by high or low host availability. Data are means (n = 6) ± s.e.m.
Consistent with previous correlative and experimental studies [16–19], our experiment shows that limited host availability in spatially structured environments selects for prudent, slow-transmitting parasites. Most theory and data suggest that this also results in low virulence/slow time to kill for obligate killers; but here we show low transmission resulting from fast-killing viruses. It is important to emphasize that our experimental design (specifically, selection on standing variation) has constrained the outcome to lower transmission being achieved by faster time to kill. Indeed, previous experimental work demonstrating that parasites can evolve to have lower transmission when infection occurs locally [16] reported a correlated reduction in competitive ability, which is consistent with a slower rather than faster time to kill. The other experimental study reporting a similar finding only measured infectivity and not correlated life-history traits [17]. However, our results suggest that selection for low transmission may favour faster killing as opposed to slow, because fast or slow killing can achieve the same optimal level of transmission. Analogous arguments can be applied to the evolution of high and low virulence strategies if the strategies have equivalent impacts on parasite transmission and host population growth rate, which may be the case if infected hosts are unable to reproduce following infection and recovery.
In addition to the direct fitness benefit of low transmission being a more-efficient host exploitation strategy in viscous populations, there is an indirect fitness benefit that low transmission reduces competition between kin [15]. We show both processes can have independent and important effects, although host availability appears to be more significant than relatedness in this case. As within our previous study [36], we found that faster time to kill was favoured in mixed versus single clone (low versus high relatedness) infections where parasite growth is less constrained. However, when growth is more constrained by host availability, relatedness had no effect: fast killing was always selected. These results highlight how consequences of population structure, in this case local host availability, can interact in their effect on parasite life-history evolution. In summary, our key result is that fast-killing parasites can be selected in spatially structured populations because, like slow-killing parasites, they are less transmissible and hence use their hosts ‘prudently’.
Supplementary Material
Acknowledgements
We are very thankful to Troy Day, Kevin Foster and Xav Harrison for advice and discussions about our data and to two anonymous referees for their comments that improved the article.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
H.C.L. and A.B. conceived the project and designed the study. H.C.L. performed the experiments, analysed the data and wrote the first draft of the article. All authors contributed substantially to the analysis and interpretation of data and drafting the article.
Competing interests
We have no competing interests.
Funding
This work was funded by NERC, AXA Research fund, BBSRC, the Royal Society and The Royal Commission for the Exhibition of 1851.
References
- 1.Keeling MJ, et al. 2001. Dynamics of the 2001 UK foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science 294, 813–817. ( 10.1126/science.1065973) [DOI] [PubMed] [Google Scholar]
- 2.Boots M, Best A, Miller MR, White A. 2009. The role of ecological feedbacks in the evolution of host defence: what does theory tell us? Phil. Trans. R. Soc. B 364, 27–33. ( 10.1098/rstb.2008.0160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. 2005. Superspreading and the effect of individual variation on disease emergence. Nature 438, 355–359. ( 10.1038/nature04153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson PJ, Dobson AP, Newborn D. 1998. Prevention of population cycles by parasite removal. Science 282, 2256–2258. ( 10.1126/science.282.5397.2256) [DOI] [PubMed] [Google Scholar]
- 5.Tompkins DM, White AR, Boots M. 2003. Ecological replacement of native red squirrels by invasive greys driven by disease. Ecol. Lett. 6, 189–196. ( 10.1046/j.1461-0248.2003.00417.x) [DOI] [Google Scholar]
- 6.Haydon DT, et al. 2006. Low coverage vaccination strategies for the conservation of endangered species. Nature 443, 692–695. ( 10.1038/nature05177) [DOI] [PubMed] [Google Scholar]
- 7.Lafferty KD, et al. 2008. Parasites in food webs: the ultimate missing links. Ecol. Lett. 11, 533–546. ( 10.1111/j.1461-0248.2008.01174.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lion S, Boots M. 2010. Are parasites ‘prudent’ in space? Ecol. Lett. 13, 1245–1255. ( 10.1111/j.1461-0248.2010.01516.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank SA. 1996. Models of parasite virulence. Q. Rev. Biol. 71, 37–78. ( 10.1086/419267) [DOI] [PubMed] [Google Scholar]
- 10.Queller DC. 1994. Genetic relatedness in viscous populations. Evol. Ecol. 8, 70–73. ( 10.1007/BF01237667) [DOI] [Google Scholar]
- 11.Rand DA, Keeling M, Wilson HB. 1995. Invasion, stability and evolution to criticality in spatially extended, artificial host-pathogen ecologies. Proc. R. Soc. Lond. B 259, 55–63. ( 10.1098/rspb.1995.0009) [DOI] [Google Scholar]
- 12.Boots M, Sasaki A. 1999. Small worlds’ and the evolution of virulence: infection occurs locally and at a distance. Proc. R. Soc. Lond. B 266, 1933–1938. ( 10.1098/rspb.1999.0869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haraguchi Y, Sasaki A. 2000. The evolution of parasite virulence and transmission rate in a spatially structured population. J. Theor. Biol. 203, 85–96. ( 10.1006/jtbi.1999.1065) [DOI] [PubMed] [Google Scholar]
- 14.Kamo M, Sasaki A, Boots M. 2006. The role of trade-off shapes in the evolution of parasites in spatial host populations: an approximate analytical approach. J. Theor. Biol. 244, 588–596. ( 10.1016/j.jtbi.2006.08.013) [DOI] [PubMed] [Google Scholar]
- 15.Wild G, Gardner A, West SA. 2009. Adaptation and the evolution of parasite virulence in a connected world. Nature 459, 983–986. ( 10.1038/nature08071) [DOI] [PubMed] [Google Scholar]
- 16.Kerr B, Neuhauser C, Bohannan BJM, Dean AM. 2006. Local migration promotes competitive restraint in a host-pathogen 'tragedy of the commons’. Nature 442, 75–78. ( 10.1038/nature04864) [DOI] [PubMed] [Google Scholar]
- 17.Boots M, Mealor M. 2007. Local interactions select for lower pathogen infectivity. Science 315, 1284–1286. ( 10.1126/science.1137126) [DOI] [PubMed] [Google Scholar]
- 18.Herre EA. 1993. Population structure and the evolution of virulence in nematode parasites of fig wasps. Science 259, 1442–1445. ( 10.1126/science.259.5100.1442) [DOI] [PubMed] [Google Scholar]
- 19.Ewald PW. 1994. Evolution of infectious diseases. Oxford, UK: Oxford University Press. [Google Scholar]
- 20.Levin S, Pimentel D. 1981. Selection of intermediate rates of increase in parasite-host systems. Am. Nat. 117, 308–315. ( 10.1086/283708) [DOI] [Google Scholar]
- 21.Anderson RM, May RM. 1982. Coevolution of hosts and parasites. Parasitology 85, 411–426. ( 10.1017/S0031182000055360) [DOI] [PubMed] [Google Scholar]
- 22.Levin BR, et al. 1982. Evolution of hosts and parasites. In Population biology of infectious disease (eds Anderson RM, May RM). Berlin, Germany: Springer. [Google Scholar]
- 23.May RM, Anderson RM. 1983. Epidemiology and genetics in the coevolution of parasites and hosts. Proc. R. Soc. Lond. B 219, 281–313. ( 10.1098/rspb.1983.0075) [DOI] [PubMed] [Google Scholar]
- 24.de Roode JC, Yates AJ, Altizer S. 2008. Virulence-transmission trade-offs and population divergence in virulence in a naturally occurring butterfly parasite. Proc. Natl Acad. Sci. USA 105, 7489–7494. ( 10.1073/pnas.0710909105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Read JM, Keeling MJ. 2003. Disease evolution on net-works: the role of contact structure. Proc. R. Soc. Lond. B 270, 699–798. ( 10.1098/rspb.2002.2305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bremermann HJ, Thieme HR. 1989. A competitive exclusion principle for pathogen virulence. J. Math. Biol. 27, 179–190. ( 10.1007/BF00276102) [DOI] [PubMed] [Google Scholar]
- 27.Ebert D, Weisser WW. 1997. Optimal killing for obligate killers: the evolution of life histories and virulence of semelparous parasites. Proc. R. Soc. Lond. B 264, 985–991. ( 10.1098/rspb.1997.0136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson RM, May RM. 1981. The population dynamics of microparasites and their invertebrate hosts. Phil. Trans. R. Soc. Lond. B 291, 451–524. ( 10.1098/rstb.1981.0005) [DOI] [Google Scholar]
- 29.Miller LK, Lingg AJ, Bulla LA. 1983. Bacterial, viral and fungal insecticides. Science 219, 715–721. ( 10.1126/science.219.4585.715) [DOI] [PubMed] [Google Scholar]
- 30.Bonhoffer S, Nowak MA. 1994. Intra-host and inter-host selection: viral evolution of immune function impairment. Proc. Natl Acad. Sci. USA 91, 8062–8066. ( 10.1073/pnas.91.17.8062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Baalen M, Sabelis MW. 1995. The dynamics of multiple infection and the evolution of virulence. Am. Nat. 1446, 881–970. ( 10.1086/285830) [DOI] [Google Scholar]
- 32.Brown SP, Hochberg ME, Grenfell BT. 2002. Does multiple infection select for raised virulence? Trends Microbiol. 10, 401–405. ( 10.1016/S0966-842X(02)02413-7) [DOI] [PubMed] [Google Scholar]
- 33.West SA, Buckling A. 2003. Cooperation, virulence and siderophore production in bacterial parasites. Proc. R. Soc. Lond. B 270, 37–44. ( 10.1098/rspb.2002.2209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner A, West SA, Buckling A. 2004. Bacteriocins, spite and virulence. Proc. R. Soc. Lond. B 271, 1529–1535. ( 10.1098/rspb.2004.2756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowak MA, May RM. 1994. Superinfection and the evolution of parasite virulence. Proc. R. Soc. Lond. B 255, 81–89. ( 10.1098/rspb.1994.0012) [DOI] [PubMed] [Google Scholar]
- 36.Leggett HC, Benmayor R, Hodgson DJ, Buckling A. 2013. Experimental evolution of adaptive phenotypic plasticity in a parasite. Curr. Biol. 23, 1–4. ( 10.1016/j.cub.2012.11.045) [DOI] [PubMed] [Google Scholar]
- 37.Hall AR, Scanlan PD, Leggett HC, Buckling A. 2012. Multiplicity of infection does not accelerate infectivity evolution of viral parasites in laboratory microcosms. J. Evol. Biol. 25, 409–415. ( 10.1111/j.1420-9101.2011.02434.x) [DOI] [PubMed] [Google Scholar]
- 38.Ellis EL, Delbrück M. 1939. The growth of bacteriophage. J. Gen. Physiol. 22, 365–384. ( 10.1085/jgp.22.3.365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom. J. 50, 346–363. ( 10.1002/bimj.200810425) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.