Abstract
A multiplex PCR assay was developed for detection of the six types of glycopeptide resistance characterized in enterococci and for identification of Enterococcus faecium, Enterococcus faecalis, Staphylococcus aureus, and Staphylococcus epidermidis at the species level. Primers targeting the genes vanA, vanB, vanC, vanD, vanE, vanG, and ddl of E. faecium and E. faecalis and nuc of S. aureus and a chromosomal portion specific to S. epidermidis were designed to allow amplification of fragments with various sizes. This specific and sensitive technique allows detection of glycopeptide-resistant strains, in particular methicillin-resistant S. aureus, that may escape phenotype-based automated rapid methods.
Glycopeptide antibiotics are used in the treatment of infections caused by gram-positive bacteria in case of resistance or allergy to other antibiotics. Six types of glycopeptide resistance have been described in enterococci that can be distinguished on the basis of the sequence of the structural gene for the resistance ligase (vanA, vanB, vanC, vanD, vanE, and vanG). VanA-type resistance is characterized by high-level resistance to both vancomycin and teicoplanin (3, 21), whereas VanB-type strains are resistant to variable levels of vancomycin but susceptible to teicoplanin (3). VanD-type strains are characterized by resistance to moderate levels of vancomycin and teicoplanin (10, 11, 30). VanC (13), VanE (15), and VanG (8, 25) isolates exhibit low-level resistance to vancomycin only.
Glycopeptide-resistant enterococci (GRE), first reported in 1988 (21, 33), are often resistant to multiple antibiotics, have a broad geographical distribution, and have become a major cause of nosocomial infections (5). Three glycopeptide-resistant Staphylococcus aureus strains with a vanA genotype have been recently isolated in the United States (18, 26, 31), and most importantly, two of these clinical isolates were not detected by automated in vitro susceptibility testing methods (18, 32).
The first multiplex PCR-based method to identify glycopeptide resistance genotypes, as well as the enterococcal host species (12), was followed by numerous refinements (4, 17, 19, 23, 27). However, detection of the vanD (10, 29, 30), vanE (15), and vanG (8, 25) genes was not included since these genotypes have only been detected recently.
The purpose of this work was to design a simple and rapid multiplex PCR for reliable detection of the various glycopeptide resistance genes and identification of the Enterococcus faecalis, Enterococcus faecium, Enterococcus gallinarum-Enterococcus casseliflavus-Enterococcus flavescens, S. aureus, and Staphylococcus epidermidis hosts at the species level.
Several methods for rapid preparation of template DNA from gram-positive cells have been reported. Direct suspension of bacteria in a PCR mixture (27) or the boiling method (4), which involves heating at 100°C, followed by centrifugation of the cell suspension, are the simplest and fastest techniques but did not yield the expected PCR products consistently. Extraction of bacterial DNA by rapid alkaline lysis (12) was the most efficient method for multiplex PCR. Cells from 3 ml of enterococci or 1.5 ml of staphylococci from an overnight shaken culture in brain heart infusion broth were harvested (15,000 × g, 5 min); suspended in 150 μl of a solution containing 50 mM Tris hydrochloride (pH 8.0), 10 mM EDTA, and 7% sucrose with either lysozyme (10 mg/ml; Sigma Chemical, St Louis, Mo.) for enterococci or lysostaphin (5 mg/ml; AMBI UK, Trowbridge, United Kingdom) for staphylococci; and incubated at 37°C for 20 min. The resulting protoplasts were lysed with sodium dodecyl sulfate (1.25%) for 10 min on ice, and after two phenol-chloroform extractions, total DNA was recovered in the supernatant after centrifugation (15,000 × g, 5 min).
On the basis of the sequence alignment of the vanA, vanB, vanC, vanD, vanE, and vanG resistance genes, pairs of primers specific to each gene were designed to amplify internal fragments with sizes ranging from 430 to 941 bp (Table 1). The vanB primers derived from the sequences of the vanB-1, vanB-2, and vanB-3 subtypes (28), which confer indistinguishable phenotypes, allow amplification of a single 635-bp product (Table 1). Since the prototype VanD (30) and “VanD-4” (7, 11) proteins are only 85% identical but confer similar resistance phenotypes, a pair of primers able to amplify all of the vanD genes was designed (Table 1). There are three vanC genes, vanC-1, vanC-2, and vanC-3, which are, respectively, specific to the motile, intrinsically vancomycin-resistant enterococcal species E. gallinarum, E. casseliflavus, and E. flavescens (13, 27). The vanC-2 and vanC-3 genes are nearly identical (99% identity) and are usually reported as vanC-2/3 (28). A pair of degenerate primers that could amplify vanC-1 and vanC-2/3 was designed (Table 1) that enables discrimination of these three species from other enterococci.
TABLE 1.
Deoxyoligonucleotide primers used in this study
| Primera | Sequence (5′→3′) | Gene | Position(s)b | Size of PCR product (bp) | Accession no. | Reference or source |
|---|---|---|---|---|---|---|
| EA1(+) | GGGAAAACGACAATTGC | vanA | 176-192 | 732 | M97297 | 12 |
| EA2(−) | GTACAATGCGGCCGTTA | 907-891 | ||||
| EB3(+) | ACGGAATGGGAAGCCGA | vanB | 169-185 | 647 | U00456/AF550667 | This work |
| EB4(−) | TGCACCCGATTTCGTTC | 815-799 | ||||
| EC5(+) | ATGGATTGGTAYTKGTATc | vanC1/2 | 133-150/142-159 | 815/827 | AF162694/L29638 | This work |
| EC8(−) | TAGCGGGAGTGMCYMGTAAc | 947-929/968-950 | ||||
| ED1(+) | TGTGGGATGCGATATTCAA | vanD | 357-375 | 500 | AF130997/AY082011 | This work |
| ED2(−) | TGCAGCCAAGTATCCGGTAA | 856-837 | ||||
| EE1(+) | TGTGGTATCGGAGCTGCAG | vanE | 364-382 | 430 | AF430807 | This work |
| EE2(−) | ATAGTTTAGCTGGTAAC | 793-777 | ||||
| EG1(+) | CGGCATCCGCTGTTTTTGA | vanG | 68-86 | 941 | AY271782 | This work |
| EG2(−) | GAACGATAGACCAATGCCTT | 1008-989 | ||||
| DD13(+) | CACCTGAAGAAACAGGC | ddl (E. faecalis) | 206-222 | 475 | U00457 | This work |
| DD3-2(−) | ATGGCTACTTCAATTTCACG | 680-661 | ||||
| FAC1-1(+) | GAGTAAATCACTGAACGA | ddl (E. faecium) | 1-18 | 1,091 | U39790 | This work |
| FAC2-1(−) | CGCTGATGGTATCGATTCAT | 1091-1072 | ||||
| Tn1(+) | GACTATTATTGGTTGATCCACCTGd | nuc (S. aureus) | 350-363 | 218 | V01281 | 6 |
| Tn2(−) | GCCTTGACGAACTAAAGCTTCGd | 567-554 | ||||
| Se705-1(+) | ATCAAAAAGTTGGCGAACCTTTTCA | S. epidermidise | 21-45 | 125 | NAf | 24 |
| Se705-2(−) | CAAAAGAGCGTGGAGAAAAGTATCA | 145-121 |
+, sense primer; −, antisense primer.
Nucleotide numbering begins at the initiation codon of the gene.
K = G or T; M = A or C; Y = C or T.
A sequence carrying a Sau3A or a HindIII restriction site (underlined) has been incorporated into oligodeoxynucleotides Tn1 and Tn2, respectively.
Chromosomal fragment of S. epidermidis.
NA, not applicable.
Since E. faecalis and E. faecium represent more than 95% of the clinical isolates collected (5), identification of enterococci based on the amplification of a fragment internal to the ddl gene encoding a d-Ala-d-Ala ligase included only these two species. Primers complementary to the S. aureus thermonuclease nuc gene (6) and to a chromosomal fragment specific to S. epidermidis (24) were used for identification of the two species.
One to three microliters of total DNA was subjected to multiplex PCR amplification in a 100-μl reaction mixture containing 1× PCR buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.2 mg of bovine serum albumin per ml), 50 μM each deoxynucleoside triphosphate, 40 pmol of each of the 10 primer pairs (Table 1), and 2 U of Taq polymerase (QBIOgene, Montreal, Quebec, Canada). Amplification was carried out with the following thermal cycling profile: 3 min at 94°C and 30 cycles of amplification consisting of 1 min at 94°C, 1 min at 54°C, and 1 min at 72°C, with 7 min at 72°C for the final extension. DNA fragments were analyzed by electrophoresis in 0.5× Tris-borate-EDTA on a 1% agarose gel stained with ethidium bromide.
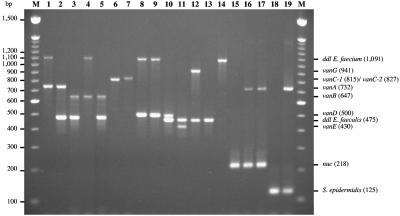
Assays using DNA of 19 phenotypically and genotypically well-characterized strains including glycopeptide-susceptible controls as a template confirmed the specificity of the PCR primers (Fig. 1). Each GRE exhibited two amplification products with the expected size between 430 and 1,092 bp (Table 1): one corresponding to the resistance genotype and the other corresponding to the host species (Fig. 1). There were no discrepancies between the results obtained by multiplex PCR and the previously characterized genotypes for resistance and identification. For VanC-type strains, a single PCR product resulting from vanC amplification was obtained with E. gallinarum vanC-1 and E. casseliflavus vanC-2-E. flavescens vanC-3 (Fig. 1). This result confirmed (i) that degenerate oligodeoxynucleotides (EC5 and EC8) specifically amplified a portion of the vanC-1 and vanC-2/3 genes and (ii) that primers specific to the ddl genes of E. faecalis and E. faecium were not able to amplify those of E. gallinarum and E. casseliflavus-E. flavescens. Furthermore, the primers used to amplify portions of the van genes did not amplify the host ddl gene. Glycopeptide-susceptible E. faecalis JH2-2 and E. faecium BM4107 produced a single PCR fragment corresponding to their respective ddl genes (Fig. 1).
FIG. 1.
Analysis by DNA amplification of glycopeptide-resistant and -susceptible enterococci and staphylococci. Lanes: 1, E. faecium BM4147 (vanA) (21); 2, E. faecalis BM4316 (vanA) (2); 3, E. faecalis V583 (vanB-1) (14); 4, E. faecium BM4524 (vanB-2) (9); 5, E. faecalis VRE45 (vanB-3) (28); 6, E. gallinarum BM4174 (vanC-1) (13); 7, E. casseliflavus ATCC 25788 (vanC-2) (34); 8, E. faecium BM4339 (vanD) (30); 9, E. faecium 10/96A (vanD-4) (7); 10, E. faecalis BM4539 (vanD) (10); 11, E. faecalis BM4405 (vanE) (15); 12, E. faecalis BM4518 (vanG) (8); 13, E. faecalis JH2-2 (susceptible) (16); 14, E. faecium BM4107 (susceptible) (22); 15, S. aureus COL (susceptible) (20); 16, S. aureus MI-VRSA (vanA) (31); 17, S. aureus PA-VRSA (vanA) (26); 18, S. epidermidis BM4577 (susceptible) (our collection); 19, S. epidermidis BM4577 mixed with pAT613 DNA (vanA) (1); M, molecular size marker (100-bp DNA ladder; Invitrogen, Groningen, The Netherlands). The size (in base pairs) of each PCR product is in parentheses on the right.
Two S. aureus strains, MI-VRSA (31) and PA-VRSA (26), with the vanA genotype have been isolated in Michigan and in Pennsylvania, respectively. The total DNAs of these strains and glycopeptide-susceptible S. aureus COL were subjected to the multiplex PCR (Fig. 1). Two fragments of 732 and 218 bp corresponding, respectively, to amplification of the vanA and nuc genes (Table 1) were obtained with the DNA of MI-VRSA and PA-VRSA, and a single 218-bp fragment was obtained with that of susceptible S. aureus COL (Fig. 1). These results confirm that, as expected, the ddl primers for identification of E. faecalis (DD13 and DD3-2) and E. faecium (FAC1-1 and FAC2-1) (Table 1) did not amplify S. aureus ddl and that the multiplex PCR can thus be used to detect the van operons in S. aureus.
Glycopeptide-susceptible S. epidermidis BM4577 was tested with primers Se705-1 and Se705-2 (Table 1) (24), and the expected PCR product of 124 bp was obtained (Fig. 1). To test if the multiplex PCR was able to detect a putative VanA-type S. epidermidis strain, DNA from plasmid pAT613, which contains the vanA gene (1), was added at one copy per genome equivalent to that of S. epidermidis BM4577 and the mixture was subjected to amplification. As expected, fragments of 730 bp corresponding to vanA and of 124 bp specific to S. epidermidis were visible (Fig. 1).
Seventy previously studied clinical isolates from our collection, including 5 of E. faecium (vanA), 4 of E. faecalis (vanA), 4 of E. faecium (vanB), 1 of E. faecalis (vanB), 4 of E. gallinarum (vanC-1), 4 of E. casseliflavus (vanC-2), 2 of E. flavescens (vanC-3), 4 of E. faecium (vanD) (10, 29), 4 of E. faecalis (vanD) (10), 4 of E. faecalis (vanE) (E. Lambert, C. McCullough, G. Coombs, F. O'Brien, J. Pearson, J. Bell, A. Berry, and K. Christiansen, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1118, 2002), 4 of E. faecalis (vanG) (8), susceptible Enterococcus durans ATCC 19432, E. durans BM4578 (vanA), and 28 susceptible strains, including 8 of E. faecalis, 8 of E. faecium, 10 of S. aureus, and 2 of S. epidermidis, were also tested. The PCR patterns obtained with DNAs from these strains were in agreement with their previously determined resistance genotypes and host species (data not shown). No amplification products were obtained with susceptible E. durans ATCC 19432.
Resistance to glycopeptides is disseminating rapidly (5, 36) and has recently spread to methicillin-resistant S. aureus (18, 26, 31). Rapid and accurate methods are thus essential for the detection of such clinical isolates and the prevention of their transmission. In addition, MIC determination is time-consuming and does not detect GRE with low-level glycopeptide resistance (36) and misidentification of E. faecalis and E. faecium can occur with commercial systems (35). Compared with the method of Patel et al., the technique we have devised does not require MspI restriction analysis to distinguish vanA from vanB (27).
The multiplex PCR developed in this study is robust, sensitive, specific, and fast. It allows simultaneous identification of the most clinically important enterococci and staphylococci at the species level and detection of the six glycopeptide resistance gene clusters described so far in human isolates. One of the limitations of the method proposed, or of other similar assays, could be the sequence variability among van genes that has occasionally been observed (7, 9, 10, 14, 28-30).
REFERENCES
- 1.Arthur, M., F. Depardieu, L. Cabanié, P. Reynolds, and P. Courvalin. 1998. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 30:819-830. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., F. Depardieu, G. Gerbaud, M. Galimand, R. Leclercq, and P. Courvalin. 1997. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J. Bacteriol. 179:97-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, M., P. Reynolds, and P. Courvalin. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401-407. [DOI] [PubMed] [Google Scholar]
- 4.Bell, J. M., J. C. Paton, and J. Turnidge. 1998. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J. Clin. Microbiol. 36:2187-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesneau, O., J. Allignet, and N. el Solh. 1993. Thermonuclease gene as a target nucleotide sequence for specific recognition of Staphylococcus aureus. Mol. Cell. Probes 7:301-310. [DOI] [PubMed] [Google Scholar]
- 7.Dalla Costa, L. M., P. E. Reynolds, H. A. Souza, D. C. Souza, M. F. Palepou, and N. Woodford. 2000. Characterization of a divergent vanD-type resistance element from the first glycopeptide-resistant strain of Enterococcus faecium isolated in Brazil. Antimicrob. Agents Chemother. 44:3444-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Depardieu, F., M. G. Bonora, P. E. Reynolds, and P. Courvalin. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931-948. [DOI] [PubMed] [Google Scholar]
- 9.Depardieu, F., P. Courvalin, and T. Msadek. 2003. A six amino acid deletion, partially overlapping the VanSB G2 ATP-binding motif, leads to constitutive glycopeptide resistance in VanB-type Enterococcus faecium. Mol. Microbiol. 50:1069-1083. [DOI] [PubMed] [Google Scholar]
- 10.Depardieu, F., M. Kolbert, H. Pruul, J. Bell, and P. Courvalin. 2004. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 48:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Depardieu, F., P. E. Reynolds, and P. Courvalin. 2003. VanD-type vancomycin-resistant Enterococcus faecium 10/96A. Antimicrob. Agents Chemother. 47:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutka-Malen, S., C. Molinas, M. Arthur, and P. Courvalin. 1992. Sequence of the vanC gene of Enterococcus gallinarum BM4174 encoding a d-alanine:d-alanine ligase related protein necessary for vancomycin resistance. Gene 112:53-58. [DOI] [PubMed] [Google Scholar]
- 14.Evers, S., D. F. Sahm, and P. Courvalin. 1993. The vanB gene of vancomycin-resistant Enterococcus faecalis V583 is structurally related to genes encoding d-Ala:d-Ala ligases and glycopeptide-resistance proteins VanA and VanC. Gene 124:143-144. [DOI] [PubMed] [Google Scholar]
- 15.Fines, M., B. Perichon, P. Reynolds, D. F. Sahm, and P. Courvalin. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob. Agents Chemother. 43:2161-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaratne, P., and C. Rutherford. 1999. Detection of clinically relevant genotypes of vancomycin-resistant enterococci in nosocomial surveillance specimens by PCR. J. Clin. Microbiol. 37:2090-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kacica, M., and L. C. McDonald. 2004. Vancomycin-resistant Staphylococcus aureus—New York, 2004. Morb. Mortal. Wkly. Rep. 53:322-323. [PubMed] [Google Scholar]
- 19.Kariyama, R., R. Mitsuhata, J. W. Chow, D. B. Clewell, and H. Kumon. 2000. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 38:3092-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornblum, J., B. J. Hartman, R. P. Novick, and A. Tomasz. 1986. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur. J. Clin. Microbiol. 5:714-718. [DOI] [PubMed] [Google Scholar]
- 21.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 22.Leclercq, R., E. Derlot, M. Weber, J. Duval, and P. Courvalin. 1989. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 33:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, J. J., C. L. Perng, T. S. Chiueh, S. Y. Lee, C. H. Chen, F. Y. Chang, C. C. Wang, and W. M. Chi. 2001. Detection and typing of vancomycin-resistance genes of enterococci from clinical and nosocomial surveillance specimens by multiplex PCR. Epidemiol. Infect. 126:357-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martineau, F., F. J. Picard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1996. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J. Clin. Microbiol. 34:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKessar, S. J., A. M. Berry, J. M. Bell, J. D. Turnidge, and J. C. Paton. 2000. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob. Agents Chemother. 44:3224-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, D., V. Urdaneta, A. Weltman, and S. Park. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 27.Patel, R., J. R. Uhl, P. Kohner, M. K. Hopkins, and F. R. Cockerill III. 1997. Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J. Clin. Microbiol. 35:703-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel, R., J. R. Uhl, P. Kohner, M. K. Hopkins, J. M. Steckelberg, B. Kline, and F. R. Cockerill III. 1998. DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob. Agents Chemother. 42:202-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perichon, B., B. Casadewall, P. Reynolds, and P. Courvalin. 2000. Glycopeptide-resistant Enterococcus faecium BM4416 is a VanD-type strain with an impaired d-alanine:d-alanine ligase. Antimicrob. Agents Chemother. 44:1346-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perichon, B., P. E. Reynolds, and P. Courvalin. 1997. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother. 41:2016-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sievert, D. M., M. L. Boulton, G. Stolman, D. Johnson, M. G. Stobierski, F. P. Downes, P. A. Somsel, J. T. Rudrik, W. J. Brown, W. Hafeez, T. Lundstrom, E. Flanagan, R. Johnson, J. Mitchell, and S. Chang. 2002. Staphylococcus aureus resistant to vancomycin. - United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 32.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. Georges. 1988. Vancomycin resistant enterococci. Lancet i:57-58. [DOI] [PubMed] [Google Scholar]
- 34.Vincent, S., R. G. Knight, M. Green, D. Sahm, and D. M. Shlaes. 1991. Vancomycin susceptibility and identification of motile enterococci. J. Clin. Microbiol. 29:2335-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willey, B. M., R. N. Jones, A. McGeer, W. Witte, G. French, R. B. Roberts, S. G. Jenkins, H. Nadler, and D. E. Low. 1999. Practical approach to the identification of clinically relevant Enterococcus species. Diagn. Microbiol. Infect. Dis. 34:165-171. [DOI] [PubMed] [Google Scholar]
- 36.Woodford, N., A. P. Johnson, D. Morrison, and D. C. E. Speller. 1995. Current perspectives on glycopeptide resistance. Clin. Microbiol. Rev. 8:585-615. [DOI] [PMC free article] [PubMed] [Google Scholar]