Abstract
Aim:
We sought to identify potential pleiotropy involving pharmacogenes.
Methods:
We tested 184 functional variants in 34 pharmacogenes for associations using a custom grouping of International Classification and Disease, Ninth Revision billing codes extracted from deidentified electronic health records of 6892 patients.
Results:
We replicated several associations including ABCG2 (rs2231142) and gout (p = 1.73 × 10-7; odds ratio [OR]: 1.73; 95% CI: 1.40–2.12); and SLCO1B1 (rs4149056) and jaundice (p = 2.50 × 10-4; OR: 1.67; 95% CI: 1.27–2.20).
Conclusion:
In this systematic screen for phenotypic associations with functional variants, several novel genotype–phenotype combinations also achieved phenome-wide significance, including SLC15A2 rs1143672 and renal osteodystrophy (p = 2.67 × 10-6; OR: 0.61; 95% CI: 0.49–0.75).
Keywords: : absorption, distribution, metabolism and excretion; electronic health records; genetic association study; pharmacogenomics; phenome-wide association study
It has long been recognized that genetic variation affects human drug response [1]. In the past decade, candidate gene and genome-wide association studies (GWAS) have identified specific genetic variants underlying the physiological response to highly prescribed medications, including warfarin [2–5], clopidogrel [6] and statins [7], to name a few. The appeal of personalized or precision medicine has prompted many clinics to implement genetic testing for these variants [8–12], often despite little [13,14] or even contrary [15] evidence regarding clinical utility. Among the consequences of this recent rise in clinical genotyping and sequencing has been the growing recognition that pharmacogenetic variants regularly display pleiotropic effects [16] incidental to the original purpose of testing [17–19].
The term pleiotropy refers to the phenomenon of cross-phenotype associations by a single genetic locus [20], mechanisms of which include protein multifunctionality and alternative DNA splicing. Early studies of the National Human Genome Research Institute GWAS Catalog revealed extensive pleiotropy among the common variants associated with complex traits. Many such examples have since been replicated and annotated in traditional epidemiologic and contemporary clinical collections [21–26].
The genetic variants in drug-metabolizing enzymes and drug transporters can be considered exemplars of pleiotropy. During their long evolutionary history, pharmacogenes have taken on multiple pharmacokinetic roles across a range of drugs, pollutants and endogenous compounds [27], and by modulating exposure to xenobiotics, pharmacogenes likely play a large role in shaping the environment's impact on disease [28]. Furthermore, genetic variants in these genes are often responsible for the bevy of phenotypes associated with inborn errors of metabolism and other clinically relevant congenital disorders [29].
To test pharmacogenetic variants for pleiotropic associations, we conducted a phenome-wide association study (PheWAS) on 184 functional variants in 34 pharmacogenes. In a PheWAS, each genetic variant is tested for association with multiple phenotypes (the phenome) as opposed to a single phenotype, as in GWAS [30–32]. We used the Illumina® Absorption, Distribution, Metabolism and Elimination (ADME) Core Panel (CA, USA) [33] to genotype 6067 European–American and 762 African–American patients linked to deidentified electronic health records (EHRs). The EHR is a source of high-dimensional clinical data, well suited for studying pleiotropic effects across a wide spectrum of clinical phenotypes [34,35].
We replicated four previously described genotype–phenotype associations, including three with disease, and identified several novel pleiotropic relationships. Among the novel results, we identified a significant association in European–Americans between a nonsynonymous SNP in the gene SLC15A2 rs1143672 and renal osteodystrophy, a disorder of mineral and bone metabolism (p = 2.67 × 10-6; odds ratio [OR]: 0.61; 95% CI: 0.49–0.75). Follow-up tests of association for rs1143672 with all pairs of phenotypes using the multivariate platform MultiPhen [36] revealed significant associations (p < 1.88 × 10-6) with either renal osteodystrophy or end-stage renal disease (ESRD) and abnormal heart sounds, skin neoplasm (of uncertain behavior), joint pain and back pain. These results indicate that ‘unmeasured phenotypes’, which are not well represented in single phenotype analyses, likely contribute to an array of diseases recorded by International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) codes. Coupled together, these PheWAS and MultiPhen results suggest a complex relationship between genetic variants and outcomes denoted by billing codes in this patient population. These results represent a catalog of potential effects from genotype–xenobiotic and other environmental interactions.
Methods
Study population & genotyping
The study population described here was derived from BioVU, the Vanderbilt University Medical Center biorepository linked to deidentified EHRs. The establishment of this opt-out clinical collection, including ethical and legal issues, has been detailed elsewhere [37]. In brief, DNA is extracted from discarded blood drawn for routine clinical care in an outpatient setting. DNA samples are linked to a deidentified version of the EHR termed the synthetic derivative. Data contained within the synthetic derivative are considered limited datasets as defined by the Health Insurance Portability and Accountability Act and are in accordance with provisions of Title 45, Code of Federal Regulations, part 46 (45 CFR 46) that define criteria for ‘nonhuman subjects’ research.
The DNA samples included in this study were genotyped on the Illumina® ADME Core Panel at Vanderbilt University's Center for Human Genetics Research (CHGR) DNA Resources Core. These samples were genotyped as part of various studies performed in BioVU by Vanderbilt Electronic Systems for Pharmacogenetic Assessment (Supplementary Table 1). The ADME module calling software utilizes predefined boundaries for calling genotypes preventing the introduction of errors related to batch effects in the data. We calculated quality control metrics with the genetic analysis software PLINK version 1.07 [38].
We removed ten and six SNPs from the European and African–American datasets, respectively, that deviated from our Hardy–Weinberg equilibrium threshold (p < 0.001). Compared with the other genes on the ADME Core Panel, CYP2D6 showed a considerable burden of deviations of Hardy–Weinberg equilibrium expectations. Call rates of nine markers (GSTM1 rs1065411, CYP2D6 rs1080985, UGT2B15 rs1902023, CYP2C8 rs11572103, CYP2A6 rs28399444, CYP2A6 rs28399454, CYP2A6 rs1801272, CYP2A6 rs28399433 and CYP2D6 rs28371706) below 0.95 were flagged and included in the analyses. All remaining markers with an allele frequency >0.01 (71 markers for European–Americans and 74 markers for African–Americans) were included in analyses. Duplicates (n = 25) and samples with genotyping thresholds below 0.10 (n = 171) were removed from subsequent analysis.
Population stratification
Race/ethnicity in BioVU is administratively assigned (third-party reported) and available as a structured field in the EHR. Previous studies have demonstrated that this identifier is highly correlated with ancestry in European–Americans and African–Americans in BioVU [39,40]. We applied STRUCTURE 2.2 to the ADME Core genotype data to identify outliers where genetic ancestry is discordant with third-party reported race/ethnicity [41]. Model-based clustering on the African–American and European–American samples was performed using the admixture model. STRUCTURE was run with 10,000 burn-in iterations, 50,000 simulation cycles and assigned K = 2. A total of 241 samples were assigned a probability of <90% that the race/ethnicity matches his/her genetic ancestry, and these samples were removed from further analyses.
Phenome derivation from ICD-9 codes
For this study we used ICD-9-CM codes from the deidentified BioVU EHR. ICD-9-CM codes are a hierarchical classification system of diseases used by healthcare for billing and administrative purposes [42]. ICD-9-CM codes were translated with the database management system MySQL into 1368 ‘phecodes’, which group disease codes in different clinical settings into a single code (e.g., ‘Type 1 diabetes’ and ‘hypertension’). The ICD-9-CM phecode translation table used in this study is available for download at [43,25].
Phecodes were used to define case–control status as follows. To be categorized a ‘case’ for any single phecode, the individual must have had at least two phecodes in his or her record. For any single phecode, to be a control, the phecode had to be absent from his or her patient record. Given the hierarchical nature of ICD-9-CM codes, subjects were excluded from both cases and controls for a particular phecode if he/she had only a single instance of that phecode or a related code. Codes were considered related if they existed in the same section grouping in the ICD-9-CM hierarchy, which groups related conditions. Phecodes with fewer than 35 cases were excluded from the analysis. A total of 808 and 333 codes in the European–American and African–American datasets, respectively, remained for analysis.
Statistical methods
The PheWAS was performed with a script designed in R, which tests exhaustively all genotype–phecode combinations. Statistical tests were modeled using logistic regression assuming an additive genetic model adjusted for age (age at first code for cases and the maximum age recorded in the EHR for controls) and sex. We declared phenome-wide significance at a Bonferroni-corrected phenome-wide significance in European–Americans (p < 6.18 × 10- 5) and African–Americans at (p < 1.50 × 10- 4).
The paired trait analysis was performed with the R package MultiPhen [36]. In a MultiPhen analysis, the genetic variant is held as the dependent variable and a joint test of association is performed with a linear combination of phenotypes as predictor variables. The genotype data are treated as allele count and is modeled with ordinal regression. Here, we exhaustively tested pairwise combinations of phecodes that met sample size criteria in joint tests of associations with a single variant. Pairs of codes were included if there were at least 50 individuals where both codes were present; 50 individuals where both codes were absent and the measured correlation phi (φ) was below 0.95. Reported p-values refer to the joint p-value recorded in the MultiPhen output. Regression models in MultiPhen were adjusted for the ages of the samples at the two traits and sex. Bonferroni-corrected significance for the MultiPhen analysis was declared at p < 1.88 × 10-6 (0.05 corrected for 26,599 paired traits) and p < 2.46 × 10-5 (0.05 corrected for 2035 paired traits) in the European–American and African–American analyses, respectively.
Data visualization
Data visualization of the Sun Plots was performed with the PheWAS Viewer [44].
Results
Study population
Samples used in this study were collected by Vanderbilt University Medical Center's DNA biorepository (BioVU), located in Nashville, TN, USA [37]. The clinical characteristics of the study population are shown in Table 1. A total of 7266 DNA samples were genotyped using the Illumina® ADME Core Panel, which has been used for previous pharmacogenetic studies in BioVU (Supplementary Table 1) [45]. After quality control (see ‘Methods’), the majority of the samples were European–American (n = 6067) and the remainder African–American (n = 762). Overall, 48% of the samples were female, slightly less than the 52% observed in BioVU overall [46]. The mean body mass index of this PheWAS study population was in the overweight category (28.34 kg/m2), and the average age at last EHR record was 55 years (Table 1).
Table 1. . Clinical characteristics of phenome-wide association study study population.
| Variable | Sample size, percent, or mean (SD) |
|---|---|
| European–Americans |
6067 |
| African–Americans |
762 |
| Female |
48.1 |
| Body mass index, kg/m2 |
28.34 (7.19) |
| Age at last record in years | 55.02 (19.59) |
SD: Standard deviation.
After excluding markers and PheWAS phenotypes (‘phecodes’) that did not meet quality control standards (see ‘Methods’), we performed tests of association for 808 and 333 phecodes in European and African–Americans, respectively (Figure 1, Supplementary Table 2 & 3). A Bonferroni-corrected phenome-wide significance threshold was used to declare novel significant associations (p < 6.18 × 10-5 for European–Americans and p < 1.50 × 10-4 in African–Americans), while previously reported associations were deemed significant at p < 0.05.
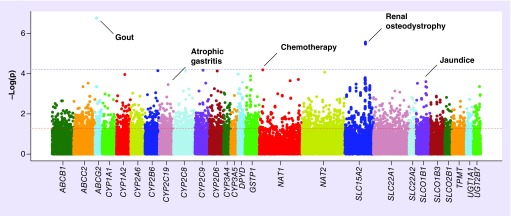
Figure 1. . Manhattan plot of phenome-wide association study results in European–Americans.
ADME Core variants grouped by gene are plotted on the X-axis against the -log10 of the p-value on the y-axis. The red and blue dashed lines show significance thresholds of 0.05 and phenome-wide significance (6.19 × 10-5), respectively. Annotations of the phecodes are provided for selected associations that met significance as a novel or replicated association. Width differences between genes reflect SNP coverage for individual genes on the ADME Core Panel passing quality control.
ADME: Absorption, distribution, metabolism, and elimination.
Replicating associations
The most significant association among European–Americans was between the pharmacogene ABCG2 rs2231142 and gout, a common disease caused by hyperuricemia (p = 1.73 × 10-7; OR: 1.73; 95% CI: 1.41–2.12; Table 2). Also known as Q141K, rs2231142 is a missense variant associated with gout as identified by a GWAS performed in the Framingham and Rotterdam cohorts [47], and subsequently replicated in multiple populations [48]. The minor allele (A) frequency was 0.17 in cases (n = 363) and 0.11 in controls (n = 5528). The association between rs2231142 and gout in African–Americans was not significant (p = 0.86; OR: 1.10; 95% CI: 0.37–3.24), but statistical power was limited by the minor allele frequency (0.03) in cases and controls. Nonetheless, direction of genetic effect (odds ratio) observed among African–Americans was consistent with the result in European–Americans as well as with previous reports in the literature (Table 2) [47]. Direction of effect and coded allele frequency (CAF) were also consistent with previous reports in African–Americans [48].
Table 2. . Replications identified from phenome-wide association study in European–Americans and corresponding results in African–Americans.
| PheCode | SNP | Gene | Coded allele | Controls, n | Cases, n | Control, CAF | Case, CAF | OR (95% CI) | p-value |
|---|---|---|---|---|---|---|---|---|---|
|
(A) European–Americans | |||||||||
| Gout |
rs2231142 |
ABCG2 |
A |
5528 |
363 |
0.11 |
0.17 |
1.73 (1.41–2.12) |
1.73 × 10-7 |
| Jaundice |
rs4149056 |
SLCO1B1 |
C |
5058 |
154 |
0.15 |
0.25 |
1.67 (1.33–2.11) |
2.35 × 10-4 |
| Tobacco use disorder |
rs28399433 |
CYP2A6 |
G |
4882 |
1132 |
0.03 |
0.04 |
0.81 (0.66–0.90) |
0.04 |
| Atrophic gastritis |
rs4244285 |
CYP2C19 |
A |
3366 |
77 |
0.15 |
0.25 |
1.95 (1.43–2.65) |
3.45 × 10-4 |
| Chemotherapy |
rs4986782 |
NAT1 |
A |
4664 |
956 |
0.02 |
0.03 |
1.85 (1.37–2.50) |
6.45 × 10-5 |
|
(B) African–Americans | |||||||||
| Gout |
rs2231142 |
ABCG2 |
A |
617 |
64 |
0.03 |
0.03 |
1.20 (0.48–2.97) |
0.74 |
| Jaundice |
rs4149056 |
SLCO1B1 |
C |
579 |
22 |
0.03 |
0.02 |
0.71 (0.12–3.97) |
0.75 |
| Tobacco use disorder |
rs28399433 |
CYP2A6 |
G |
525 |
163 |
0.10 |
0.07 |
0.67 (0.41–1.12) |
0.13 |
| Atrophic gastritis |
rs4244285 |
CYP2C19 |
A |
381 |
3 |
0.17 |
0.00 |
0.00 (0.00–∞) |
0.99 |
| Chemotherapy | rs4986782 | NAT1 | A | 594 | 75 | 0.003 | 0.00 | 0.00 (0.00–∞) | 0.98 |
CAF: Coded allele frequency; OR: Odds ratio.
We identified an association between SLCO1B1 rs4149056 and jaundice (p = 2.50 × 10-4; OR: 1.67; 95% CI: 1.27–2.20). In previous studies of European populations, rs4149056 has been associated with elevated serum bilirubin concentrations [49], a known cause of jaundice. The CAF was 0.23 for cases (n = 154) and 0.15 for controls (n = 5058). The association between rs4149056 and jaundice was not significant among African–Americans (Table 2), again possibly due to low statistical power as the African–American study population had fewer cases of jaundice (n = 22) in comparison to the European–Americans (n = 154).
Two CYP2C19 alleles from the ADME Core list were included in this analysis. The rs4244285 A allele (CYP2C19*2) has been implicated in poor drug metabolism, and the rs12248560 T allele (CYP2C19*17) with ultrarapid metabolism. Several epidemiological studies have reported an association between CYP2C19 poor metabolizer alleles (defined as at least *2 or *3) and the development of multiple forms of gastrointestinal cancer [50]. In this European–American cohort, rs4244285 was associated with gastric cancer at p = 0.03 (59 cases; OR: 1.74; 95% CI: 1.03–2.93) and with atrophic gastritis at p = 3.50 × 10-4 (77 cases and 3366 controls; OR: 1.95; 95% CI: 1.35–2.81). Atrophic gastritis is a necessary precursor to gastric adenocarcinoma, and the severity of atrophy is a major indicator of cancer risk [51]. The CYP2C19 rs12248560 T allele was also associated with hepatic cancer in European–Americans at p = 6.00 × 10-3 (65 cases; OR: 1.84; 95% CI: 1.19–2.85). The African–American dataset had too few cases of atrophic gastritis (n = 3) and stomach cancer (n = 0) to test for association with CYP2C19.
‘Slow acetylator’ variants of the pharmacogene NAT1 have been implicated in several cancers, often in the context of exposure to polycyclic aromatic hydrocarbons [52]. The most common of the slow acetylator variants, rs4986782, is referred to as NAT1*14B in the pharmacology literature [53]. We identified an association between rs4986782 and the phecode for chemotherapy, a probable indicator of the presence of nonspecific cancer (956 cases and 4664 controls; p = 6.45 × 10- 5; OR: 1.85; 95% CI: 1.37–2.50). However, we noted that the chemotherapy phecode fails to exclude a number of specific cancer types from the control group. After expanding the range of exclusions from controls (all neoplastic ICD-9-CM codes 140–239), evidence for replication remained (p = 8.68 × 10-5; OR: 1.98; 95% CI: 1.48–2.64). Furthermore, we detected associations with two cancer and cancer-related phenotypes at p < 0.01: cervical cancer (37 cases; p = 7.54 × 10-3; OR: 5.54; 95% CI: 1.58–19.94) and myeloid leukemia (47 cases; p = 1.95 × 10-3; OR: 3.82; 95% CI: 1.58–8.90). We were underpowered to attempt replication in the African–American population (MAF = 0.003).
Lastly, we identified an association between CYP2A6 rs28399433 (coded allele G; CYP2A6*2) and the phecode for tobacco use disorder (1132 cases and 4882 controls; p = 0.03; OR: 0.81; 95% CI: 0.66–0.99). Several studies have reported that CYP2A6*2 and other genotypes associated with poor nicotine metabolizer status correlate with a decrease in cigarettes per day [54]. These results validate a previous study that demonstrated that ICD-9-CM codes can be markers of smoking behavior in EHRs [55].
Novel associations among European–Americans
One novel association met this study's phenome-wide significance threshold in European–Americans. Four SLC15A2 variants (rs1143672, rs2293616, rs1143671 and rs2257212) in strong linkage disequilibrium (r2 > 0.99 in this dataset and in HapMap3 CEU samples) were associated with renal osteodystrophy, a disorder of mineral and bone metabolism that increases risk to fractures and joint problems often as a complication of chronic kidney disease (205 cases and 3811 controls among European–Americans). All SLC15A2 variants had identical minor allele frequencies in the combined population (CAF = 0.47; Supplementary Figure 1). The four variants in SLC15A2 on the ADME Core Panel are exonic; three encode nonsynonymous changes, one synonymous (rs2293616). The most significant association was with rs1143672 (p = 2.67 × 10-6, OR: 0.61; 95% CI: 0.49–0.75), which exhibited pleiotropic effects on renal- and bone-related traits such as osteoporosis, renal failure, diabetic nephropathy, among others (Figure 2). For the 79 African–American cases and 308 controls, the association between rs1143672 and renal osteodystrophy was not significant (p = 0.12). However, the direction and magnitude of effect among African–American cases compared with controls was consistent with results for European–Americans. We also observed nominally significant associations with SLC15A2 rs1143672 and other advanced renal disease traits in the African–Americans, such as Type 1 (cases = 31; p = 0.01; OR: 0.49) and Type 2 diabetic nephropathy (cases = 101; p = 0.03; OR: 0.70).
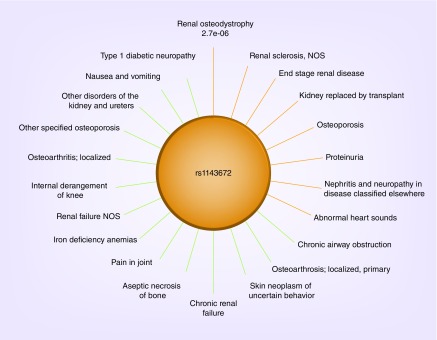
Figure 2. . Sun plots of SLC15A2 rs1143672.
The most significant genotype–phenotype association is at 12 o'clock and p-values increase in a clockwise fashion around the circle. Bars are colored in red for p-values less than 0.001. The remaining p-values are < 0.05.
This study defined renal osteodystrophy by one ICD-9-CM code: 588.0 (Supplementary Figure 2). The coding definition excluded two ICD-9-CM groups: ‘nephritis, nephrotic syndrome and nephrosis’ (580–589) and ‘other diseases of urinary system’ (590–599). However, a substantial proportion (11.2%) of patients in this study had undergone a kidney transplant [45,56]. Because of the strong correlation between renal osteodystrophy diagnoses and kidney transplant in this dataset (rφ= 0.82, p > 10-20), we retested the association between SLC15A2 rs1143672 and renal osteodystrophy after adjustment for kidney transplant status and found that the association was not significant (p = 0.22). This adjusted model suggests that SLC15A2 is broadly associated with advanced kidney disease, and that the association with renal osteodystrophy may be the result of mediated pleiotropy [20].
Novel associations among African–Americans
In total, seven associations met the phenome-wide significance threshold in the African–American dataset (p < 1.50 × 10-4, Supplementary Table 3). The strongest association was between SLC22A2 rs316019 and cardiac arrhythmias (p = 2.79 × 10-5; OR: 2.76; 95% CI: 1.71–4.42). This variant was also associated with the phecode for heart transplant (p = 1.36 × 10-4; OR: 3.25; 95% CI: 1.96–5.40). The allele frequency of the SLC22A2 rs316019 coded (T) allele was 0.28 in cases (n = 64) and 0.13 in controls (n = 437). We also detected an association between SLC22A1 rs628031 and the phecode for cough (cases = 257; p = 1.6 × 10-4; OR: 0.59; 95% CI: 0.47–0.74). These associations were not significant in the European–American cohort (p > 0.05). The remaining phenome-wide associations in the African–American analysis were for phecodes that had relatively low case numbers (n < 41; Supplementary Table 3).
Paired trait analysis
We performed multivariate tests of association between SLC15A2 and all pairs of phecodes. We excluded pairs of codes with correlations >0.95, and limited this analysis to those pairs for which there were at least 50 patients with both codes present and at least 50 patients with both codes absent. This resulted in 26,599 pairs of codes for the European–Americans and 2035 pairs of codes for the African–Americans, leading to Bonferroni-corrected significance thresholds of p = 1.88 × 10-6 and p = 2.46 × 10-5, respectively. All four SLC15A2 variants expectedly produced similar results. Results for rs1143672, the most significant SNP in the PheWAS, are presented here.
In European–Americans, four paired traits passed the significance threshold after correction for multiple testing: abnormal heart sounds and ESRD (p = 5.73 × 10-7); skin neoplasm of uncertain behavior and ESRD (p = 1.54 × 10-7); pain in joint (p = 3.26 × 10-7) and renal osteodystrophy and back pain and renal osteodystrophy (p = 7.65 × 10-7) (Table 3). All significant pairs included at least one advanced renal disease related trait. Although no paired traits passed the significance threshold after correction for multiple testing in African–Americans, the strongest paired association with SLC15A2 included an advanced renal disease phecode: iron deficiency anemias and chronic renal failure (p = 1.01 × 10-3). These traits were correlated (rφ= 0.57) and relatively prevalent in this dataset (cases for iron deficiency anemia: n = 102; cases for chronic renal failure: n = 258). Notably, the iron deficiency anemias and chronic renal failure pairing was also marginally significant in the paired-trait analysis of European–Americans (p = 7.62 × 10-3).
Table 3. . Significant associations detected in paired-trait analysis for SLC15A2 rs1143672.
| Trait 1 (n) | Trait 2 (n) | Joint p-value | rφ |
|---|---|---|---|
| Abnormal heart sounds (290) |
ESRD (558) |
5.11 × 10-7 |
0.12 |
| Renal osteodystrophy (205) |
Pain in joint (1918) |
3.36 × 10-7 |
0.10 |
| Renal osteodystrophy (205) |
Back pain (1330) |
4.66 × 10-7 |
0.06 |
| Skin neoplasm of uncertain behavior (393) | ESRD (558) | 1.77 × 10-7 | 0.06 |
We performed joint tests of association with pairs of phecodes and the four variants in SLC15A2. Analyses were limited to pairs of codes with at least 50 individuals with both codes present; 50 individuals where both codes were absent and the correlation, rφ, was less than 0.95. Regression models in MultiPhen were adjusted for the ages of the samples at the two traits and sex. Bonferroni-corrected significance for the MultiPhen analysis in European–Americans was declared at p < 1.88 × 10- 6 (26,599 paired traits). Median correlation (rφ) of the paired traits is given for each joint test shown. p-values for tests of correlation are <0.001.
ESRD: End-stage renal disease.
A Q–Q plot of p-values revealed significant inflation for European–Americans (Supplementary Figure 3). In contrast, there was no detectable inflation for African–Americans (Supplementary Figure 3). When we performed the same multivariate analysis on other variants that approached phenome-wide significance in the single-trait analysis (SLC22A2 rs316019, DYPD rs1801265, NAT2 rs1799931, CYP2B6 rs8192709, CYP2D6 rs5030656, CYP2D6 rs35742686 and CYP2D6 rs5030655), we also did not observe a similar inflation of p-values (Supplementary Figure 4). Given the expected low type-1 error rate of MultiPhen [36], we can conclude that the inflation we observed for the SLC15A2 paired-trait analysis was likely driven by true pleiotropic effects across a spectrum of phecodes, specifically those associated with high morbidity, such as kidney transplant (Supplementary Figure 5).
Discussion
Here, we report the results of a PheWAS of the ADME Core variants in an EHR-linked biorepository. As expected, given the known pleiotropy of pharmacogenes, we detected diverse signals across the phenomic spectrum. We replicated several known associations in European–Americans and detected potentially novel pleiotropic associations in both European–Americans and African–Americans, broadening the general catalog of human pleiotropy and further highlighting the complexity in disentangling genotype–phenotype relationships.
ADME PheWAS replicates single trait associations
We robustly reproduced two associations between ADME genes and physiological traits: ABCG2 and gout and SLCO1B1 and jaundice [47,49]. The remaining replications are signals driven by the impact of ADME variants on the metabolism of xenobiotics (CYP2A6 on nicotine, CYP2C19 and NAT1 on carcinogens). A recent meta-analysis of candidate gene studies confirmed the association between CYP2C19 poor-metabolizer genotypes and gastric cancer [50]. Previous studies focused on Asian populations, but to our knowledge this is the first report the signal has been reproduced in individuals of European descent (stomach cancer: p < 0.04, OR: 1.69). In addition, we found an association with atrophic gastritis (p = 3.50 × 10-4, OR: 1.99), which can be associated with Helicobacter pylori infections or autoimmune conditions, and is associated with an increased risk of gastric cancer. Interestingly, we detected a significant signal between the gain-of-function CYP2C19*17 allele and hepatic cancer (p-value = 6.00 × 10-3; OR: 1.84). This finding corroborates a recent gene expression study of hepatic cancer, which found increased CYP2C19 mRNA expression in hepatocarinoma tissue [57]. These findings together suggest that CYP2C19 may have independent roles in the elimination of carcinogens and bioactivation of procarcinogens in the stomach and liver, respectively.
ADME PheWAS identifies potential novel pleiotropy
We detected a novel and biologically plausible association between SLC15A2 rs1143672 and renal osteodystrophy in European–Americans. SLC15A2 is expressed in the proximal tubule of the nephron [58] and encodes PEPT2, a proton-peptide cotransporter responsible for the absorption di- and tripeptides produced by luminal pepidases. PEPT2 also serves as a transporter for beta-lactam antibiotics, cephalexin as well as other drugs with peptide-like structures [59]. A haplotype analysis of the 14 most common SNPs in SLC15A2 revealed that >90% of the exonic variation in the gene was captured by two major haplotypes: *1 and *2 [59]. The SLC15A2*2 haplotype is tagged by the SLC15A2 rs1143672 genotyped here [59]. The *1 and *2 haplotypes are different in their uptake of dipeptides in that the *2 haplotype, which was associated with protection from renal osteodytrophy in this PheWAS, has significantly lower affinity than the *1 haplotype [59].
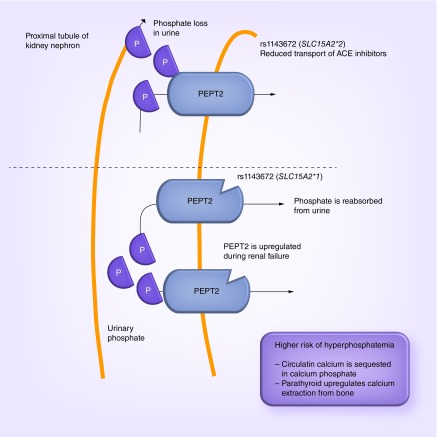
The pleiotropic effect of SLC15A2 on bone and renal disease observed here is likely mediated by a shared etiology [60]. The kidneys maintain proper levels of calcium and phosphorus in the blood, which are crucial compounds in the maintenance of bone hormone levels. The kidneys accomplish this feat by converting vitamin D into calcitriol, which helps the body absorb dietary calcium and phosphorus into the bones. Calcitriol also regulates production of the parathyroid hormone, another important compound in the maintenance of circulating calcium. These functions are disturbed in patients with kidney disease and can lead to a decline in bone mass (e.g., renal osteodystrophy). Furthermore, studies of 5/6 nephrectomized rats, a model of human chronic renal failure, demonstrated that PEPT2 is upregulated resulting in an increase in reabsorption of small peptides in this diseased state [61]. The statistical association data here coupled with the functional data previously published suggest that kidney disease patients with the SLC15A2*2 haplotype have lower affinity and thus less uptake of peptides and/or drugs in the reabsorption process of the kidney's proximal tubule compared with patients with the SLC15A2*1 haplotype. Indeed, PEPT2 is known to transport ACE inhibitors [62], a drug often prescribed to patients with chronic kidney disease to lower blood pressure to prevent further kidney damage. ACE inhibitors should be used with caution in patients with high potassium levels as these drugs may further increase the patient's potassium levels leading to hyperkalemia. It may be that kidney disease patients in this study with the SLC15A2*2 haplotype experience lower uptake of ACE inhibitors, altering their blood concentration resulting in increased urinary loss of phosphate and a decreased risk of hyperparathyroidism secondary to hyperphosphatemia (thought to be the cause of renal osteodystrophy) (Figure 3). This novel PheWAS result may be an example of a weak pharmaocgenomic effect of SLC15A2 on ACE inhibitors in the general population that is amplified in patients with existing kidney disease due to increased expression of the gene. More generally, these PheWAS results suggest that a pharmacogenomic effect of genetic variants in SLC15A2 may be induced by kidney disease, which has downstream risks of bone disease that are greatly amplified during the aging process.
Figure 3. . Proposed physiologic model of the effects of SLC15A2 rs1143672.
In nonhuman models of chronic kidney disease, PEPT2 is upregulated leading to dysregulation of proper levels of calcium and phosphorus in the blood. We propose the SLC15A2*2 haplotype effect observed here statistically is induced by kidney failure. That is, kidney disease patients with the SLC15A2*2 haplotype have lower affinity and thus less uptake of peptides and ACE inhibitors in the reabsorption process of the kidney's proximal tubule compared with patients with the SLC15A2*1 haplotype. The lower affinity likely results in increased urinary loss of phosphate and a decreased risk of hyperparathyroidism secondary to hyperphosphatemia (thought to be the cause of renal osteodystrophy).
Published data from the electronic MEdical Records & GEnomics (eMERGE) Network PheWAS catalog [63] tentatively corroborate the association results observed in this PheWAS. In the eMERGE network analysis, a PheWAS was performed on 3144 SNPs in the National Human Genome Research Institute GWAS catalog that met genome-wide significance (p < 5 × 10-8) [25]. SLC15A2 rs4285028, a variant in the 3′ region of SLC15A2 and previously identified as risk factor for multiple sclerosis [64], was included in the eMERGE network analysis [25]. While the eMERGE network dataset does not represent a suitable replication cohort given the nominal correlation between the variants SLC15A2 rs4285028 and SLC15A2 rs1143672 (1000 Genomes Project CEU: r2 = 0.23; D′ = 1) and the relatively low sample sizes of advanced renal traits (ESRD n = 132, renal osteodystrophy n = 92, kidney replaced by transplant n = 116), the dataset does provide an opportunity to examine pleiotropy at the gene level with the traits available. The eMERGE network PheWAS reported that SLC15A2 rs4285028 was associated with calcium/phosphorus disorders (European–American, n = 496; p = 0.02; OR: 1.19), fracture of hand or wrist (cases = 691; p = 0.02; OR: 0.85), and calculus of kidney (n = 628, p = 0.03; OR: 1.15) [25]. The regulation of calcium and phosphorous levels is a shared factor in the etiology of these traits, a finding that supports a bone mineral-advanced renal disease pleiotropic effect of SLC15A2.
The results the paired trait analysis of SLC15A2 rs1143672 in MultiPhen revealed Bonferonni-corrected significant associations with two advanced kidney disease related traits: ESRD and renal osteodystrophy (Table 3). MultiPhen utilizes multiple discrete traits to detect genetic associations with ‘unmeasured phenotypes’ hidden in single phenotype analyses [36]. The improvement of MultiPhen over PheWAS or GWAS is most evident when the discrete traits are not in the same direction as the correlation between the two phenotypes. As previously described [36], MultiPhen is powerful when a variant affects only one of two highly correlated phenotypes, or a variant affects negatively correlated phenotypes in the same direction or positively correlated phenotypes in opposite directions.
The significant pairing of ESRD and skin neoplasm of uncertain behavior supports the model that specific SLC15A2 rs1143672 alleles are enriched in the kidney transplant subpopulation beyond expectation. Immunosuppressant agents, cyclosporine and tacrolimus, are regularly prescribed to organ transplant recipients to prevent the onset of graft versus host disease. Skin cancer is a common side effect of these drugs; as such, kidney transplant patients are at increased risk for its development [65]. This common adverse side effect of immunosuppressants may explain the modest association with SLC15A2 rs1143672 and skin cancer observed in the PheWAS (p = 1.31 × 10-3; OR: 0.79). We suspect that kidney transplantation is likely unmeasured phenotype driving the significant paired association, skin neoplasm of uncertain behavior and ESRD, detected by MultiPhen.
In BioVU's kidney transplant population, codes for chronic kidney failure may vary and/or lose sensitivity, especially among those patients who entered BioVU post-transplant. In this heterogeneous population, with respect to the timing of entry into BioVU and specific kidney disease coding, one could expect a weakening of associations with specific renal disease codes and an ESRD risk variant in the PheWAS. Instead, the relative independence of skin cancer and ESRD codes in the kidney transplant population still achieved significance for an association with an ESRD risk variant by MultiPhen. Overall, the paired associations observed between rs1143672 and skin cancer and ESRD could be interpreted as further evidence that the SLC15A2 represents an undiscovered genetic factor in the onset of end-stage kidney disease.
The other significant paired associations with SLC15A2 rs1143672 are more difficult to interpret. The association with abnormal heart sounds is interesting, as cardiovascular disease is a leading cause of death among patients with ESRD [66]. It is plausible that since kidney patients have too much fluid and increased vascular disease, they would more often have abnormal heart sounds on exam. The paired associations with renal osteodystrophy included back pain and pain in joint, two physical symptoms which are reflective of the bone-mineral density disorder. This finding may be interpreted as the presence of physical symptoms in the billing codes that can improve association signals with primary phenotypes by adding additional layers of phenotypic dimension. That is, renal osteodystrophy may be undiagnosed or uncoded in patients with kidney disease resulting in an independent signal of joint pain and back pain with SLC15A2.
ADME PheWAS in African–Americans
In general, the small sample size for African–Americans reduced statistical power and our ability to replicate or generalize the findings detected in the PheWAS performed among European–Americans (Table 2). African–Americans had much fewer cases of gout (n = 65), jaundice (n = 22) and atrophic gastritis (n = 3) compared with European–Americans in this dataset. Also impacting statistical power was the difference in CAFs between the two populations. The allele frequencies of NAT1 rs4986782 (CAF = 0.003) and CYP2A6 rs1801272 (CAF = 0.005) in the African–American dataset were much lower than in European–Americans.
In contrast with standard PheWAS, the paired trait analysis was able to generalize a pattern of association with SLC15A2 and advanced renal disease observed in European–Americans to African–Americans. The most significant paired association, iron deficiency anemia and chronic renal failure, supports a suspected role for SLC15A2 in ESRD in African–Americans. Anemia is actively monitored in patients with advanced kidney disease [67]. Cells in the kidney make the hormone erythropoietin, which stimulates the bone marrow to make new red blood cells that on average last about 120 days. As the kidneys fail, patients make less of the hormone and anemia ensues. In addition, kidney failure is a state of inflammation, which affects iron stores and usage. Therefore, many patients with kidney disease are at risk for iron deficiencies and require iron infusions to help with the anemia. We suspect that chronic renal failure codes and iron deficiency anemia codes are correlated with ESRD in this patient population, driving the association with SLC15A2 rs1143672.
Limitations & strengths
This present study has numerous limitations and strengths. The present study population is a composite of samples genotyped for pharmacogenetic studies performed in BioVU and genotyped on a fixed-content array designed for pharmacogenomics [45]. Consequently, this patient population is a highly medicated and disease-burdened group in comparison with the general population. Given the role of ADME genes, we suspect some of the results observed here may be driven by gene–xenobiotic interactions abundant in this patient population that were not considered in this analysis. As previously mentioned, we were statistically underpowered to replicate associations observed in European–Americans in the African–American patient population. Many of the common ADME Core variants in the European–American dataset were rare in the African–American dataset, and vise-versa (Table 2). Also, BioVU in general contains fewer African–Americans than European–Americans (˜13 African–Americans for every 100 European–Americans), resulting in fewer case and control counts available for analysis among African–Americans. Finally, imprecise phenotyping may have also lowered the statistical power of this study. ICD-9-CM codes may not be reflective of the actual clinical outcome of the patient.
Another major limitation is the fact that the present study does not include an independent replication dataset. Despite corrections for multiple testing, it is possible that the novel findings observed here could represent false positive associations. Indeed, the Q–Q plots of the resulting PheWAS p-values showed moderate inflation (Supplementary Figure 6). However, this property of the p-values was attenuated by the removal of specific genes with strong signals of association in the PheWAS (Supplementary Figure 7). Given the strong correlation across phecodes in this dataset (Supplementary Figures 5 & 8), the modest correlation of p-values in the single-trait analysis is unsurprising and does not necessarily signify the presence of spurious results. Also, although not a technical replication given the low level of linkage disequilibrium between tested variants, the eMERGE network PheWAS for variants in SLC15A2 corroborate our overall observations and conclusions that these data support a bone mineral-advanced renal disease pleiotropic effect of SLC15A2.
Replication is notoriously difficult for pharmacogenomic studies [68]. SLC15A2 has not yet been implicated in published GWAS for kidney-related quantitative traits, chronic kidney disease or ESRD. However, it should be noted that existing studies in the literature do not mirror the study population presented here. For example, existing GWAS of kidney-related quantitative traits such as serum phosphorous levels [69] and serum calcium [70] were studies of the traits within the normal range. Also, a recent large GWAS of kidney function and decline focused on change in kidney function regardless of chronic kidney disease status at baseline as measured by the estimated glomerular filtration rate at different clinic visits [71]. The pharmacogenomic sample presented here is unique in many ways including its EHR basis and the oversampling of kidney disease patients (˜29% had renal osteodystrophy) and kidney transplant (16% of the study population) [45]. Existing EHRs linked to biorepositories with GWAS data such as those available in the eMERGE I Network also do not mirror the sample described here given that the individual study sites selected samples for genotyping based on other phenotypes (e.g., dementia, cataracts, electrocardiographic traits, peripheral artery disease and Type 2 diabetes) [72]. Thus, the bias for extreme kidney disease in this sample cannot be recapitulated in the existing GWAS datasets such as those available by the eMERGE I Network.
Despite the limitations, the present study findings are bolstered by biological plausibility based on published functional in vitro and in vivo data. The present study also benefited from the unique phenotypic data the EHR provides compared with other epidemiological cohorts. Many of the traits captured by the EHR and included in this PheWAS require a depth only available in a clinical setting. This phenotypic depth analyzed in conjunction with the genetic variants reveals genotype–phenotype complexities that can in turn be used to refine definitions of disease in coded in the clinic.
Conclusion
These results expand our understanding of the diverse roles of ADME genes and serve as a powerful tool for hypothesis generation. The novel association between SLC15A2 and renal disease warrants further investigation as these data offer the first clues into the genetic etiology of a complex trait at the interface of bone and kidney diseases. Our results also underscore the rich clinical data uniquely available to an EHR-derived phenome for detailed studies of pleiotropy across the human genome–phenome. With the paired trait approach, we were able to utilize more phenotypic data compared with previous phenome-wide approaches. Further methodological improvements on multivariate analyses in reverse genetic approaches are warranted.
Executive summary.
Background
Pleiotropy refers to cross-phenotype associations by a single genetic locus. Pleiotropy can arise from a variety of mechanisms, and recent studies have show that pleiotropy is common among genome-wide association study (GWAS)-identified variants.
Systematic searches for cross-phenotype associations have not yet been performed for functional pharmacogenomic variants, many of which are genotyped in the clinic to inform prescription practices without considering the consequences of pleiotropic effects.
We conducted a phneome-wide association study (PheWAS) on 184 functional variants in 34 pharmacogenes assayed by the Illumina® Absorption, Distribution, Metabolism, and Elimination (ADME) Core Panel for 6067 European–Americans and 762 African–Americans linked to deidentified eletronic health records. Among these patients, a total of 808 and 333 phenotypes (termed ‘phecodes’) were available among European–Americans and African–Americans for this PheWAS.
Findings
We replicated several previously reported genotype-phenotype associations and identified potentially novel pleitropic associations.
Among European–Americans, replicated associations included ABCG2 rs2231142 and gout and SLCO1B1 and rs4149056 and jaundice.
One novel association was identified after correction for multiple testing: SLC15A2 rs1143672 and renal osteodystrophy in European–Americans. Adjustment for kidney transplant status (oversampled in this patient population) attenuated the association, suggesting the association between rs1143672 and renal osteodystrophy is a result of mediated pleiotropy. Paired trait analysis between SLC15A2 rs1143672 and all pairs of phecodes identified four paired traits after correction for multiple testing. All significant pairs included at least one advanced renal disease related trait.
African–Americans in general were underpowered to replicate previously reported associations. Several significant potentially novel pleiotropic associations were identified among African–Americans, including SLC22A2 rs316019 and cardiac arrhythmias and SLC22A1 rs628031 and cough.
Conclusion
Replication of previously reported associations further validates the high-throughput approach of PheWAS.
We identified potentially novel pleiotropic associations, notably SLC15A2 rs1143672 (and three variants in linkage disequilibrium with it) among European–Americans. The association identified in this PheWAS has biologic plausibility. SLC15A2 is expressed in the proixmal tubule of the nephron and encodes PEPT2, a proton-peptide co-transpoter. Previously published functional studies have shown that the SLC15A2*2 haplotype, tagged by rs1143672, has lower affinity and less uptake of peptides and/or drugs such as ACE inhibitors commonly prescribed in patients with chronic kidney disease. Published studies of 5/6 nephrectomized rats, a model of human chronic renal failure, demonstrated that PEPT2 is upregulated resulting in an increase in reabsorption of small peptides in this diseased state. Taken together, the observed statistical association may be representative of a weak pharmacogenomic effect of SLC15A2 on ACE inhibitors in the general population that is amplied in patients with existing kidney disease.
Replication for this and other pharmacogenomic studies in general is difficult due to lack of comparable datasets.
Interpreting cross-phenotype associations is statistically challenging and requires further functional follow-up for confirmation.
Supplementary Material
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Motulsky AG. Drug reactions, enzymes, and biochemical reactions. JAMA. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 2.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N. Engl. J. Med. 2005;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 3.Cooper GM, Johnson JA, Langaee TY, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112(4):1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111(8):4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuldiner AR, O'Connell JR, Bliden KP, et al. Association of cytochrome p450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302(8):849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.SEARCH Collaborative Group. Link E, Parish S, et al. SLCO1B1 variants and statin-induced myopathy – a genomewide study. N. Engl. J. Med. 2008;359(8):789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 8.Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the vanderbilt PREDICT project. Clin. Pharmacol. Ther. 2012;92(1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shuldiner AR, Palmer K, Pakyz RE, et al. Implementation of pharmacogenetics: the University of Maryland personalized anti-platelet pharmacogenetics program. Am. J. Med. Genet. Part C. 2014;166(1):76–84. doi: 10.1002/ajmg.c.31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am. J. Med. Genet. Part C. 2014;166(1):56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Donnell PH, Danahey K, Jacobs M, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care – initial results of the University of Chicago “1,200 Patients Project”. Am. J. Med. Genet. Part C. 2014;166(1):68–75. doi: 10.1002/ajmg.c.31385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time – using genomic data to individualize treatment protocol. Mayo Clin. Proc. 2014;89(1):25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott SA, Sangkuhl K, Stein CM, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 2013;94(3):317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilke RA, Ramsey LB, Johnson SG, et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin. Pharmacol. Ther. 2012;92(1):112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 2013;369(24):2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kocarnik JM, Fullerton SM. Returning pleiotropic results from genetic testing to patients and research participants. JAMA. 2014;311(8):795–796. doi: 10.1001/jama.2014.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westbrook MJ, Wright MF, Van Driest SL, et al. Mapping the incidentalome: estimating incidental findings generated through clinical pharmacogenomics testing. Genet. Med. 2013;15(5):325–331. doi: 10.1038/gim.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson AD, Bhimavarapu A, Benjamin EJ, et al. CLIA-tested genetic variants on commercial SNP arrays: potential for incidental findings in genome-wide association studies. Genet. Med. 2010;12(6):355–363. doi: 10.1097/GIM.0b013e3181e1e2a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burke W, Matheny Antommaria AH, Bennett R, et al. Recommendations for returning genomic incidental findings? We need to talk! Genet. Med. 2013;15(11):854–859. doi: 10.1038/gim.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 2013;14(7):483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pendergrass SA, Brown-Gentry K, Dudek S, et al. Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genet. 2013;9(1):e1003087. doi: 10.1371/journal.pgen.1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivakumaran S, Agakov F, Theodoratou E, et al. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 2011;89(5):607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pendergrass SA, Brown-Gentry K, Dudek SM, et al. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype–phenotype relationships and pleiotropy discovery. Genet. Epidemiol. 2011;35(5):410–422. doi: 10.1002/gepi.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013;31(12):1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell SL, Hall JB, Goodloe RJ, et al. Investigating the relationship between mitochondrial genetic variation and cardiovascular-related traits to develop a framework for mitochondrial phenome-wide association studies. BioData Min. 2014;7:6. doi: 10.1186/1756-0381-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer. 2006;6(12):947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- 28.Nebert DW, Gonzalez FJ. P450 genes: structure, evolution, and regulation. Annu. Rev. Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- 29.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360(9340):1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 30.Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat. Rev. Genet. 2016;17(3):129–45. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 31.Tyler AL, Crawford DC, Pendergrass SA. The detection and characterization of pleiotropy: discovery, progress, and promise. Brief Bioinform. 2016;17(1):13–22. doi: 10.1093/bib/bbv050. [DOI] [PubMed] [Google Scholar]
- 32.Pendergrass SA, Verma A, Okula A, et al. Phenome-wide association studies: embracing complexity for discovery. Hum. Hered. 2015;(3):111–123. doi: 10.1159/000381851. [DOI] [PubMed] [Google Scholar]
- 33.Oetjens MT, Denny JC, Ritchie MD, et al. Assessment of a pharmacogenomic marker panel in a polypharmacy population identified from electronic medical records. Pharmacogenomics. 2013;14(7):735–744. doi: 10.2217/pgs.13.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am. J. Hum. Genet. 2011;89(4):529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Reilly PF, Hoggart CJ, Pomyen Y, et al. MultiPhen: joint model of multiple phenotypes can increase discovery in GWAS. PLoS ONE. 2012;7(5):e34861. doi: 10.1371/journal.pone.0034861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumitrescu L, Ritchie MD, Brown-Gentry K, et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet. Med. 2010;12(10):648–650. doi: 10.1097/GIM.0b013e3181efe2df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall JB, Dumitrescu L, Dilks HH, Crawford DC, Bush WS. Accuracy of administratively-assigned ancestry for diverse populations in an electronic medical record-linked biobank. PLoS ONE. 2014;9(6):e99161. doi: 10.1371/journal.pone.0099161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67(1):170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bramer GR. International statistical classification of diseases and related health problems. Tenth revision. World Health Stat. Q. 1988;41(1):32–36. [PubMed] [Google Scholar]
- 43.PheWAS. http://phewascatalog.org
- 44.Pendergrass SA, Dudek SM, Crawford DC, Ritchie MD. Visually integrating and exploring high throughput Phenome-Wide Association Study (PheWAS) results using PheWAS-View. BioData Min. 2012;5(1):5. doi: 10.1186/1756-0381-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowton E, Field JR, Wang S, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci. Transl. Med. 2014;6(234) doi: 10.1126/scitranslmed.3008604. 234cm3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford DC, Goodloe R, Farber-Eger E, et al. Leveraging epidemiologic and clinical collections for genomic studies of complex traits. Hum. Hered. 2015;79(3):137–46. doi: 10.1159/000381805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dehghan A, Kottgen A, Yang Q, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372(9654):1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Spencer KL, Voruganti VS, et al. Association of functional polymorphism rs2231142 (Q141K) in ABCG2 gene with serum uric acid and gout in four US populations: the Population Architecture using Genomics and Epidemiology (PAGE) Study. Am. J. Epidemiol. 2013;177(9):923–932. doi: 10.1093/aje/kws330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson AD, Kavousi M, Smith AV, et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum. Mol. Genet. 2009;18(14):2700–2710. doi: 10.1093/hmg/ddp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Song K, Chen Z, Yu Y. Poor metabolizers at the cytochrome P450 2C19 loci is at increased risk of developing cancer in Asian populations. PLoS ONE. 2013;8(8):e73126. doi: 10.1371/journal.pone.0073126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Vries AC, van Grieken NCT, Looman CWN, et al. Gastric cancer risk in patients with premalignant gastric lesions: a nationwide cohort study in The Netherlands. Gastroenterology. 2008;134(4):945–952. doi: 10.1053/j.gastro.2008.01.071. [DOI] [PubMed] [Google Scholar]
- 52.Hein DW, Doll MA, Fretland AJ, et al. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol. Biomarkers Prev. 2000;9(1):29–42. [PubMed] [Google Scholar]
- 53.Millner LM, Doll MA, Cai J, States JC, Hein DW. Phenotype of the most common “slow acetylator” arylamine N-acetyltransferase 1 genetic variant (NAT1*14B) is substrate-dependent. Drug Metab. Dispos. 2012;40(1):198–204. doi: 10.1124/dmd.111.041855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiley LK, Shah A, Xu H, Bush WS. ICD-9 tobacco use codes are effective identifiers of smoking status. J. Am. Med. Inform. Assoc. 2013;20(4):652–658. doi: 10.1136/amiajnl-2012-001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birdwell KA, Grady B, Choi L, et al. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet. Genomics. 2012;22(1):32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu M, Chen S, Wu X. Differences in cytochrome P450 2C19 (CYP2C19) expression in adjacent normal and tumor tissues in Chinese cancer patients. Med. Sci. Monit. 2006;12(5):BR174–BR178. [PubMed] [Google Scholar]
- 58.Smith DE, Pavlova A, Berger UV, et al. Tubular localization and tissue distribution of peptide transporters in rat kidney. Pharm. Res. 1998;15(8):1244–1249. doi: 10.1023/a:1011996009332. [DOI] [PubMed] [Google Scholar]
- 59.Pinsonneault J, Nielsen CU, Sadee W. Genetic variants of the human H+/dipeptide transporter PEPT2: analysis of haplotype functions. J. Pharmacol. Exp. Ther. 2004;311(3):1088–1096. doi: 10.1124/jpet.104.073098. [DOI] [PubMed] [Google Scholar]
- 60.Hruska KA, Teitelbaum SL. Renal osteodystrophy. N. Engl. J. Med. 1995;333(3):166–174. doi: 10.1056/NEJM199507203330307. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi K, Masuda S, Nakamura N, et al. Upregulation of H+-peptide cotransporter PEPT2 in rat remnant kidney. Am. J. Physiol. 2001;281(6):F1109–F1116. doi: 10.1152/ajprenal.0346.2000. [DOI] [PubMed] [Google Scholar]
- 62.Rubio-Aliaga I, Daniel H. Mammalian peptide transporters as targets for drug delivery. Trends Pharmacol. Sci. 2002;23(9):434–440. doi: 10.1016/s0165-6147(02)02072-2. [DOI] [PubMed] [Google Scholar]
- 63.PheWAS. http://phewas.mc.vanderbilt.edu/
- 64.Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harwood CA, Mesher D, McGregor JM, et al. A surveillance model for skin cancer in organ transplant recipients: a 22-year prospective study in an ethnically diverse population. Am. J. Transplant. 2013;13(1):119–129. doi: 10.1111/j.1600-6143.2012.04292.x. [DOI] [PubMed] [Google Scholar]
- 66.House AA. Cardio–renal syndrome type 4: epidemiology, pathophysiology and treatment. Semin. Nephrol. 2012;32(1):40–48. doi: 10.1016/j.semnephrol.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 67.Levey AS, Coresh J, Balk E, et al. National Kidney Foundation practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Ann. Intern. Med. 2003;139(2):137–147. doi: 10.7326/0003-4819-139-2-200307150-00013. [DOI] [PubMed] [Google Scholar]
- 68.Daly AK. Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 2010;11(4):241–246. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 69.Kestenbaum B, Glazer NL, Kottgen A, et al. Common genetic variants associate with serum phosphorus concentration. J. Am. Soc. Nephrol. 2010;21(7):1223–1232. doi: 10.1681/ASN.2009111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Seaghdha CM, Wu H, Yang Q, et al. Meta-analysis of genome-wide association studies identifies six new loci for serum calcium concentrations. PLoS Genet. 2013;9(9):e1003796. doi: 10.1371/journal.pgen.1003796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorski M, Tin A, Garnaas M, et al. Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int. 2015;87(5):1017–1029. doi: 10.1038/ki.2014.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCarty C, Chisholm R, Chute C, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics. 2011;4(1):13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.