Abstract
Human metapneumovirus (hMPV) is one of the major pathogens of respiratory illness. Reinfection with hMPV occurs frequently throughout life. We describe an infant who was infected with two different hMPV strains during a period of only 1 month.
CASE REPORT
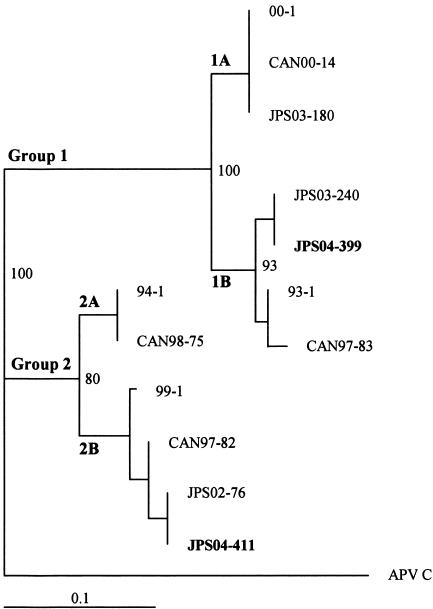
A 9-month-old girl was admitted to Hokkaido Social Insurance Hospital (Sapporo, Japan) on 27 April 2004 because of fever, nasal congestion, coughing, and vomiting caused by the coughing, which had persisted for 4 days. Her medical history showed that she had been admitted to the hospital four times with a diagnosis of bronchitis. On initial physical examination, her temperature was 37.2°C and her respiratory rate was 30/min, with bilateral rhonchi on lung auscultation. A complete blood count showed 9,000 leukocytes/mm3 (30% neutrophils, 64% lymphocytes, 5% monocytes, and 1% eosinocytes). C-reactive protein was 1.1 mg/dl. Nasopharyngeal culture showed normal flora. A rapid antigen detection test for human respiratory syncytial virus (hRSV) was negative. Her temperature decreased on the day of admission. Her condition was rapidly improved by supportive therapy, and she was discharged on day 4 after admission. On the day of discharge, a human metapneumovirus (hMPV) sequence (JPS04-399; GenBank accession number AY653168) belonging to group 1 was detected in a nasopharyngeal swab sample by reverse transcription-PCR (RT-PCR) (Fig. 1). A subsequent nasopharyngeal swab sample taken 6 days after discharge was negative for hMPV by RT-PCR. The final clinical diagnosis was bronchitis.
FIG. 1.
Phylogenetic analysis of hMPV fusion nucleotide sequences. A tree was constructed by the neighbor-joining method with 100 bootstraps and random sequence addition. The length of each horizontal line represents the number of substitutions per site. Bootstrap values over 70% are shown. APV C (avian pneumovirus subgroup C; GenBank accession number AF187152) was the outgroup and was used to root the tree. The hMPV sequence analysis included the following (GenBank accession number): 94-1 (AF371342), 99-1 (AF371344), 00-1 (AF371337), and 93-1 (AF371341), isolated in The Netherlands (18); CAN00-14 (AY145299), CAN97-83 (AY297749), CAN98-75 (AY297748), and CAN-97-82 (AY145295), isolated in Canada (1); and JPS03-180 (AY530092), JPS03-240 (AY530095), JPS02-76 (AY530089), JPS04-399 (AY653168), and JPS04-411 (AY653175), isolated in Sapporo, Japan (8, 13).
On 13 May 2004, 12 days after discharge, she was brought to the hospital again because of exacerbation of coughing, nasal congestion, and wheezing. On physical examination, her temperature was 39.6°C and her respiratory rate was 40/min. Bilateral wheezing and rhonchi were heard on lung auscultation. An otoscopic examination revealed bilateral otitis media. A chest X-ray indicated interstitial and alveolar infiltrates. A complete blood count showed 10,930 leukocytes/mm3 (14% neutrophils, 80% lymphocytes, 5% monocytes, and 1% eosinocytes). C-reactive protein was 0.28 mg/dl. On the day after admission, an hMPV sequence (JPS04-411; GenBank accession number AY653175) belonging to group 2 (Fig. 1) was detected in a nasopharyngeal swab sample by RT-PCR. Nasopharyngeal culture showed normal flora. A rapid antigen detection test for hRSV was negative. The clinical diagnosis was wheezy bronchitis and pneumonia. Her condition was gradually improved by supportive therapy. Her high fever persisted for 3 days, and then a low-grade fever continued for 4 days. The duration of wheezing was 7 days. She was discharged on day 10 after the second admission.
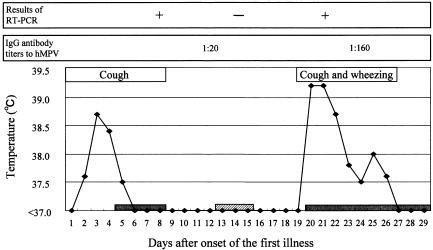
hMPV-specific immunoglobulin G (IgG) antibody was detected in two serum samples by an indirect immunofluorescence assay. On days 12 and 24 after the onset of the first illness, titers of IgG antibody against hMPV were 1:20 and 1:160, respectively (Fig. 2). This rise in the IgG antibody titer and the presence of the group 1 hMPV sequence indicated that the first respiratory tract infection was caused by the group 1 hMPV. Since the titer of IgG antibody against hMPV was still low on day 12 after the onset of the first illness (1:20), it is conceivable that the first illness was a primary infection with hMPV. In the second respiratory tract infection, we detected the group 2 hMPV sequence after the disappearance of the group 1 hMPV sequence was confirmed (Fig. 2). Therefore, it was likely that the second illness was caused by the group 2 hMPV. Since the incubation period of hMPV has been estimated to be 4 to 6 days (8), she might have been exposed to group 2 hMPV on days 13 to 15 after the onset of the first illness (Fig. 2).
FIG. 2.
Time lines of clinical and virological findings. Filled bars indicate the periods of hospitalization. The hatched bar indicates the estimated period when the patient was re-exposed to hMPV.
RT-PCR and sequencing.
The RT-PCR method used in this study was described previously (8). cDNA was synthesized from total RNA extracted from each nasopharyngeal swab sample. A forward primer with a sequence of 5′-CTGTTCCATTGGCAGCAATA-3′ and a reverse primer with a sequence of 5′-TCAAAGCTGCTTGACACTGG-3′ were used to detect the group 1 F protein gene. A forward primer with a sequence of 5′-AATCGGGTTGGAATCATCAA-3′ and a reverse primer with a sequence of 5′-GCTGTTCACCTTCAACTTTGC-3′ were used to detect the group 2 F protein gene. The PCR mixture consisted of 100 μmol of each deoxyribonucleotide, 1.0 U of AmpliTaq Gold, 50 mmol of potassium chloride per liter, 10 mmol of Tris-HCl (pH 8.3) per liter, 1.5 mmol of magnesium chloride per liter, 0.01% (wt/vol) gelatin, 10 pmol of each primer, and cDNA in a volume of 25 μl. The PCR conditions were as follows: 94°C for 9 min, followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. Sequencing of the 80 nucleotide bases of the PCR product was performed by using a BigDye terminator cycle sequencing ready reaction kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.) with an ABI Prism 310 genetic analyzer (Perkin-Elmer Applied Biosystems). Phylogenetic trees were constructed with CLUSTAL W software and the Phylip 3.6 (alfa3) program (DNADist and Neighbor software).
Detection of antibody to hMPV.
An indirect immunofluorescence assay was carried out as a serologic test for hMPV with LLC-MK2 cells infected with hMPV (JPS02-76; GenBank accession number AY530089) as described previously (8, 19). In brief, these infected cells were fixed to glass slides with acetone and incubated for 30 min at 37°C with serial twofold dilutions of serum samples, beginning at 1:10. After washing, the slides were incubated for 30 min at 37°C with fluorescein isothiocyanate-conjugated rabbit anti-human IgG (Dako, Glostrup, Denmark) at 1:40. They were then washed and examined by fluorescence microscopy. This assay could not differentiate a group-specific antibody of hMPV (8).
Discussion.
hMPV and hRSV are grouped into the same subfamily, Pneumovirinae, of the family Paramyxoviridae (7, 18). Both viruses are separated into two groups by genetic differences mainly in the G and F glycoproteins (1, 2, 13, 15, 17). Since the viral genome, clinical manifestations, and epidemiology of hMPV are similar to those of hRSV (4, 16, 19, 20), we speculate on the risk of reinfection of our patient with hMPV on the basis of our knowledge of the risk of reinfection with hRSV.
Reinfection with hRSV occurs throughout life, implying that the host immune response induced by natural infection provides incomplete protection for a limited period of time, probably not lasting until the subsequent epidemic season (6). The immune response includes production of neutralizing antibodies and T-cell-specific immunity. The F and G proteins appear to be the most important for induction of protective humoral immunity (6, 12, 14). Several studies have shown that the risk of reinfection is inversely related to the levels of neutralizing antibodies to hRSV and that reinfection illnesses are generally mild (9, 10, 11). The cellular immune response to hRSV infection also plays a role in the clearance of the virus. The N protein (nucleoprotein), F protein, and M2 protein of hRSV are targets for hRSV-specific T cells in humans (6, 5). However, the hRSV-specific T-cell responses induced during primary hRSV infection do not completely protect against subsequent reinfection (6).
Our patient developed wheezy bronchitis and pneumonia caused by reinfection with group 2 hMPV only 19 days after the onset of the first illness. Illness from reinfection with hRSV has been reported to be generally mild (11). However, the second illness caused by reinfection with group 2 hMPV was more severe than the illness caused by primary infection with group 1 hMPV in our patient. Since we did not perform viral isolation by culture methods, we could not completely rule out the possibility of coinfection with viruses other than hRSV. However, we suspected that the respiratory infections might be associated with hMPV infection.
When she was reinfected with group 2 hMPV, the level of IgG antibody against hMPV was very low (approximately 1:20). Although antigenic diversity probably has some influence on sequential hMPV infection, she might not have had sufficient immunity for protection against subsequent infection within the short period. Therefore, the reinfection in our case occurred like a primary infection under the condition of almost no immunological memory, regardless of the antigenic difference between groups of hMPV.
Our previous serological study has shown that reinfection with hMPV frequently occurs among children because more than half of the children infected with hMPV have IgG antibodies against hMPV in the acute phase of illness. In this case report, by using sequence analysis, we showed that reinfection with hMPV frequently occurs even in early childhood under the condition of simultaneous cocirculation of different hMPV strains (3, 8, 19). Further studies are needed to clarify the risk of reinfection with hMPV.
Acknowledgments
This study was partially supported by a grant-in-aid for exploratory research (14657179 [2002]) from the Ministry of Education, Culture, Sports, Science and Technology and by a grant-in-aid for the 21st century Center of Excellence for Zoonosis Control from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
We thank Yasuto Suzuki, Yoshikazu Kinugawa, and Yoshiko Tateno of the Hokkaido Social Insurance Hospital for providing nasopharyngeal swab and serum samples. We also thank Kunihiko Kobayashi, emeritus professor of Hokkaido University, for giving us suggestions and Stewart Chisholm for proofreading the manuscript.
REFERENCES
- 1.Bastien, N., S. Normand, T. Taylor, D. Ward, T. C. Peret, G. Boivin, L. J. Anderson, and Y. Li. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 93:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., G. De Serres, S. Cote, R. Gilca, Y. Abed, L. Rochette, M. G. Bergeron, and P. Dery. 2003. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 9:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherrie, A. H., K. Anderson, G. W. Wertz, and P. J. Openshaw. 1992. Human cytotoxic T cells stimulated by antigen on dendritic cells recognize the N, SH, F, M, 22K, and 1b proteins of respiratory syncytial virus. J. Virol. 66:2102-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domachowske, J. B., and H. F. Rosenberg. 1999. Respiratory syncytial virus infection: immune response, immunopathogenesis, and treatment. Clin. Microbiol. Rev. 12:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Easton, A. J., J. B. Domachowske, and H. F. Rosenberg. 2004. Animal pneumoviruses: molecular genetics and pathogenesis. Clin. Microbiol. Rev. 17:390-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebihara, T., R. Endo, H. Kikuta, N. Ishiguro, H. Ishiko, M. Hara, Y. Takahashi, and K. Kobayashi. 2004. Human metapneumovirus infection in Japanese children. J. Clin. Microbiol. 42:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glezen, W. P., L. H. Taber, A. L. Frank, and J. A. Kasel. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 140:543-546. [DOI] [PubMed] [Google Scholar]
- 10.Hall, C. B., E. E. Walsh, K. C. Schnabel, C. E. Long, K. M. McConnochie, S. W. Hildreth, and L. J. Anderson. 1990. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J. Infect. Dis. 162:1283-1290. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, F. W., A. M. Collier, W. A. Clyde, Jr., and F. W. Denny. 1979. Respiratory-syncytial-virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N. Engl. J. Med. 300:530-534. [DOI] [PubMed] [Google Scholar]
- 12.Hendry, R. M., J. C. Burns, E. E. Walsh, B. S. Graham, P. F. Wright, V. G. Hemming, W. J. Rodriguez, H. W. Kim, G. A. Prince, K. McIntosh, R. M. Chnock, and B. R. Murphy. 1988. Strain-specific serum antibody responses in infants undergoing primary infection with respiratory syncytial virus. J. Infect. Dis. 157:640-647. [DOI] [PubMed] [Google Scholar]
- 13.Ishiguro, N., T. Ebihara, R. Endo, X. Ma, H. Kikuta, H. Ishiko, and K. Kobayashi. 2004. High genetic diversity of the attachment (G) protein of human metapneumovirus. J. Clin. Microbiol. 42:3406-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, P. R., R. A. Olmsted, G. A. Prince, B. R. Murphy, D. W. Alling, E. E. Walsh, and P. L. Collins. 1987. Antigenic relatedness between glycoproteins of human respiratory syncytial virus subgroups A and B: evaluation of the contributions of F and G glycoproteins to immunity. J. Virol. 61:3163-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peiris, J. S., W. H. Tang, K. H. Chan, P. L. Khong, Y. Guan, Y. L. Lau, and S. S. Chiu. 2003. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg. Infect. Dis. 9:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peret, T. C., Y. Abed, L. J. Anderson, D. D. Erdman, and G. Boivin. 2004. Sequence polymorphism of the predicted human metapneumovirus G glycoprotein. J. Gen. Virol. 85:679-686. [DOI] [PubMed] [Google Scholar]
- 18.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Hoogen, B. G., G. J. van Doornum, J. C. Fockens, J. J. Cornelissen, W. E. Beyer, R. de Groot, A. D. Osterhaus, and R. A. Fouchier. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 188:1571-1577. [DOI] [PubMed] [Google Scholar]
- 20.Williams, J. V., P. A. Harris, S. J. Tollefson, L. L. Halburnt-Rush, J. M. Pingsterhaus, K. M. Edwards, P. F. Wright, and J. E. Crowe, Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 350:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]