Abstract
The fluorogenic TaqMan reverse transcriptase PCR (RT-PCR) assay was developed for detecting each of the dengue virus (DV) types 1 to 4. DV genome was detected in all the 35 serum samples from confirmed dengue cases by the TaqMan RT-PCR, although it was not detected in 13 and 21% by conventional type-specific and cross-reactive RT-PCR, respectively.
Dengue virus (DV) infections occur in most tropical and subtropical areas of the world (10). The number of travelers to areas where dengue is endemic has been increasing. Consequently, the number of imported dengue cases has also been increasing. A sensitive and specific assay to detect and discriminate a wide range of isolates of DV types 1 to 4 is needed. A fluorogenic probe-based assay has advantages over the conventional reverse transcriptase PCR (RT-PCR) (1, 3, 7, 8, 15). In the present study, we developed an improved serotype-specific fluorogenic (TaqMan) RT-PCR assay for detecting dengue virus genome. We then examined the newly developed TaqMan RT-PCR for sensitivity and specificity in comparison with conventional RT-PCR, by using serum samples from Japanese dengue patients.
DV types 1, 3, and 4, isolated from Japanese dengue fever patients, were propagated in C6/36 cells. DV type 2, Trinidad strain TR1751, was propagated in suckling mouse brain. West Nile virus, New York and Eg101 strains, Kunjin virus, K47382 and OR393 strains, Japanese encephalitis virus, Gar01 strain, and yellow fever virus, 17D strain, were propagated in Vero cells and used in flavivirus cross-reactivity studies.
One serum sample each was collected from 35 Japanese dengue patients. These patients visited countries where DV was endemic, including India, Sri Lanka, Bangladesh, Thailand, Cambodia, Laos, Singapore, Malaysia, East Timor, Indonesia, the Philippines, Tahiti, and Samoa during the years of 1994 to 2003. Some patients visited more than one country. Of 35 patients, 18 patients were infected with DV type 1, 8 with DV type 2, 6 with DV type 3, and 3 with DV type 4. DV infection was confirmed by immunoglobulin M capture enzyme-linked immunosorbent assay, conventional RT-PCR, or plaque titration assays, as reported by Yamada et al. (21, 22) and Nawa et al. (13).
All the envelope (E) gene nucleotide sequences available in GenBank were retrieved and aligned using CLUSTAL X (version 1.8) (17) to prepare optimized primers and probes for the TaqMan RT-PCR. We used the E gene because it had been best analyzed among dengue genes (2, 4, 5, 6, 9, 14, 16, 18, 19). The strains retrieved from GenBank are shown in Table 1. Primer and probe sequences were selected using the Primer Express software (PE Applied Biosystems, Foster City, Calif.).
TABLE 1.
Oligonucleotide primers and probes used in the conventional RT-PCR and TaqMan RT-PCR
| Assay | Serotype | Identification | Sequence (5′-3′) | Size (bp) | Gene | No. of strainsc |
|---|---|---|---|---|---|---|
| Conventional RT-PCR with cross-reactive primers | Dus (forward) | TCAATATGCTGAAACGCGCGAGAAACCG | 511 | C-PrM. | ||
| Duc (reverse) | TTGCACCAACAGTCAATGTCTTCAGGTTC | |||||
| Conventional RT-PCR with type-specific primers | 1 | D1s (forward) | GGACTGCGTATGGAGTTTTG | 490 | E-NS1 | |
| D1c (reverse) | ATGGGTTGTGGCCTAATCAT | |||||
| 2 | D2S (forward) | GTTCCTCTGCAAACACTCCA | 230 | E | ||
| D2c (reverse) | GTGTTATTTTGATTTCCTTG | |||||
| 3 | D3s (forward) | GTGCTTACACAGCCCTATTT | 320 | E-NS1 | ||
| D3c (reverse) | TCCATTCTCCCAAGCGCCTG | |||||
| 4 | D4s (forward) | CCATTATGGCTGTGTTGTTT | 398 | NS2a-NS2b | ||
| D4c (reverse) | CTTCATCCTGCTTCACTTCT | |||||
| TaqMan RT-PCR with type- specific primers and probes | 1 | D1MGBEn469s (forward) | GAACATGGRACAAYTGCAACYAT | 67 | E | 76d |
| D1MGBEn493pa (probe) | ACACCTCAAGCTCC | |||||
| D1MGBEn536r (reverse) | CCGTAGTCDGTCAGCTGTATTTCA | |||||
| 2 | D2MGBEn493s (forward) | ACACCACAGAGTTCCATCACAGA | 68 | E | 203e | |
| D2MGBEn545pa (probe) | CGATGGARTGCTCTC | |||||
| D2MGBEn568r (reverse) | CATCTCATTGAAGTCNAGGCC | |||||
| 3 | D3MGBEn1s (forward) | ATGAGATGYGTGGGAGTRGGAAAC | 70 | E | 59f | |
| D3MGBEn27pa (probe) | AGATTTTGTGGAAGGYCT | |||||
| D3MGBEn71r (reverse) | CACCACDTCAACCCACGTAGCT | |||||
| 4 | D4TEn711s (forward) | GGTGACRTTYAARGTHCCTCAT | 75 | E | 30g | |
| D4TEn734pb (probe) | CCAAGAGACAGGATGTGACAGTGCTRGGATC | |||||
| D4TEn786c (reverse) | WGARTGCATRGCTCCYTCCTG |
Labeled at the 5′ end with the FAM reporter dye and at the 3′ end with the quencher dye TAMRA.
MGB probes were labeled at the 5′ end with the FAM reporter dye and were not labeled at the 3′ end with the quencher dye TAMRA.
Retrieved from GenBank to design primers and probes.
AF425639, AF514876, AF514878, AF514883, AF514885, AJ574760, AY206457, AB111064-78, AF226685, AF298807-8, AF311956-58, AF425609-38, AF513110, AJ438941, AY145121-23, AY153755, D00501-05, M23027, M87512, S64849, U88535-37, and X76219.
A91810, AF004019-20, AF01041, AF022434-41, AF038402-3, AF093674, AF100459-69, AF19661, AF163096, AF195032-44, AF19678-88, AF204177-78, AF231715-20, AF264053-4, AF276619, AF295694-709, AF359579, AF363069-92, AF398106-14, AF410345-79, AF469175-6, AF489932, AF509530, AY037116, AY044442, AY079923-24, D00345-6, D10514, AB111448-54, L04561, L10040-55, M15075, M19197, M20558, M24444-51, M29095, M84727-28, U87411-12, X15214, X15433-34, X54319, and X65240.
AF147456-60, AF317645, AF349753, AF533079, AY038605, AY135419, AY145712-30, AB111080-84, L11422-42, L11619-20, L41125, and NC_001475.
AF231722-25, AF289029, AF326573, AF375822, AB111085-88, NC_002640, and U18425-42.
Viral RNA was isolated using a QIAamp viral RNA kit (QIAGEN, Valencia, Calif.). The conventional RT-PCR was performed with a TITAN One-Tube RT-PCR kit (Roche Molecular Biochemicals, Indianapolis, Ind.), according to the method of Yamada et al. (21). The cross-reactive and type-specific primers for DV types 1 to 4 were previously reported and evaluated by Morita et al. (11, 12) and Yamada et al. (21) (Table 1). In the TaqMan assay, 5 μl of RNA isolated from a serum sample was mixed with 100 pmol of each primer and 15 pmol of a probe in a 25-μl reaction volume, using a TaqMan RT-PCR Ready-Mix kit (PE Applied Biosystems). The samples were subjected to amplification in an ABI Prism 7000 Sequence Detection System instrument (PE Applied Biosystems). The TaqMan RT-PCR assay consisted of a 30-min RT step at 48°C and 45 cycles of the PCR step (95°C for 15 s and 57°C for 60 s).
PCR products were directly detected by monitoring the increase in fluorescence of a dye-labeled oligonucleotide probe with an ABI Prism 7000 sequence detector. Assay specificity was evaluated by testing serotype-specific probe and primer sets against virus panels that included DV types 1 to 4, West Nile virus (New York strain, 1.0 × 106 PFU/tube; Eg101 strain, 5.4 × 104 PFU/tube), Kunjin virus (K47382 strain, 1.8 × 107 PFU/tube; OR393 strain, 1.6 × 108 PFU/tube), Japanese encephalitis virus (7.6 × 105 PFU/tube), and yellow fever virus (6.0 × 103 PFU/tube). The detection threshold of each TaqMan assay was determined using 10-fold serial dilutions of RNA extracted from stock DV. The threshold was defined as 0.5. This corresponds to the level at which specific viral RNA was consistently detected and at which no nonspecific viral RNA was detected. The threshold cycle value (Ct) was defined as the number of amplification cycles at the threshold. Ct remained within a standard curve having a minimum correlation coefficient of 0.98.
First, the sensitivities of the TaqMan RT-PCR assay for DV types 1 to 4 was evaluated. Reference samples of DV types 1 to 4, which were previously quantified, were diluted 10-fold and used for sensitivity assays. Sensitivities were 1.2 PFU/tube for DV type 1 (Ct = 40.7), 0.1 PFU/tube for type 2 (Ct = 39.2), 1.0 PFU/tube for type 3 (Ct = 38.2), and 0.1 PFU/tube for type 4 (Ct = 38.8). The primer and probe pairs were also tested for virus specificity and did not detect other DV types or other flaviviruses. These results indicate that the newly developed TaqMan RT-PCR assays for DV types 1 to 4 are highly specific.
DV genome was detected in all the 35 serum samples (100%), when tested by the TaqMan RT-PCR. When tested by conventional RT-PCR, DV genome was detected in 27 of 31 serum samples (87%) with type-specific primers and in 26 of 33 samples (79%) with dengue cross-reactive primers (Table 2). These results suggest that sensitivity of TaqMan RT-PCR is higher than those of conventional RT-PCRs (P < 0.02).
TABLE 2.
Virus gene detection of serum samples from confirmed dengue patients by TaqMan RT-PCR and conventional RT-PCR
| Ct range of TaqMan RT-PCR (cycles) | Detection by:
|
||
|---|---|---|---|
| TaqMan RT-PCR | Conventional RT-PCR
|
||
| Type specific | Cross-reactive | ||
| <25 | 10/10 (100)a | 10/10 (100) | 9/9 (100) |
| 25-29.9 | 7/7 (100) | 6/7 (86) | 6/6 (100) |
| 30-34.9 | 7/7 (100) | 6/6 (100) | 7/7 (100) |
| 35-39.9 | 6/6 (100) | 3/4 (75) | 3/6 (50) |
| ≥40 | 5/5 (100) | 2/4 (50) | 1/5 (20) |
| Total | 35/35 (100) | 27/31 (87) | 26/33 (79) |
Positive samples/number of tested samples (%).
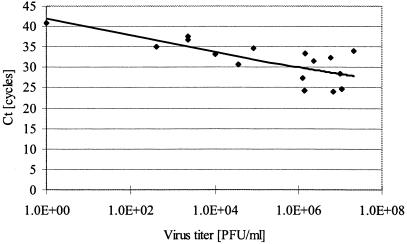
We next examined the relationship between virus titers and Ct values by analyzing serum samples from DV type 1-infected patients. There was an inverse correlation between virus titer and Ct (Fig. 1). Additionally, seven serum samples from nondengue patients were tested by TaqMan RT-PCR. DV genome was not detected in any of these samples (data not shown).
FIG. 1.
A log-scale plot of the virus threshold cycles (Ct) versus the virus titer for dengue virus type 1. Correlation coefficient, 0.502 (6-carboxyfluorescein [FAM] filter).
We detected DV genome in all the 35 tested serum samples from dengue patients by TaqMan RT-PCR, while 13% (type specific) and 21% (cross-reactive) were determined to be negative by the conventional RT-PCR. The newly developed TaqMan RT-PCR is, therefore, a useful tool for testing acute-phase serum samples from clinically suspected dengue cases. We did not observe any cross-reactions to other dengue types or other flaviviruses. This indicates that the E gene displays a high degree of sequence variation among flaviviruses, which enables us to construct primers and probes with high specificity and sensitivity. To our knowledge, a TaqMan RT-PCR system with a high specificity and high sensitivity has not been reported using other target regions. For example, Warrilow et al. (20) reported a group assay to detect all four dengue serotypes, using the 3′ noncoding region as the target area, but the sensitivity was not high. Callahan et al. also document a group assay with two sets of primers and probes, using the NS5 and C regions as the target area (1). Although they succeeded in detecting the DV types 1 to 4 with a high sensitivity using group assays, they did not obtain high sensitivities in type-specific assays. Additionally, since their group assay requires two sets of primers and probes, its advantage over a type-specific approach is reduced. Furthermore, in contrast to our results, the sensitivity of their type-specific TaqMan assay was not consistent with plaque titration results. Unfortunately, neither we nor Callahan et al. (1) converted the sensitivity units from PFU per milliliter to RNA copies. We will create a stable internal positive control for quantization, using plasmids containing the targeted viral nucleotide sequence, and further standardize the assay in the future.
Acknowledgments
We thank the doctors of the clinics and hospitals for providing us with serum samples for laboratory diagnosis of dengue. We also thank David W. Smith, Arbovirus Research and Surveillance Group, Department of Microbiology, University of Western Australia, for providing the initial stock of Kunjin virus (K47382 and OR393 strains).
This work was supported by the grant for the Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labour, and Welfare, Japan.
REFERENCES
- 1.Callahan, J. D., S. J. Wu, A. Dion-Schultz, B. E. Mangold, L. F. Peruski, D. M. Watts, K. R. Porter, G. R. Murphy, W. Suharyono, C. C. King, C. G. Hayes, and J. J. Temenak. 2001. Development and evaluation of serotype- and group-specific fluorogenic reverse transcriptase PCR (TaqMan) assays for dengue virus. J. Clin. Microbiol. 39:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crill, W. D., and J. T. Roehrig. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75:7769-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten, C., S. Gottig, S. Schilling, M. Asper, M. Panning, H. Schmitz, and S. Gunther. 2002. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 40:2323-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster, J. E., S. N. Bennett, H. Vaughan, V. Vorndam, W. O. McMillan, and C. V. Carrington. 2003. Molecular evolution and phylogeny of dengue type 4 virus in the Caribbean. Virology 306:126-134. [DOI] [PubMed] [Google Scholar]
- 5.Goncalvez, A. P., A. A. Escalante, F. H. Pujol, J. E. Ludert, D. Tovar, R. A. Salas, and F. Liprandi. 2002. Diversity and evolution of the envelope gene of dengue virus type 1. Virology 303:110-119. [DOI] [PubMed] [Google Scholar]
- 6.Lanciotti, R. S., D. J. Gubler, and D. W. Trent. 1997. Molecular evolution and phylogeny of dengue-4 viruses. J. Gen. Virol. 78:2279-2284. [DOI] [PubMed] [Google Scholar]
- 7.Lanciotti, R. S., A. J. Kerst, R. S. Nasci, M. S. Godsey, C. J. Mitchell, H. M. Savage, N. Komar, N. A. Panella, B. C. Allen, K. E. Volpe, B. S. Davis, and J. T. Roehrig. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 38:4066-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laue, T., P. Emmerich, and H. Schmitz. 1999. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan automated amplification system. J. Clin. Microbiol. 37:2543-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leitmeyer, K. C., D. W. Vaughn, D. M. Watts, R. Salas, I. Villalobos de Chacon, C. Ramos, and R. Rico-Hesse. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 73:4738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monath, T. P. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA 91:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morita, K., T. Maemoto, S. Honda, K. Onishi, M. Murata, M. Tanaka, and A. Igarashi. 1994. Rapid detection of virus genome from imported dengue fever and dengue hemorrhagic fever patients by direct polymerase chain reaction. J. Med. Virol. 44:54-58. [DOI] [PubMed] [Google Scholar]
- 12.Morita, K., M. Tanaka, and A. Igarashi. 1991. Rapid identification of dengue virus serotypes by using polymerase chain reaction. J. Clin. Microbiol. 29:2107-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nawa, M., K. Yamada, T. Takasaki, T. Akatsuka, and I. Kurane. 2000. Serotype-cross-reactive IgM responses in dengue virus infections determined by enzyme-linked immunosorbent assays. Clin. Diagn. Lab. Immunol. 7:774-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rico-Hesse, R., L. M. Harrison, R. A. Salas, D. Tovar, A. Nisalak, C. Ramos, J. Boshell, M. T. de Mesa, R. M. Nogueira, and A. T. da Rosa. 1997. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 230:244-251. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie, S. A., S. Long, G. Smith, A. Pyke, and T. B. Knox. 2004. Entomological investigations in a focus of dengue transmission in Cairns, Queensland, Australia, by using the sticky ovitraps. J. Med. Entomol. 41:1-4. [DOI] [PubMed] [Google Scholar]
- 16.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246:317-328. [DOI] [PubMed] [Google Scholar]
- 17.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uzcategui, N. Y., G. Comach, D. Camacho, M. Salcedo, M. Cabello de Quintana, M. Jimenez, G. Sierra, R. Cuello de Uzcategui, W. S. James, S. Turner, E. C. Holmes, and E. A. Gould. 2003. Molecular epidemiology of dengue virus type 3 in Venezuela. J. Gen. Virol. 84:1569-1575. [DOI] [PubMed] [Google Scholar]
- 19.Wang, E., H. Ni, R. Xu, A. D. Barrett, S. J. Watowich, D. J. Gubler, and S. C. Weaver. 2000. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 74:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warrilow, D., J. A. Northill, A. Pyke, and G. A. Smith. 2002. Single rapid TaqMan fluorogenic probe based PCR assay that detects all four dengue serotypes. J. Med. Virol. 66:524-528. [DOI] [PubMed] [Google Scholar]
- 21.Yamada, K., T. Takasaki, M. Nawa, and I. Kurane. 2002. Virus isolation as one of the diagnostic methods for dengue virus infection. J. Clin. Virol. 24:203-209. [DOI] [PubMed] [Google Scholar]
- 22.Yamada, K., T. Takasaki, M. Nawa, S. Yabe, and I. Kurane. 2003. Antibody responses determined for Japanese dengue fever patients by neutralization and hemagglutination inhibition assays demonstrate cross-reactivity between dengue and Japanese encephalitis viruses. Clin. Diagn. Lab. Immunol. 10:725-728. [DOI] [PMC free article] [PubMed] [Google Scholar]